The Salmonella enterica Plasmidome as a Reservoir of Antibiotic Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Growth Conditions
2.2. DNA Preparation and Sequencing
2.3. Library Preparation and Oxford Nanopore MinION Sequencing
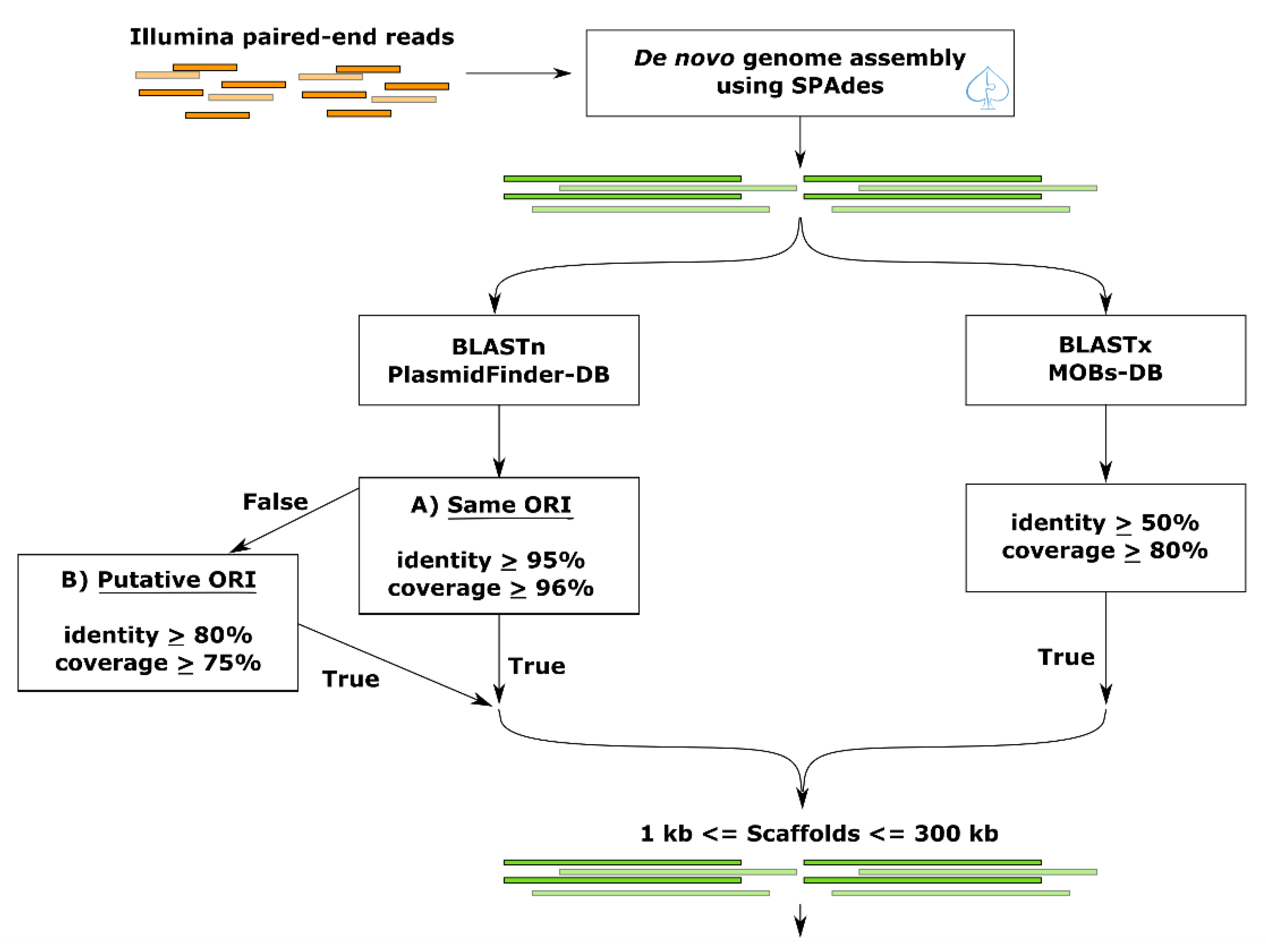
2.4. Bio-informatic Analysis and Databases
2.5. Plasmid-Gather Pipeline
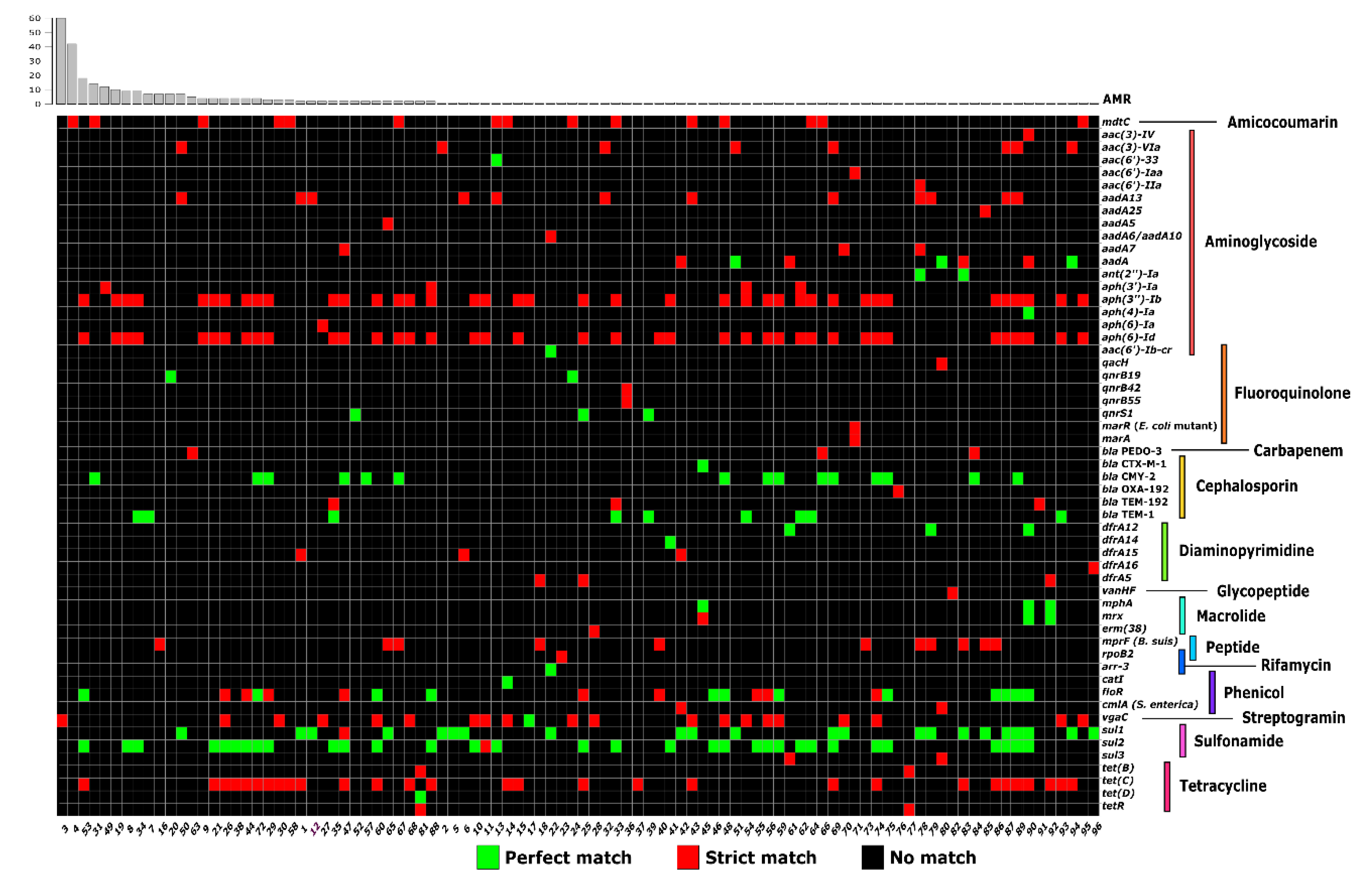
2.6. Antibiotic Resistance Gene Analysis
2.7. SISTR
2.8. Plasmid Fragments Recovery (Post-Recovery)
3. Results
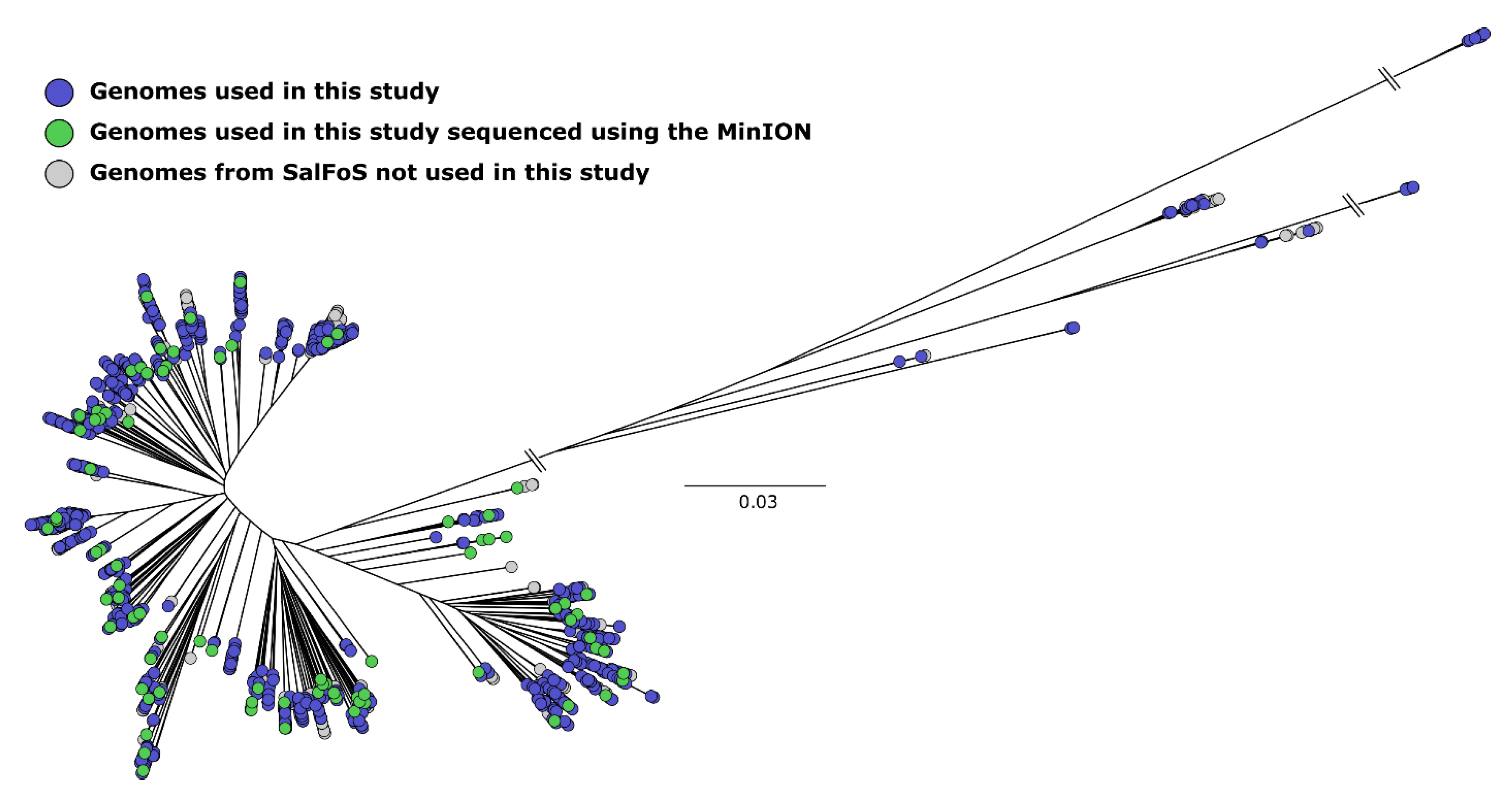
3.1. Construction of a Bioinformatics Pipeline for Plasmid Identification
3.2. Antibiotic Resistance Genes of the S. enterica Plasmidome
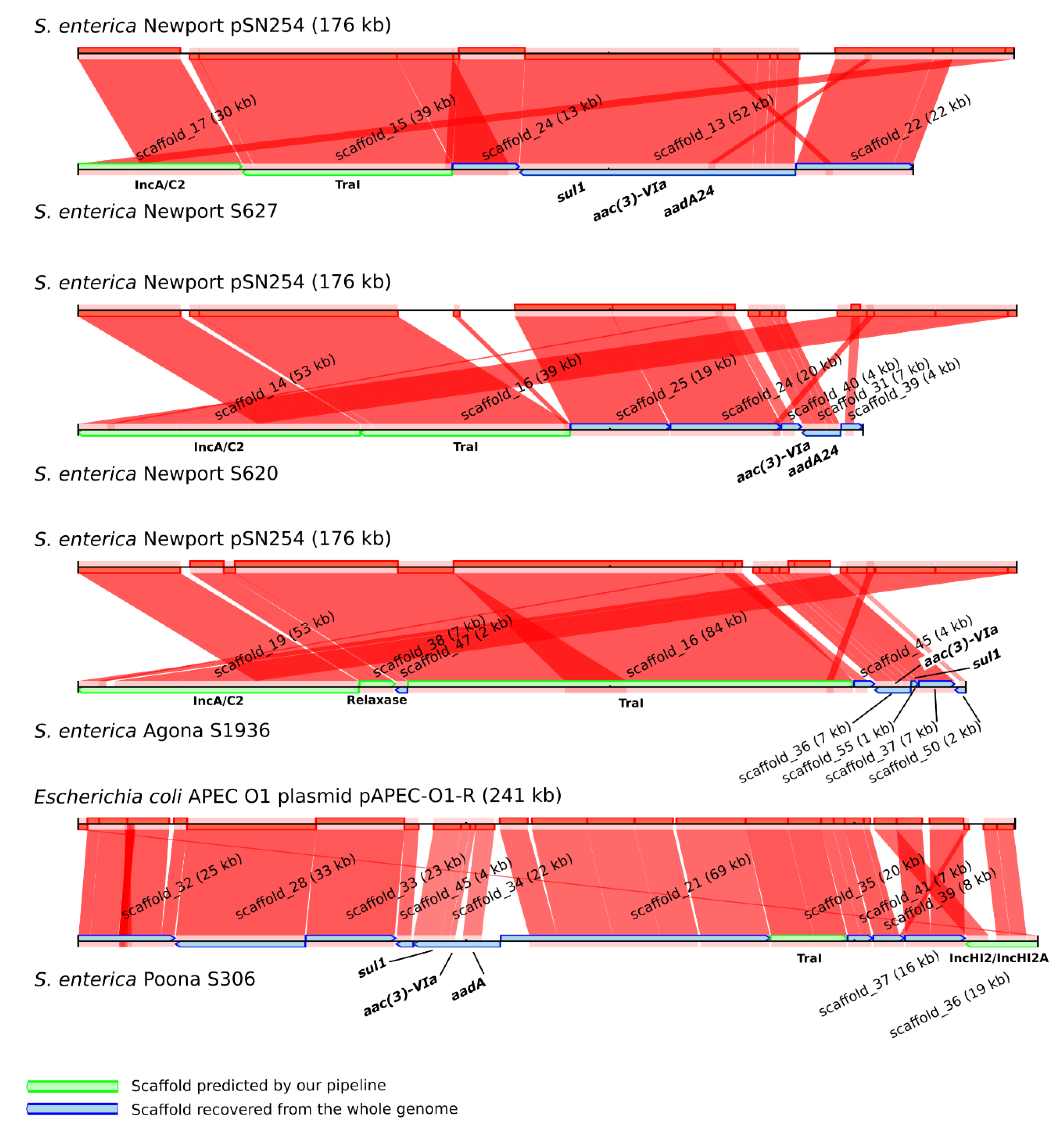
3.3. Increasing Recovery of Plasmid Scaffolds Using a Reference
3.4. The Resistome of S. enterica Plasmids and Chromosomes
3.5. Analysis of Plasmid Content Using Long-Read DNA Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grimont, P.A.; Weill, F.-X. Antigenic Formulae of the Salmonella Servovars, 9th ed.; Institut Pasteur: Paris, France, 2007; Volume 13. [Google Scholar]
- World Health Organization Antimicrobial Resistance: Global Report on Surveillance; World Heath Organization: Geneva, Switzerland, 2014.
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; González-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M. Paradigms of plasmid organization. Mol. Microb. 2000, 37, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Morosini, M.I.; Blázquez, J.; Negri, M.-C.; Cantón, R.; Loza, E.; Baquero, F. Characterization of a Nosocomial Outbreak Involving an Epidemic Plasmid Encoding for TEM-27 in Salmonella enterica Subspecies enterica Serotype Othmarschen. J. Infect. Dis. 1996, 17, 1015–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cantón, R.; Gonzalez-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In SilicoDetection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Antipov, D.; Hartwick, N.; Shen, M.; Rayko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef]
- Rozov, R.; Kav, A.B.; Bogumil, D.; Shterzer, N.; Halperin, E.; Mizrahi, I.; Shamir, R. Recycler: An algorithm for detecting plasmids from de novo assembly graphs. Bioinformatics 2017, 33, 475–482. [Google Scholar] [CrossRef]
- Lanza, V.F.; De Toro, M.; Garcillán-Barcia, M.P.; Mora, A.; Blanco, J.; Coque, T.M.; De La Cruz, F. Plasmid Flux in Escherichia coli ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences. PLoS Genet. 2014, 10, e1004766. [Google Scholar] [CrossRef]
- Zhou, F.; Xu, Y. cBar: A computer program to distinguish plasmid-derived from chromosome-derived sequence fragments in metagenomics data. Bioinformatics 2010, 26, 2051–2052. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Willems, R.J.; Van Schaik, W.; Schürch, A.C. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb. Genom. 2017, 3. [Google Scholar] [CrossRef]
- Guiney, D.G.; Fierer, J.M. The Role of the spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 1–10. [Google Scholar] [CrossRef]
- Carattoli, A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2011, 13, 36–46. [Google Scholar] [CrossRef]
- Utturkar, S.M.; Klingeman, D.M.; Hurt, R.A.J.; Brown, S.D. A Case Study into Microbial Genome Assembly Gap Sequences and Finishing Strategies. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef] [PubMed]
- Emond-Rheault, J.-G.; Jeukens, J.; Freschi, L.; Kukavica-Ibrulj, I.; Boyle, B.; Dupont, M.-J.; Colavecchio, A.; Barrere, V.; Cadieux, B.; Arya, G.; et al. A Syst-OMICS Approach to Ensuring Food Safety and Reducing the Economic Burden of Salmonellosis. Front. Microbiol. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; Garcia-Fernandez, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Rychlik, I.; Gregorova, D.; Hradecka, H. Distribution and function of plasmids in Salmonella enterica. Veter. Microbiol. 2006, 112, 1–10. [Google Scholar] [CrossRef]
- Martín, I.F.; AbuOun, M.; Reichel, R.; La Ragione, R.; Woodward, M.J. Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4,5,12:i:—, Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J. Antimicrob. Chemother. 2014, 69, 2098–2101. [Google Scholar] [CrossRef] [PubMed]
- García-Ferná Ndez, A.; Chiaretto, G.; Bertini, A.; Villa, L.; Fortini, D.; Ricci, A.; Carattoli, A. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum b-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 2008, 61, 1229–1233. [Google Scholar] [CrossRef]
- Kaldhone, P.R.P.R.; Han, J.; Deck, J.; Khajanchi, B.K.; Nayak, R.; Foley, S.L.; Ricke, S.C. Evaluation of the Genetics and Functionality of Plasmids in Incompatibility Group I1-Positive Salmonella enterica. Foodborne Pathog. Dis. 2018, 15, 168–176. [Google Scholar] [CrossRef]
- Han, J.; Pendleton, S.J.; Deck, J.; Singh, R.; Gilbert, J.; Johnson, T.J.; Sanad, Y.M.; Nayak, R.; Foley, S.L. Impact of co-carriage of IncA/C plasmids with additional plasmids on the transfer of antimicrobial resistance in Salmonella enterica isolates. Int. J. Food Microbiol. 2018, 271, 77–84. [Google Scholar] [CrossRef]
- Johnson, T.; Wannemeuhler, Y.M.; Scaccianoce, J.A.; Johnson, S.J.; Nolan, L.K. Complete DNA Sequence, Comparative Genomics, and Prevalence of an IncHI2 Plasmid Occurring among Extraintestinal Pathogenic Escherichia coli Isolates. Antimicrob. Agents Chemother. 2006, 50, 3929–3933. [Google Scholar] [CrossRef]
- Welch, T.J.; Fricke, W.F.; McDermott, P.F.; White, D.G.; Rosso, M.-L.; Rasko, D.A.; Mammel, M.K.; Eppinger, M.; Rosovitz, M.; Wagner, D.; et al. Multiple Antimicrobial Resistance in Plague: An Emerging Public Health Risk. PLoS ONE 2007, 2, e309. [Google Scholar] [CrossRef]
- Cao, G.; Allard, M.W.; Hoffmann, M.; Monday, S.R.; Muruvanda, T.; Luo, Y.; Payne, J.; Rump, L.; Meng, K.; Zhao, S.; et al. Complete Sequences of Six IncA/C Plasmids of Multidrug-Resistant Salmonella enterica subsp. enterica Serotype Newport. Genome Announc. 2015, 3, e00027-15. [Google Scholar] [CrossRef]
- Call, D.; Singer, R.S.; Meng, D.; Broschat, S.L.; Orfe, L.H.; Anderson, J.M.; Herndon, D.R.; Kappmeyer, L.; Daniels, J.B.; Besser, T.E. bla CMY-2-Positive IncA/C Plasmids from Escherichia coli and Salmonella enterica Are a Distinct Component of a Larger Lineage of Plasmids. Antimicrob. Agents Chemother. 2009, 54, 590–596. [Google Scholar] [CrossRef]
- Wasyl, D.; Kern-Zdanowicz, I.; Domanska-Blicharz, K.; Zając, M.; Hoszowski, A. High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying blaCTX-M-25 gene. Veter. Microbiol. 2015, 175, 85–91. [Google Scholar] [CrossRef]
- Silva, C.; Calva, E.; Calva, J.J.; Wiesner, M.; Fernández-Mora, M.; Puente, J.L.; Vinuesa, P. Complete Genome Sequence of a Human-Invasive Salmonella enterica Serovar Typhimurium Strain of the Emerging Sequence Type 213 Harboring a Multidrug Resistance IncA/C Plasmid and a blaCMY-2-Carrying IncF Plasmid. Genome Announc. 2015, 3, e01323-15. [Google Scholar] [CrossRef]
- Fricke, W.F.; Welch, T.J.; McDermott, P.F.; Mammel, M.K.; Leclerc, J.E.; White, D.G.; Cebula, T.A.; Ravel, J. Comparative Genomics of the IncA/C Multidrug Resistance Plasmid Family. J. Bacteriol. 2009, 191, 4750–4757. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Miriagou, V.; Bertini, A.; Loli, A.; Colinon, C.; Villa, L.; Whichard, J.M.; Rossolini, G. Replicon Typing of Plasmids Encoding Resistance to Newer β-Lactams. Emerg. Infect. Dis. 2006, 12, 1145–1148. [Google Scholar] [CrossRef]
- Fang, L.-X.; Li, X.-P.; Deng, G.-H.; Li, S.-M.; Yang, R.-S.; Wu, Z.-W.; Liao, X.-P.; Sun, J.; Liu, Y.-H. High Genetic Plasticity in Multidrug-Resistant Sequence Type 3-IncHI2 Plasmids Revealed by Sequence Comparison and Phylogenetic Analysis. Antimicrob. Agents Chemother. 2018, 62, e02068-17. [Google Scholar] [CrossRef]
- Han, J.; Lynne, A.M.; David, D.E.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.M.; Foley, S.L. DNA Sequence Analysis of Plasmids from Multidrug Resistant Salmonella enterica Serotype Heidelberg Isolates. PLoS ONE 2012, 7, e51160. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Fedorka-Cray, P.J.; Frye, J.G.; Meinersmann, R.J. Inc A/C Plasmids Are Prevalent in Multidrug-Resistant Salmonella enterica Isolates. Appl. Environ. Microbiol. 2009, 75, 1908–1915. [Google Scholar] [CrossRef]
- Bleicher, A.; Schöfl, G.; Rodicio, M.D.R.; Saluz, H.P. The plasmidome of a Salmonella enterica serovar Derby isolated from pork meat. Plasmid 2013, 69, 202–210. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Kutilova, I.; Medvecky, M.; Hrabák, J.; Dolejska, M. Characterization of the Complete Nucleotide Sequences of IncA/C2 Plasmids Carrying In809-Like Integrons from Enterobacteriaceae Isolates of Wildlife Origin. Antimicrob. Agents Chemother. 2017, 61, e01093-17. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Rehman, M.A.; Yin, X.; Persaud-Lachhman, M.G.; Diarra, M.S. First Detection of a Fosfomycin Resistance Gene, fosA7, in Salmonella enterica Serovar Heidelberg Isolated from Broiler Chickens. Antimicrob. Agents Chemother. 2017, 61, e00410-17. [Google Scholar] [CrossRef]
- Levy, S.B. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 1992, 36, 695–703. [Google Scholar] [CrossRef]
- Witte, W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 2000, 16, 19–24. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M.C. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Boil. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Marti, H.; Kim, H.; Joseph, S.J.; Dojiri, S.; Read, T.D.; Dean, D. Tet(C) Gene Transfer between Chlamydia suis Strains Occurs by Homologous Recombination after Co-infection: Implications for Spread of Tetracycline-Resistance among Chlamydiaceae. Front. Microbiol. 2017, 8, 313. [Google Scholar] [CrossRef]
- Dugan, J.; Rockey, D.D.; Jones, L.; Andersen, A.A. Tetracycline Resistance in Chlamydia suis Mediated by Genomic Islands Inserted into the Chlamydial inv-Like Gene. Antimicrob. Agents Chemother. 2004, 48, 3989–3995. [Google Scholar] [CrossRef]
- Furushita, M.; Shiba, T.; Maeda, T.; Yahata, M.; Kaneoka, A.; Takahashi, Y.; Torii, K.; Hasegawa, T.; Ohta, M. Similarity of Tetracycline Resistance Genes Isolated from Fish Farm Bacteria to Those from Clinical Isolates. Appl. Environ. Microbiol. 2003, 69, 5336–5342. [Google Scholar] [CrossRef] [PubMed]
- Mulvey, M.R.; Boyd, D.A.; Baker, L.; Mykytczuk, O.; Reis, E.M.F.; Asensi, M.D.; Rodrigues, D.P.; Ng, L. Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing novel OXA-type B-lactamase (blaoxa-53) and 6’-N-aminoglycoside acetyltransferase genes. J. Antimicrob. Chemother. 2004, 54, 354–359. [Google Scholar] [CrossRef] [PubMed]
- L’Abée-Lund, T.M.; Sørum, H. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 2002, 47, 172–181. [Google Scholar] [CrossRef]
- Frech, G.; Schwarz, S. Molecular analysis of tetracycline resistance in Salmonella enterica subsp. enterica serovars Typhimurium, Enteritidis, Dublin, Choleraesuis, Hadar and Saintpaul: Construction and application of specific gene probes. J. Appl. Microbiol. 2000, 89, 633–641. [Google Scholar] [CrossRef]
- Miko, A.; Pries, K.; Schroeter, A.; Helmuth, R. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 2005, 56, 1025–1033. [Google Scholar] [CrossRef]
- Thai, T.H.; Hirai, T.; Lan, N.T.; Shimada, A.; Ngoc, P.T.; Yamaguchi, R. Antimicrobial resistance of Salmonella serovars isolated from beef at retail markets in the north Vietnam. J. Veter. Med. Sci. 2012, 74, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 2002, 49, 25–30. [Google Scholar] [CrossRef]
- Sundin, G.W.; Bender, C.L. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 1996, 5, 133–143. [Google Scholar] [CrossRef]
- Van Overbeek, L.S.; Wellington, E.M.H.; Egan, S.; Smalla, K.; Heuer, H.; Collard, J.M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; Van Elsas, J.D. Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 2002, 42, 277–288. [Google Scholar] [CrossRef]
- Public Health Agency of Canada. Canadian Antimicrobial Resistance Surveillance System Report Ontario; Public Health Agency of Canada: Ottawa, ON, Canada, 2016; ISBN 6139572991. [Google Scholar]
- Gunell, M.; Kotilainen, P.; Jalava, J.; Huovinen, P.; Siitonen, A.; Hakanen, A. In Vitro Activity of Azithromycin against Nontyphoidal Salmonella enterica. Antimicrob. Agents Chemother. 2010, 54, 3498–3501. [Google Scholar] [CrossRef]
- Mandalakas, A.M.; Menzies, D. Outpatient treatment of patients with enteric fever. Lancet Infect. Dis. 2011, 11, 419–421. [Google Scholar]
- Guerrant, R.L.; Van Gilder, T.; Steiner, T.S.; Thielman, N.M.; Slutsker, L.; Tauxe, R.V.; Hennessy, T.; Griffin, P.M.; Dupont, H.; Sack, R.B.; et al. Practice Guidelines for the Management of Infectious Diarrhea. Clin. Infect. Dis. 2001, 32, 331–351. [Google Scholar] [CrossRef] [PubMed]
| Serotype | Number of Isolates (Given by SISTR V. 1.0.2) | Nine Most Frequent ORIs (if nreplicon ≥ 20) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IncFII | IncFIB | IncI1 | IncX1 | ColpVC | IncA/C2 | Col156 | IncHI2A | IncHI2 | ||
| Typhimurium | 201 | 147 | 145 | 28 | 3 | 7 | 11 | 6 | - | - |
| Enteritidis | 200 | 179 | 178 | 9 | 24 | 3 | - | 1 | - | - |
| Newport | 80 | 14 | - | 2 | 3 | - | 25 | - | 1 | 1 |
| Heidelberg | 51 | - | - | 13 | 35 | 19 | 1 | 2 | 4 | 4 |
| Oranienburg | 49 | 30 | - | - | - | 1 | - | - | - | - |
| Thompson | 44 | 2 | - | - | - | 3 | - | - | - | - |
| Muenchen | 43 | 4 | 2 | 3 | - | - | - | - | - | - |
| Infantis | 38 | - | - | 5 | - | - | 1 | - | - | - |
| Anatum | 38 | 13 | - | 5 | - | 1 | 3 | - | 1 | 1 |
| Senftenberg | 36 | - | 1 | - | - | - | - | - | - | - |
| Braenderup | 34 | 3 | 3 | - | - | 1 | - | - | - | - |
| Javiana | 28 | 2 | - | 2 | - | - | - | - | - | - |
| Saintpaul | 26 | 4 | 1 | 3 | - | 2 | - | 1 | 2 | 2 |
| Agona | 26 | 1 | - | 7 | - | 1 | 1 | - | 1 | 1 |
| I_4 [5]; 12:i:- | 25 | 17 | 17 | 6 | - | 1 | 1 | - | 1 | 1 |
| Montevideo | 24 | 1 | - | 1 | 1 | - | - | - | - | - |
| Paratyphi_B_var._Java_monophasic | 23 | 16 | - | 1 | - | 1 | - | - | - | - |
| Give | 23 | - | - | 1 | - | 1 | - | 1 | - | - |
| Gaminara | 21 | - | - | - | - | - | - | - | - | |
| Paratyphi_B_var._Java | 20 | 1 | - | 3 | - | 1 | - | - | - | - |
| Paratyphi_B | 20 | - | - | 2 | - | - | - | - | - | - |
| Kentucky | 20 | 9 | 9 | 11 | 13 | 1 | - | 1 | 3 | 3 |
| Typhi | 17 | - | 1 | - | - | - | - | - | - | - |
| Hartford | 17 | 17 | - | - | - | 1 | - | - | - | - |
| Tennessee | 16 | - | - | 1 | - | - | 1 | 1 | 1 | 1 |
| Mississippi | 16 | 1 | - | 1 | - | - | - | - | - | - |
| Mbandaka | 16 | - | 1 | - | - | 1 | - | - | 1 | 1 |
| Rubislaw | 15 | 2 | - | - | 2 | 5 | - | 3 | - | - |
| Manhattan | 15 | - | - | 2 | - | 1 | - | - | - | - |
| Dublin | 15 | 15 | - | - | 15 | - | 5 | - | - | - |
| Total of isolates | 1197 | 478 | 358 | 106 | 96 | 51 | 49 | 16 | 15 | 15 |
| Total of isolates in other serotypes | 553 | 55 | 13 | 36 | 2 | 37 | 4 | 13 | 9 | 9 |
| Total of isolates in all serotypes | 1750 | 533 | 371 | 142 | 98 | 88 | 53 | 29 | 24 | 24 |
| Count of serotypes | 153 | 50 | 18 | 43 | 10 | 37 | 12 | 16 | 17 | 17 |
| AMR Genes with a p-Value ≤ 0.001 | Prevalence (%) | Resistance Mechanism | Drug Class | Confers Resistance to |
|---|---|---|---|---|
| aac(3)-Via | 1.4 | antibiotic inactivation | aminoglycoside antibiotic | gentamicin B and C |
| aph(3’’)-Ib (strA) | 10.5 | antibiotic inactivation | aminoglycoside antibiotic | streptomycin |
| aph(6)-Id (strB) | 10.4 | antibiotic inactivation | aminoglycoside antibiotic | streptomycin |
| aadA8 | 0.8 | antibiotic inactivation | aminoglycoside antibiotic | streptomycin and spectinomycin |
| aadA13 | 2.5 | antibiotic inactivation | aminoglycoside antibiotic | streptomycin and spectinomycin |
| blaCMY-2 | 6.3 | antibiotic inactivation | cephamycin and cephalosporin | cefoxitin, cephamycin and ceftazidime |
| blaTEM-1 | 4.3 | antibiotic inactivation | penem, cephalosporin, monobactam, penam | amoxicillin, ampicillin and cefalotin |
| floR | 4.7 | antibiotic efflux | phenicol antibiotic | chloramphenicol and florfenicol |
| sul2 | 8.5 | antibiotic target replacement | sulfonamide antibiotic, sulfone antibiotic | sulfadiazine, sulfadimidine, sulfadoxine, sulfamethoxazole, sulfisoxazole, sulfacetamide, mafenide, sulfasalazine and sulfamethizole |
| tet(C) | 7.9 | antibiotic efflux | tetracycline antibiotic | tetracycline |
| tet(D) | 2.2 | antibiotic efflux | tetracycline antibiotic | tetracycline |
| mprF (Brucella suis) | 2.1 | antibiotic target alteration | peptide antibiotic | defensin |
| qnrB19 | 0.7 | antibiotic target protection | fluoroquinolone antibiotic | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emond-Rheault, J.-G.; Hamel, J.; Jeukens, J.; Freschi, L.; Kukavica-Ibrulj, I.; Boyle, B.; Tamber, S.; Malo, D.; Franz, E.; Burnett, E.; et al. The Salmonella enterica Plasmidome as a Reservoir of Antibiotic Resistance. Microorganisms 2020, 8, 1016. https://doi.org/10.3390/microorganisms8071016
Emond-Rheault J-G, Hamel J, Jeukens J, Freschi L, Kukavica-Ibrulj I, Boyle B, Tamber S, Malo D, Franz E, Burnett E, et al. The Salmonella enterica Plasmidome as a Reservoir of Antibiotic Resistance. Microorganisms. 2020; 8(7):1016. https://doi.org/10.3390/microorganisms8071016
Chicago/Turabian StyleEmond-Rheault, Jean-Guillaume, Jérémie Hamel, Julie Jeukens, Luca Freschi, Irena Kukavica-Ibrulj, Brian Boyle, Sandeep Tamber, Danielle Malo, Eelco Franz, Elton Burnett, and et al. 2020. "The Salmonella enterica Plasmidome as a Reservoir of Antibiotic Resistance" Microorganisms 8, no. 7: 1016. https://doi.org/10.3390/microorganisms8071016
APA StyleEmond-Rheault, J.-G., Hamel, J., Jeukens, J., Freschi, L., Kukavica-Ibrulj, I., Boyle, B., Tamber, S., Malo, D., Franz, E., Burnett, E., Daigle, F., Arya, G., Sanderson, K., Wiedmann, M., Slawson, R. M., Weadge, J. T., Stephan, R., Bekal, S., Gruenheid, S., ... Levesque, R. C. (2020). The Salmonella enterica Plasmidome as a Reservoir of Antibiotic Resistance. Microorganisms, 8(7), 1016. https://doi.org/10.3390/microorganisms8071016







