Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. MALDI-TOF MS Fingerprinting
2.3. DNA Extraction
2.4. Primer Design (Hexaplex PCR)
2.5. Hexaplex-PCR Fingerprinting
2.6. Rep-PCR Fingerprinting
2.7. Statistics
3. Results
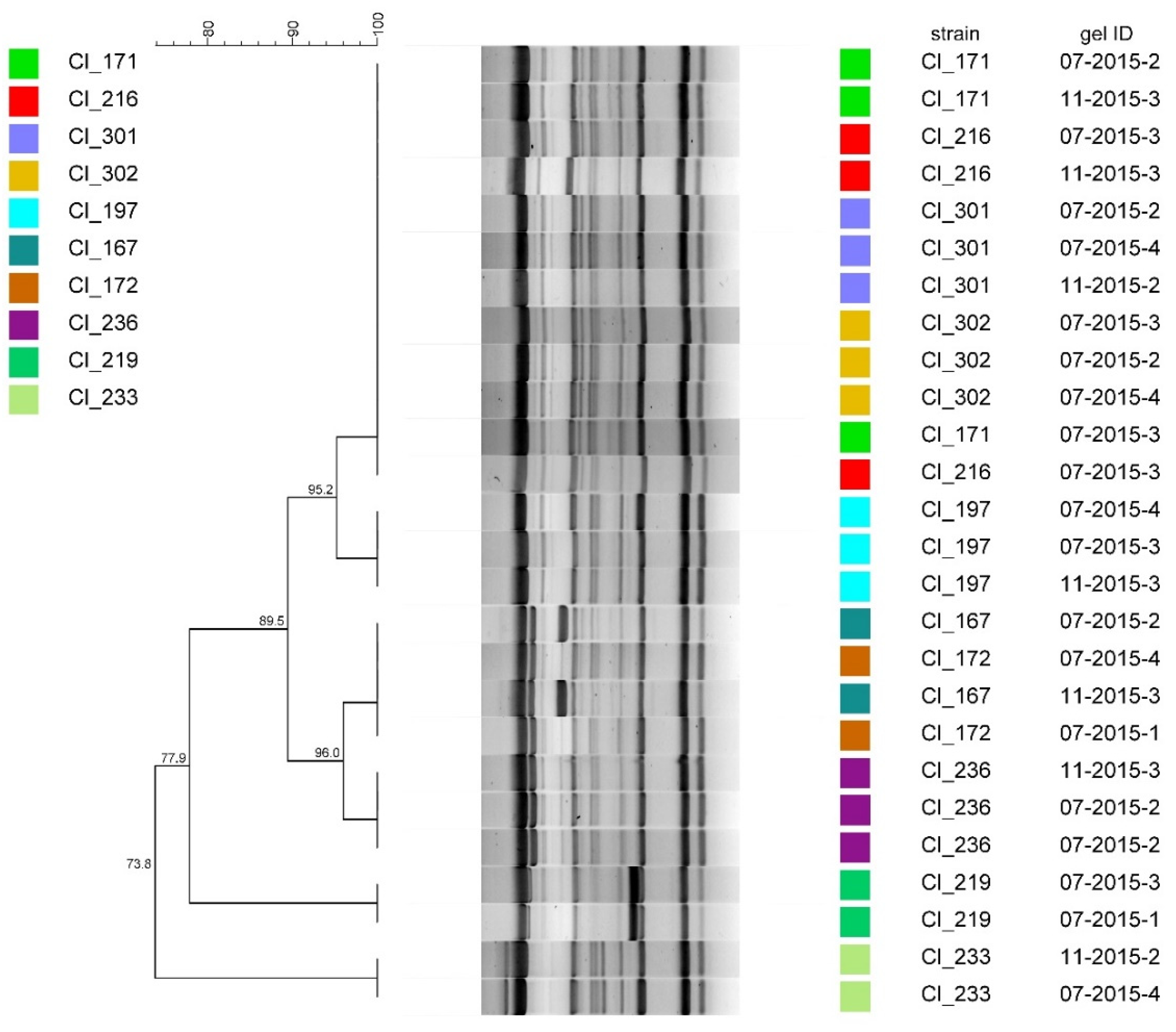
3.1. Method Reproducibility
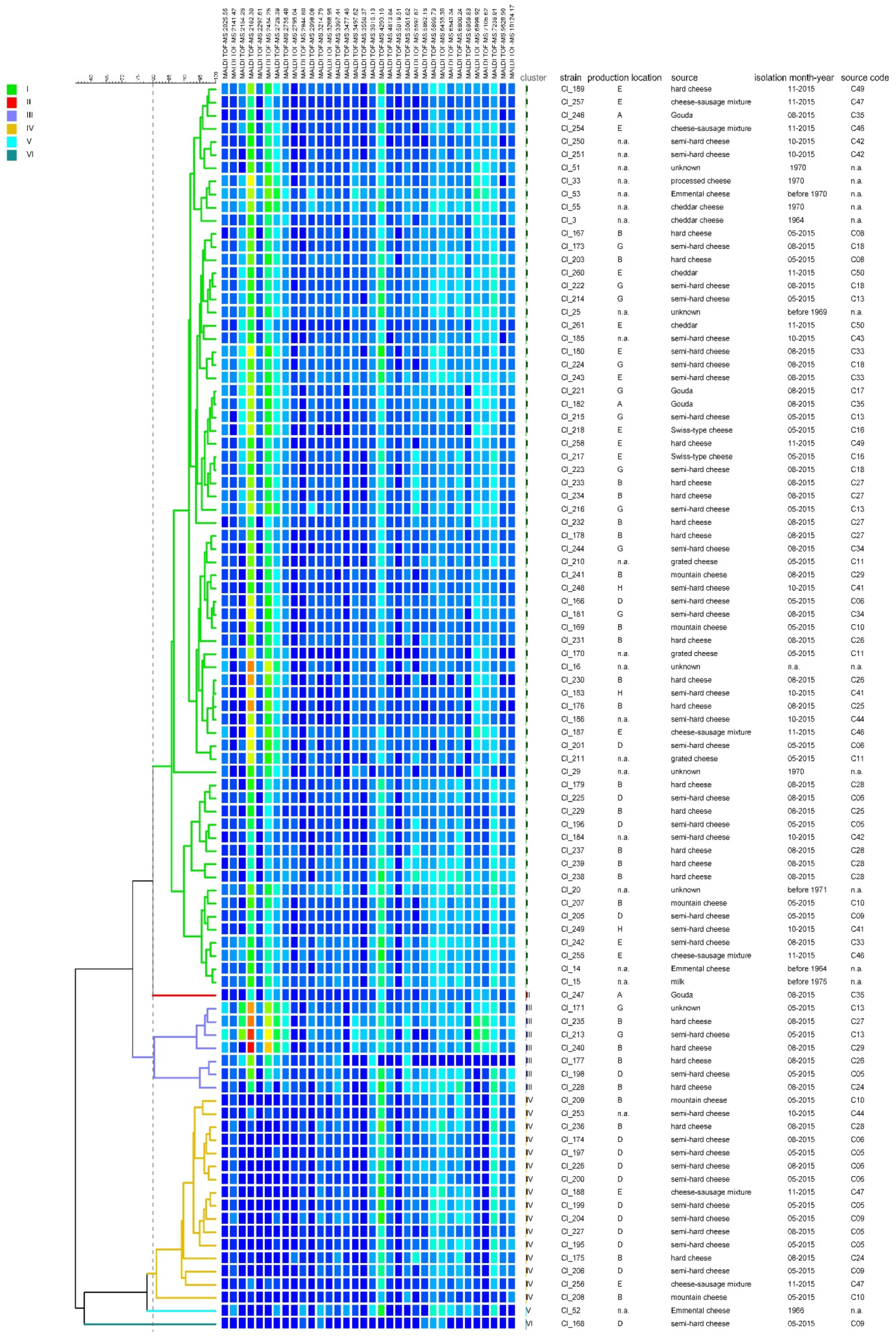
3.2. Diversity of the Test Strain Set
4. Discussion
4.1. Method Reproducibility
4.2. Diversity of the Test Strain Set
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brändle, J.; Domig, K.; Kneifel, W. Relevance and analysis of butyric acid producing clostridia in milk and cheese. Food Control 2016, 67, 96–113. [Google Scholar] [CrossRef]
- Garde, S.; Avila, M.; Gomez-Torres, N.; Nunez, M. Clostridium in Late Blowing Defect of Cheese: Detection, Prevalence, Effects and Control Strategies. In Handbook on Cheese: Production, Chemistry and Sensory Properties; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 503–518. [Google Scholar]
- Doyle, C.J.; Gleeson, D.; Jordan, K.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int. J. Food Microbiol. 2015, 197, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Ingham, S.C.; Hassler, J.R.; Tsai, Y.-W.; Ingham, B.H. Differentiation of lactate-fermenting, gas-producing Clostridium spp. isolated from milk. Int. J. Food Microbiol. 1998, 43, 173–183. [Google Scholar] [CrossRef]
- Le Bourhis, A.-G.; Saunier, K.; Dore, J.; Carlier, J.-P.; Chamba, J.-F.; Popoff, M.-R.; Tholozan, J.-L.; Dore, J. Development and Validation of PCR Primers To Assess the Diversity of Clostridium spp. in Cheese by Temporal Temperature Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2005, 71, 29–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lycken, L.; Borch, E. Characterization of Clostridium spp. Isolated from Spoiled Processed Cheese Products. J. Food Prot. 2006, 69, 1887–1891. [Google Scholar] [CrossRef] [PubMed]
- Julien, M.-C.; Dion, P.; Lafrenière, C.; Antoun, H.; Drouin, P. Sources of Clostridia in Raw Milk on Farms. Appl. Environ. Microbiol. 2008, 74, 6348–6357. [Google Scholar] [CrossRef] [Green Version]
- Garde, S.; Arias, R.; Gaya, P.; Nuñez, M. Occurrence of Clostridium spp. in ovine milk and Manchego cheese with late blowing defect: Identification and characterization of isolates. Int. Dairy J. 2011, 21, 272–278. [Google Scholar] [CrossRef]
- Arias, C.; Oliete, B.; Seseña, S.; Jiménez, L.; Pérez-Guzmán, M.; Arias, R. Importance of on-farm management practices on lactate-fermenting Clostridium spp. spore contamination of Manchega ewe milk: Determination of risk factors and characterization of Clostridium population. Small Rumin. Res. 2013, 111, 120–128. [Google Scholar] [CrossRef]
- Feligini, M.; Brambati, E.; Panelli, S.; Ghitti, M.; Sacchi, R.; Capelli, E.; Bonacina, C. One-year investigation of Clostridium spp. occurrence in raw milk and curd of Grana Padano cheese by the automated ribosomal intergenic spacer analysis. Food Control 2014, 42, 71–77. [Google Scholar] [CrossRef]
- Bassi, D.; Puglisi, E.; Cocconcelli, P.S. Understanding the bacterial communities of hard cheese with blowing defect. Food Microbiol. 2015, 52, 106–118. [Google Scholar] [CrossRef]
- Driehuis, F.; Hoolwerf, J.; Rademaker, J.L.W. Concurrence of spores of Clostridium tyrobutyricum, Clostridium beijerinckii and Paenibacillus polymyxa in silage, dairy cow faeces and raw milk. Int. Dairy J. 2016, 63, 70–77. [Google Scholar] [CrossRef]
- Turchi, B.; Pero, S.; Torracca, B.; Fratini, F.; Mancini, S.; Galiero, A.; Pedonese, F.; Nuvoloni, R.; Cerri, D. Occurrence of Clostridium spp. in ewe’s milk: Enumeration and identification of isolates. Dairy Sci. Technol. 2016, 96, 693–701. [Google Scholar] [CrossRef] [Green Version]
- Brändle, J.; Fraberger, V.; Berta, J.; Puglisi, E.; Jami, M.; Kneifel, W.; Domig, K. Butyric acid producing clostridia in cheese—Towards the completion of knowledge by means of an amalgamate of methodologies. Int. Dairy J. 2018, 86, 86–95. [Google Scholar] [CrossRef]
- Komori, K.; Ohkubo, Y.; Katano, N.; Motoshima, H. One year investigation of the prevalence and diversity of clostridial spores in raw milk from the Tokachi area of Hokkaido. Anim. Sci. J. 2018, 90, 135–139. [Google Scholar] [CrossRef]
- Borreani, G.; Ferrero, F.; Nucera, D.; Casale, M.; Piano, S.; Tabacco, E. Dairy farm management practices and the risk of contamination of tank milk from Clostridium spp. and Paenibacillus spp. spores in silage, total mixed ration, dairy cow feces, and raw milk. J. Dairy Sci. 2019, 102, 8273–8289. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Vanoni, L.; Silvetti, T.; Morandi, S.; Brasca, M. Identification of Clostridium beijerinckii, Cl. butyricum, Cl. sporogenes, Cl. tyrobutyricum isolated from silage, raw milk and hard cheese by a multiplex PCR assay. J. Dairy Res. 2012, 79, 318–323. [Google Scholar] [CrossRef]
- Brändle, J.; Heinzle, L.; Fraberger, V.; Berta, J.; Zitz, U.; Schinkinger, M.; Stocker, W.; Kneifel, W.; Domig, K. Novel approach to enumerate clostridial endospores in milk. Food Control 2018, 85, 318–326. [Google Scholar] [CrossRef]
- Bava, L.; Colombini, S.; Zucali, M.; Decimo, M.; Morandi, S.; Silvetti, T.; Brasca, M.; Tamburini, A.; Crovetto, G.M.; Sandrucci, A. Efficient milking hygiene reduces bacterial spore contamination in milk. J. Dairy Res. 2017, 84, 322–328. [Google Scholar] [CrossRef]
- Stolle, A.; Sperner, B.; Schalch, B.; Eisgruber, H. Comparison of protocols for pulsed-field gel electrophoresis of Clostridia. Electrophoresis 2001, 22, 1585–1589. [Google Scholar] [CrossRef]
- Li, W.; Raoult, D.; Fournier, P.-E. Bacterial strain typing in the genomic era. FEMS Microbiol. Rev. 2009, 33, 892–916. [Google Scholar] [CrossRef] [Green Version]
- Garde, S.; Gaya, P.; Arias, R.; Nuñez, M. Enhanced PFGE protocol to study the genomic diversity of Clostridium spp. isolated from Manchego cheeses with late blowing defect. Food Control 2012, 28, 392–399. [Google Scholar] [CrossRef]
- Nishihara, M.; Takahashi, H.; Sudo, T.; Kyoi, D.; Kawahara, T.; Ikeuchi, Y.; Fujita, T.; Kuda, T.; Kimura, B.; Yanahira, S. Multilocus variable-number of tandem repeat analysis (MLVA) for Clostridium tyrobutyricum strains isolated from cheese production environment. Int. J. Food Microbiol. 2014, 190, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, J.; González, M.J.; Olivera, J.A.; Burgueño, J.; Juliano, P.; Fox, E.M.; Reginensi, S.M. Seasonal occurrence and molecular diversity of clostridia species spores along cheesemaking streams of 5 commercial dairy plants. J. Dairy Sci. 2016, 99, 3358–3366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emele, M.F.; Možina, S.S.; Lugert, R.; Bohne, W.; Masanta, W.O.; Riedel, T.; Groß, U.; Bader, O.; Zautner, A.E. Proteotyping as alternate typing method to differentiate Campylobacter coli clades. Sci. Rep. 2019, 9, 4244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, A.; Heinold, S.; Brunisholz, R.; Schmidt, H.; Drissner, D. Application of MALDI-TOF mass spectrometry and specific PCR for tracking of E. coli O157:H− strain 431/97 in Batavia lettuce. Chem. Biol. Technol. Agric. 2019, 6, 2. [Google Scholar] [CrossRef]
- Gekenidis, M.-T.; Studer, P.; Wüthrich, S.; Brunisholz, R.; Drissner, D. Beyond the Matrix-Assisted Laser Desorption Ionization (MALDI) Biotyping Workflow: In Search of Microorganism-Specific Tryptic Peptides Enabling Discrimination of Subspecies. Appl. Environ. Microbiol. 2014, 80, 4234–4241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illikoud, N.; Rossero, A.; Chauvet, R.; Courcoux, P.; Pilet, M.-F.; Charrier, T.; Jaffrès, E.; Zagorec, M. Genotypic and phenotypic characterization of the food spoilage bacterium Brochothrix thermosphacta. Food Microbiol. 2019, 81, 22–31. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2012, 32, 188–217. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rand, W.M. Objective Criteria for the Evaluation of Clustering Methods. J. Am. Stat. Assoc. 1971, 66, 846–850. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Carriço, J.A.; Silva-Costa, C.; Cristino, J.M.; Pinto, F.R.; De Lencastre, H.; Almeida, J.S.; Ramirez, M. Illustration of a Common Framework for Relating Multiple Typing Methods by Application to Macrolide-Resistant Streptococcus pyogenes. J. Clin. Microbiol. 2006, 44, 2524–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, V.J.; Zlosnik, J.E.; Yau, Y.C.W.; Speert, D.P.; Aaron, S.D.; Guttman, D.S. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3341–3350. [Google Scholar] [CrossRef] [PubMed]
- Arbeit, R.D.; Arbique, J.C.; Beall, B.; Critchley, I.A.; Marsik, F.J.; Michaud, S.; Steward, C.; Tenover, F.C.; Trees, D.L. Molecular Methods for Bacterial Strain Typing; Approved Guideline; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2007. [Google Scholar]
- Spinali, S.; Van Belkum, A.; Goering, R.V.; Girard, V.; Welker, M.; Van Nuenen, M.; Pincus, D.H.; Arsac, M.; Durand, G. Microbial Typing by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry: Do We Need Guidance for Data Interpretation? J. Clin. Microbiol. 2015, 53, 760–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, P.R.; Haystead, T.A.J. Molecular Biologist’s Guide to Proteomics. Microbiol. Mol. Biol. Rev. 2002, 66, 39–63. [Google Scholar] [CrossRef] [Green Version]
- Reil, M.; Erhard, M.; Kuijper, E.J.; Kist, M.; Zaiss, H.; Witte, W.; Gruber, H.; Borgmann, S. Recognition of Clostridium difficile PCR-ribotypes 001, 027 and 126/078 using an extended MALDI-TOF MS system. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1431–1436. [Google Scholar] [CrossRef] [Green Version]
- Pinto, T.C.A.; Costa, N.S.; Castro, L.F.S.; Ribeiro, R.L.; Botelho, A.C.N.; Neves, F.P.G.; Peralta, J.M.; Teixeira, L.M. Potential of MALDI-TOF MS as an alternative approach for capsular typing Streptococcus pneumoniae isolates. Sci. Rep. 2017, 7, 45572. [Google Scholar] [CrossRef] [Green Version]
- Ghyselinck, J.; Van Hoorde, K.; Hoste, B.; Kim, H.; De Vos, P. Evaluation of MALDI-TOF MS as a tool for high-throughput dereplication. J. Microbiol. Methods 2011, 86, 327–336. [Google Scholar] [CrossRef]
- Pfrunder, S.; Grossmann, J.; Hunziker, P.; Brunisholz, R.; Gekenidis, M.-T.; Drissner, D. Bacillus cereus Group-Type Strain-Specific Diagnostic Peptides. J. Proteome Res. 2016, 15, 3098–3107. [Google Scholar] [CrossRef] [Green Version]
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control 2020, 113, 107188. [Google Scholar] [CrossRef]
- Christiansen, P.; Petersen, M.; Kask, S.; Møller, P.; Nielsen, E.; Vogensen, F.; Ardö, Y.; Petersen, M. Anticlostridial activity of Lactobacillus isolated from semi-hard cheeses. Int. Dairy J. 2005, 15, 901–909. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequence (5′ → 3′) | Concentration [µM] |
|---|---|---|
| CTM1 F | ATTACTCAAGCCGCCAAT | 0.08 |
| CTM1 R | CGGGTCATGGAATACTGAA | 0.08 |
| CTM5 F | GGAAAAGAACTTCCAGGAAT | 0.25 |
| CTM5 R | CTGTATCCGAAAATCCTCATTA | 0.25 |
| CTM6 F | CGACAACAGCGATATAACAAA | 0.10 |
| CTM6 R | GCCTTTTCACCATTTCCAT | 0.10 |
| CTM8 F | GGACTATGTATACAGCTGGAT | 0.12 |
| CTM8 R | ACCACCGCCAGTTAATAT | 0.12 |
| CTM12 F | CTGCTGACAAACTTGAAGAA | 0.18 |
| CTM12 R | TCTTGGTGATCCAAATGAAATT | 0.18 |
| CTM17 F | TTCATGGATGGAAGCAGT | 0.08 |
| CTM17 R | GGCACTGGATATTTCAGATAT | 0.08 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burtscher, J.; Küller, F.; Dreier, M.; Arias-Roth, E.; Drissner, D.; Domig, K.J. Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques. Microorganisms 2020, 8, 1057. https://doi.org/10.3390/microorganisms8071057
Burtscher J, Küller F, Dreier M, Arias-Roth E, Drissner D, Domig KJ. Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques. Microorganisms. 2020; 8(7):1057. https://doi.org/10.3390/microorganisms8071057
Chicago/Turabian StyleBurtscher, Johanna, Franziska Küller, Matthias Dreier, Emmanuelle Arias-Roth, David Drissner, and Konrad J. Domig. 2020. "Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques" Microorganisms 8, no. 7: 1057. https://doi.org/10.3390/microorganisms8071057
APA StyleBurtscher, J., Küller, F., Dreier, M., Arias-Roth, E., Drissner, D., & Domig, K. J. (2020). Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques. Microorganisms, 8(7), 1057. https://doi.org/10.3390/microorganisms8071057





