Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
2.2. Generation of S. typhimurium 14028 Mutant Strains
2.3. Generation of Bacterial Luciferase Reporters and Other Plasmid Vectors
2.4. Luciferase Reporter Assays
3. Results
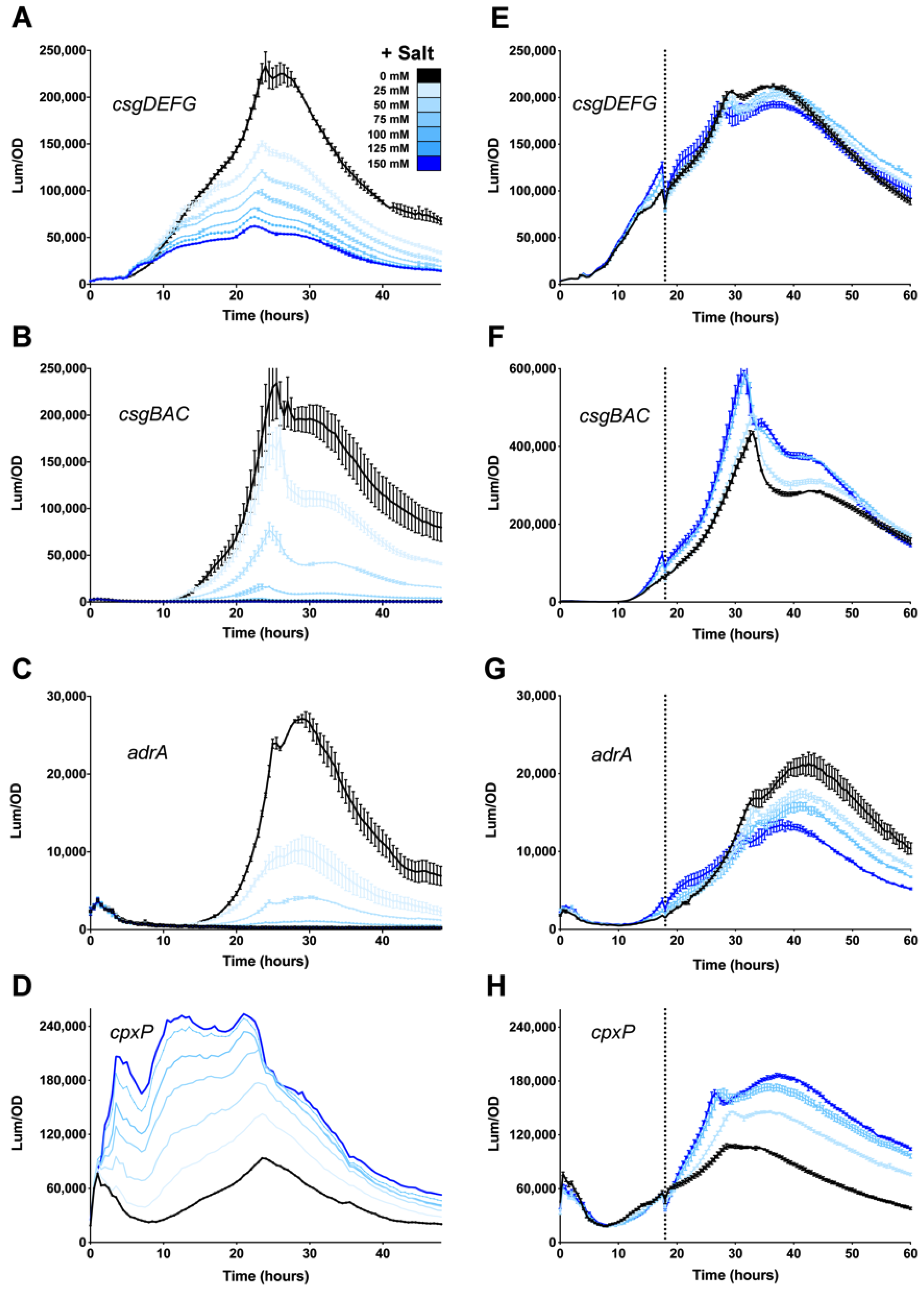
3.1. Osmolarity Has No Effect Once csgD Transcription Is Activated
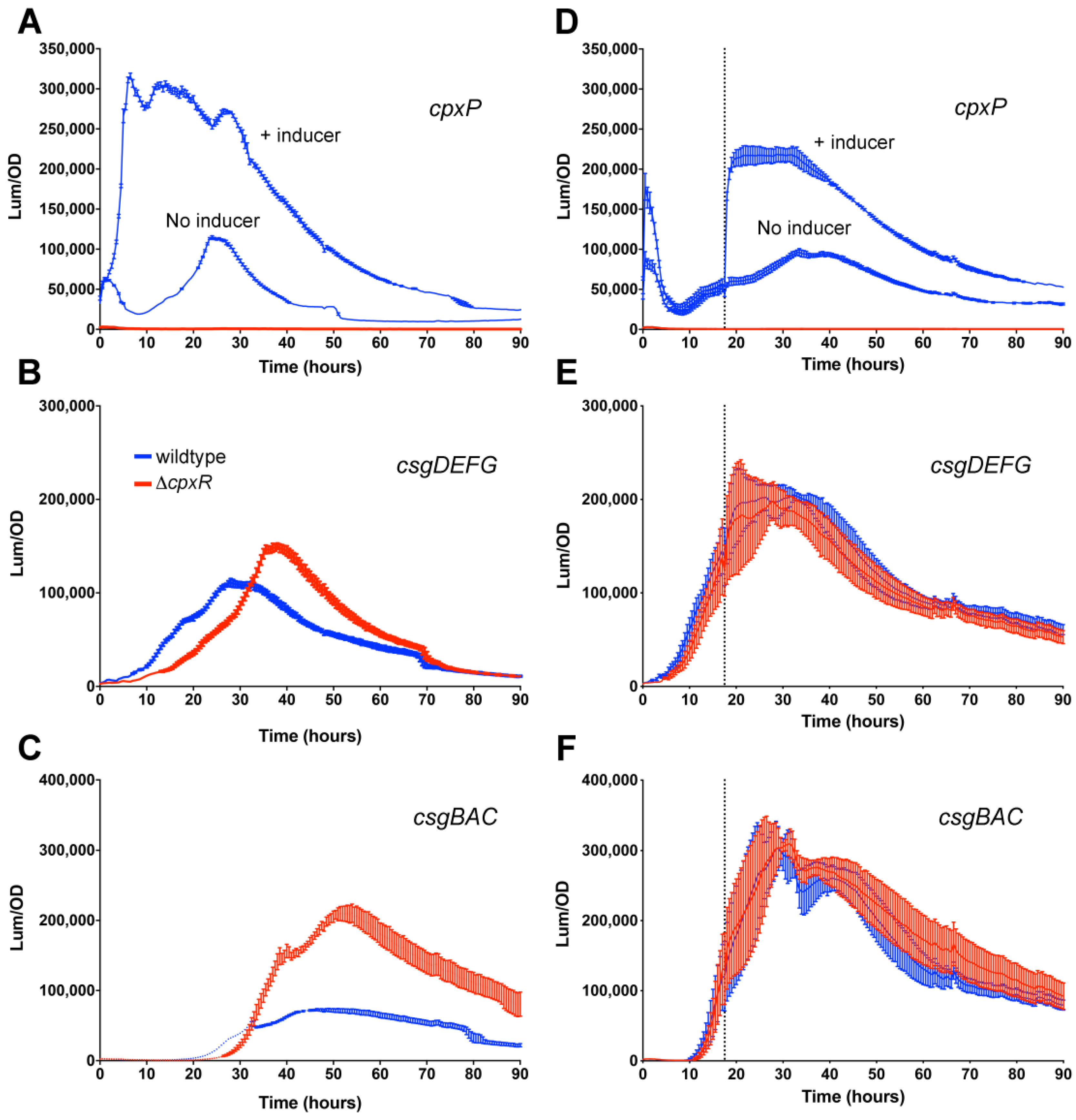
3.2. Repressive Effect of CpxR on csgD Transcription Is Alleviated During Growth
3.3. Temperature and Glucose Repress csgD Expression
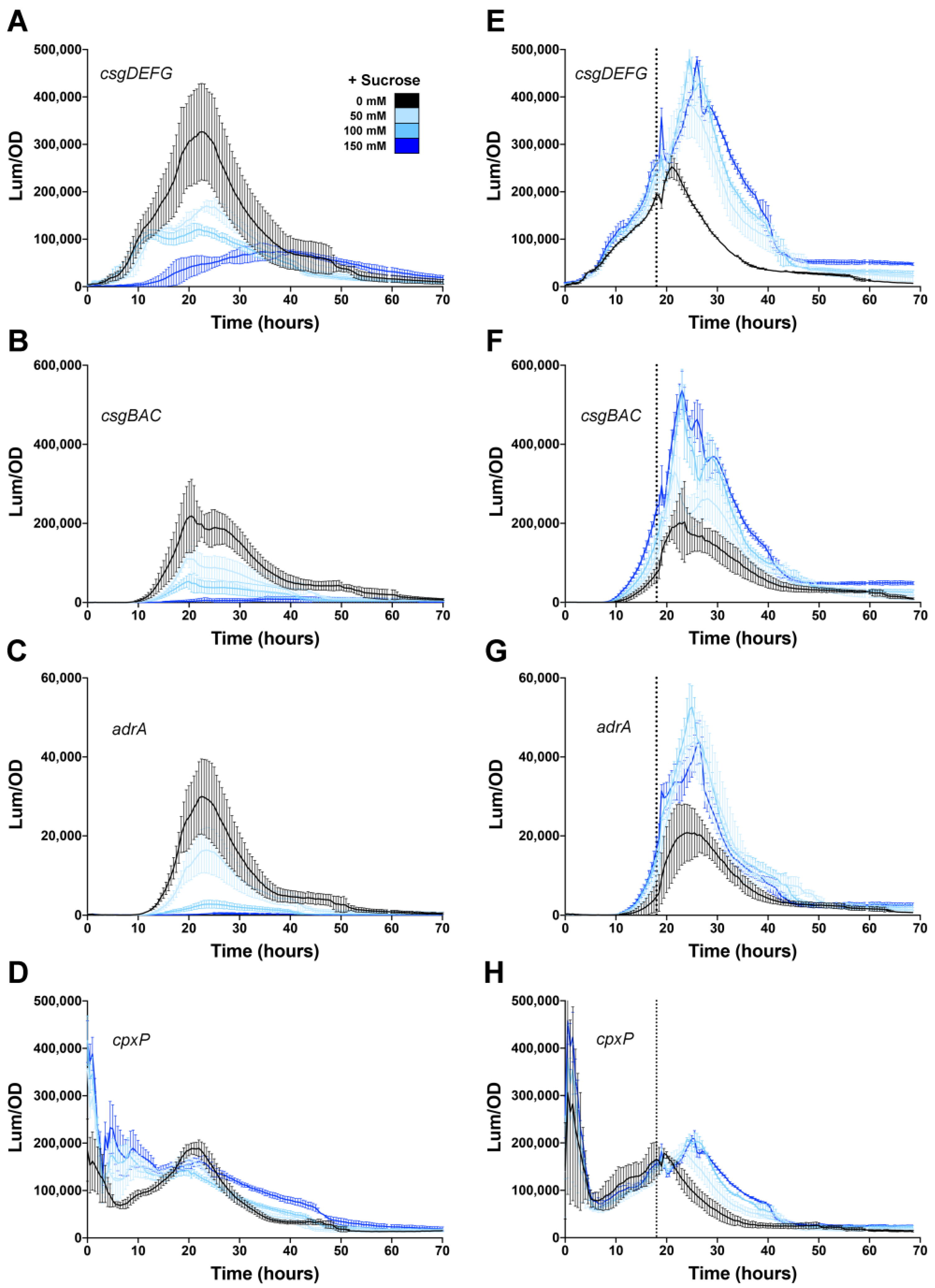
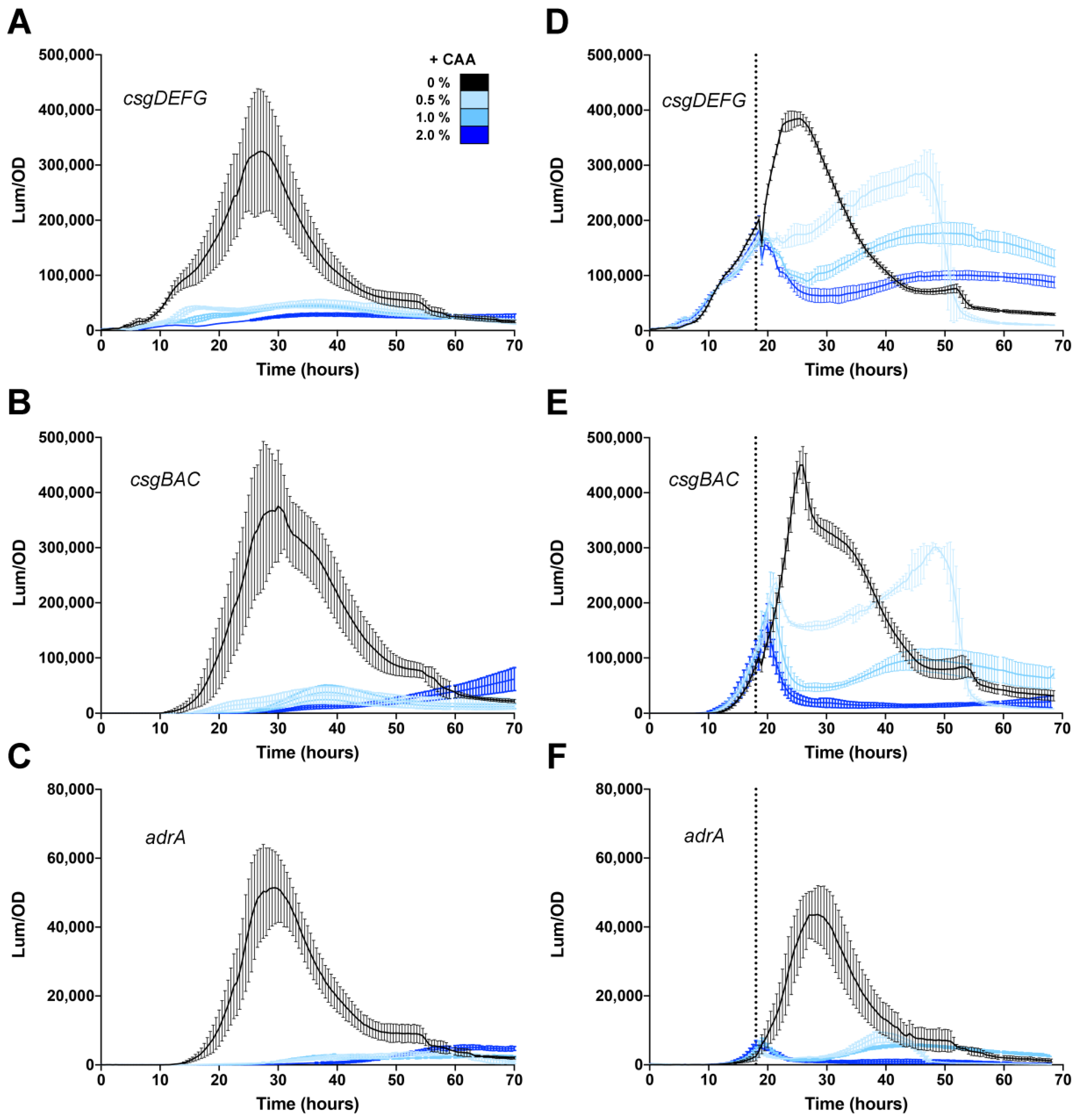
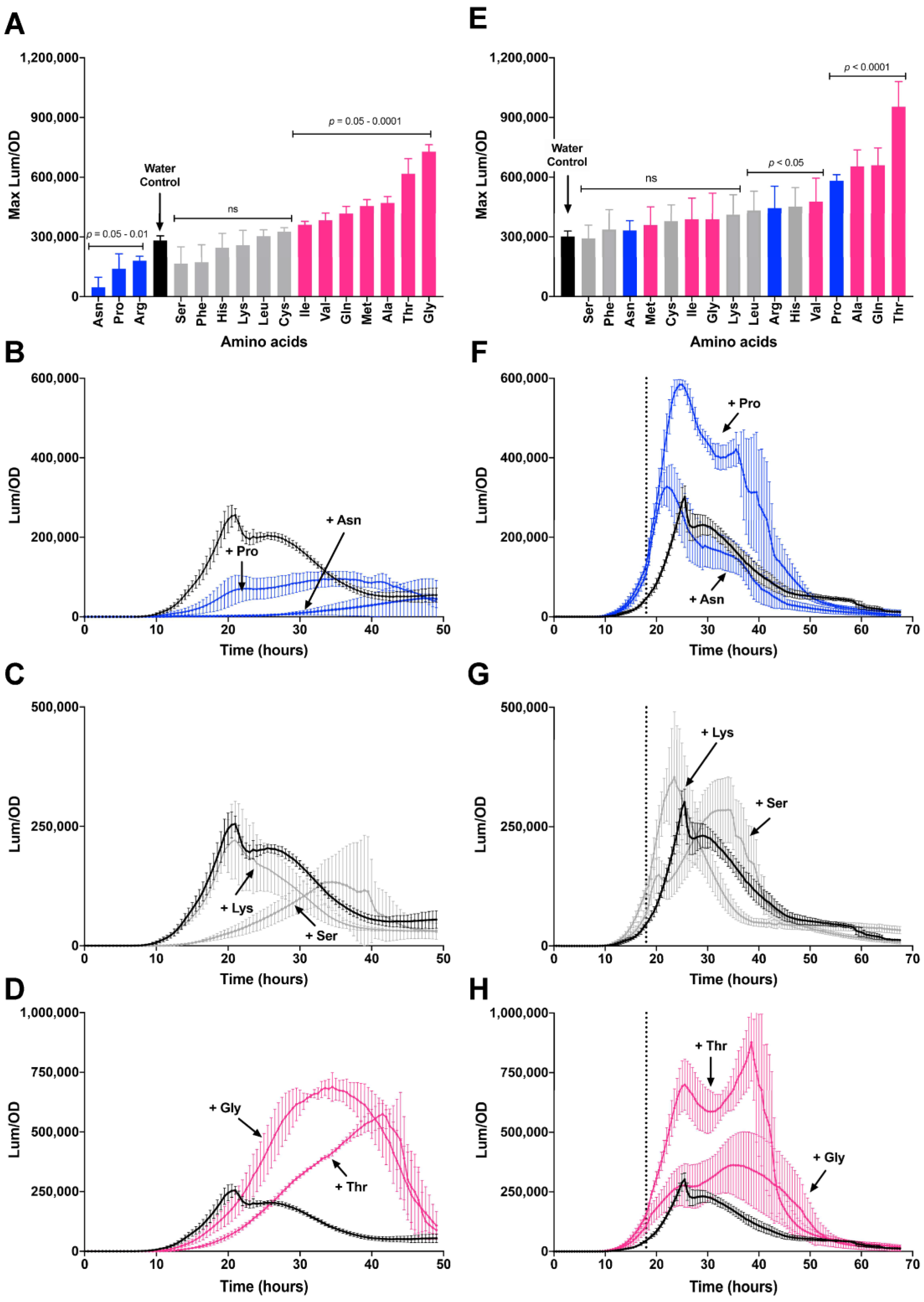
3.4. Effect of Casamino Acids on Biofilm Formation
3.5. Differing Effects of Individual Amino Acids on csgD Gene Expression
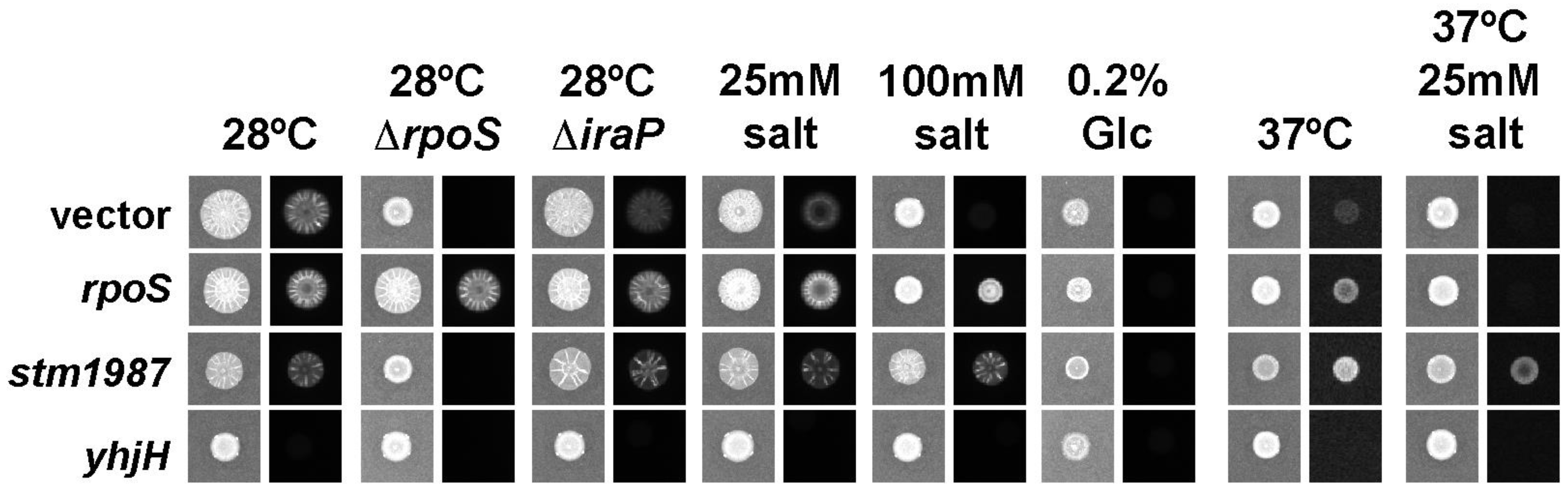
3.6. Regulation of Rdar Morphotype on Agar-Containing Media
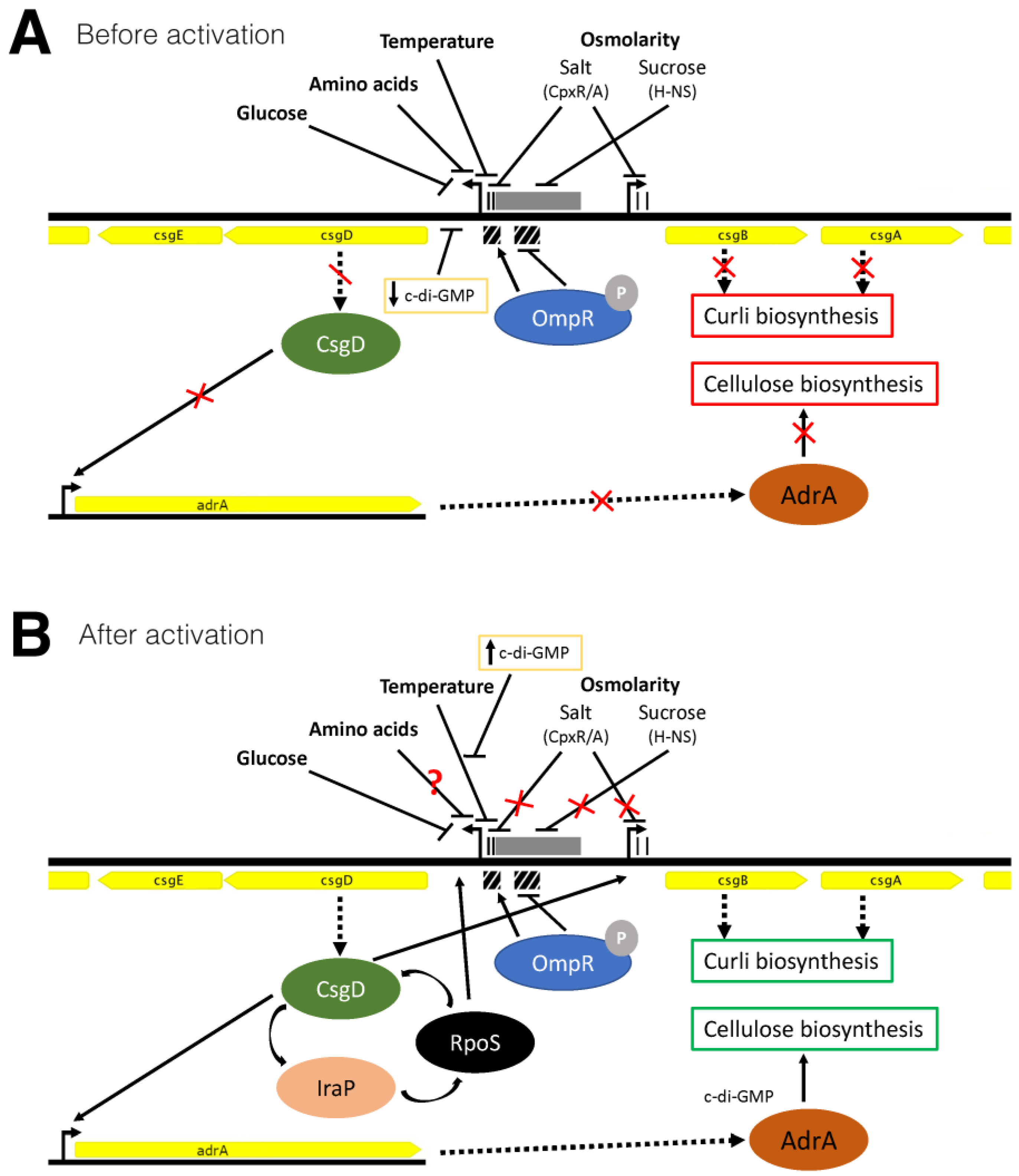
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. Correction: World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PloS Med. 2015, 12, e1001940. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; The International Collaboration on Enteric Disease “Burden of Illness” Studies. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Buckle, G.C.; Walker, C.L.F.; Black, R.E. Typhoid fever and paratyphoid fever: Systematic review to estimate global morbidity and mortality for 2010. J. Glob. Health 2012, 2, 010401. [Google Scholar] [CrossRef] [PubMed]
- McClelland, M.; Sanderson, K.E.; Clifton, S.W.; Latreille, P.; Porwollik, S.; Sabo, A.; Meyer, R.; Bieri, T.; Ozersky, P.; McLellan, M.; et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004, 36, 1268–1274. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef]
- Hassan, R.; Buuck, S.; Noveroske, D.; Medus, C.; Sorenson, A.; Laurent, J.; Rotstein, D.; Schlater, L.; Freiman, J.; Douris, A.; et al. Multistate Outbreak of Salmonella Infections Linked to Raw Turkey Products—United States, 2017–2019. Morb. Mortal. Wkly. Rep. 2019, 68, 1045–1049. [Google Scholar] [CrossRef]
- Morton, V.K.; Kearney, A.; Coleman, S.; Viswanathan, M.; Chau, K.; Orr, A.; Hexemer, A. Outbreaks of Salmonella illness associated with frozen raw breaded chicken products in Canada, 2015–2019. Epidemiol. Infect. 2019, 147, e254. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; Keersmaecker, S.C.J.D. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Proctor, M.E.; Hamacher, M.; Tortorello, M.L.; Archer, J.R.; Davis, J.P. Multistate Outbreak of Salmonella Serovar Muenchen Infections Associated with Alfalfa Sprouts Grown from Seeds Pretreated with Calcium Hypochlorite. J. Clin. Microbiol. 2001, 39, 3461–3465. [Google Scholar] [CrossRef]
- Werber, D.; Dreesman, J.; Feil, F.; van Treeck, U.; Fell, G.; Ethelberg, S.; Hauri, A.M.; Roggentin, P.; Prager, R.; Fisher, I.S.; et al. International outbreak of Salmonella Oranienburg due to German chocolate. BMC Infect. Dis. 2005, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Waldner, L.L.; MacKenzie, K.D.; Köster, W.; White, A.P. From Exit to Entry: Long-term Survival and Transmission of Salmonella. Pathogens 2012, 1, 128–155. [Google Scholar] [CrossRef] [PubMed]
- White, A.P.; Gibson, D.L.; Kim, W.; Kay, W.W.; Surette, M.G. Thin Aggregative Fimbriae and Cellulose Enhance Long-Term Survival and Persistence of Salmonella. J. Bacteriol. 2006, 188, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- Apel, D.; White, A.P.; Grassl, G.A.; Finlay, B.B.; Surette, M.G. Long-term survival of Salmonella enterica serovar Typhimurium reveals an infectious state that is underrepresented on laboratory media containing bile salts. Appl. Environ. Microb. 2009, 75, 4923–4925. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.D.; Palmer, M.B.; Köster, W.L.; White, A.P. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017, 4, 138. [Google Scholar] [CrossRef]
- Finn, S.; Condell, O.; McClure, P.; Amézquita, A.; Fanning, S. Mechanisms of survival, responses and sources of Salmonella in low-moisture environments. Front. Microbiol. 2013, 4, 331. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Winfield, M.D.; Groisman, E.A. Role of Nonhost Environments in the Lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microb. 2003, 69, 3687–3694. [Google Scholar] [CrossRef]
- Singletary, L.A.; Karlinsey, J.E.; Libby, S.J.; Mooney, J.P.; Lokken, K.L.; Tsolis, R.M.; Byndloss, M.X.; Hirao, L.A.; Gaulke, C.A.; Crawford, R.W.; et al. Loss of Multicellular Behavior in Epidemic African Nontyphoidal Salmonella enterica Serovar Typhimurium ST313 Strain D23580. MBio 2016, 7, e02265. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Wang, Y.; Musicha, P.; Hansen, E.G.; Palmer, M.B.; Herman, D.J.; Feasey, N.A.; White, A.P. Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains. PloS Genet. 2019, 15, e1008233. [Google Scholar] [CrossRef]
- Crawford, R.W.; Rosales-Reyes, R.; Ramírez-Aguilar, M.; Chapa-Azuela, O.; Alpuche-Aranda, C.; Gunn, J.S. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. USA 2010, 107, 4353–4358. [Google Scholar] [CrossRef]
- Römling, U.; Bokranz, W.; Rabsch, W.; Zogaj, X.; Nimtz, M.; Tschäpe, H. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 2003, 293, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Rohde, M.; Olsen, A.; Normark, S.; Reinkoster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Dieltjens, L.; Appermans, K.; Lissens, M.; Lories, B.; Kim, W.; Van der Eycken, E.V.; Foster, K.R.; Steenackers, H.P. Inhibiting bacterial cooperation is an evolutionarily robust anti-biofilm strategy. Nat. Commun. 2020, 11, 107. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- White, A.P.; Weljie, A.M.; Apel, D.; Zhang, P.; Shaykhutdinov, R.; Vogel, H.J.; Surette, M.G. A global metabolic shift is linked to Salmonella multicellular development. PLoS ONE 2010, 5, e11814. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 2015, 13, 497–508. [Google Scholar] [CrossRef]
- Serra, D.O.; Richter, A.M.; Klauck, G.; Mika, F.; Hengge, R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 2013, 4, e00103-13. [Google Scholar] [CrossRef]
- Gerstel, U.; Römling, U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 2003, 154, 659–667. [Google Scholar] [CrossRef]
- Gerstel, U.; Romling, U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 2001, 3, 638–648. [Google Scholar] [CrossRef]
- Beloin, C.; Roux, A.; Ghigo, J.-M. Escherichia coli biofilms. Curr. Top. Microbiol. 2008, 322, 249–289. [Google Scholar] [CrossRef]
- Kolter, R.; Siegele, D.A.; Tormo, A. The Stationary Phase of The Bacterial Life Cycle. Annu. Rev. Microbiol. 1993, 47, 855–874. [Google Scholar] [CrossRef] [PubMed]
- Hengge-Aronis, R. Signal Transduction and Regulatory Mechanisms Involved in Control of the σS (RpoS) Subunit of RNA Polymerase. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef]
- Arnqvist, A.; Olsén, A.; Normark, S. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 1994, 13, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Gualdi, L.; Tagliabue, L.; Landini, P. Biofilm formation-gene expression relay system in Escherichia coli: Modulation of sigmaS-dependent gene expression by the CsgD regulatory protein via sigmaS protein stabilization. J. Bacteriol. 2007, 189, 8034–8043. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.-M.; Lazzaroni, J.-C.; Lejeune, P.; Dorel, C. CpxR/OmpR Interplay Regulates Curli Gene Expression in Response to Osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Brombacher, E.; Vidal, O.; Ambert, A.; Lejeune, P.; Landini, P.; Dorel, C. Complex Regulatory Network Controls Initial Adhesion and Biofilm Formation in Escherichia coli via Regulation of the csgD Gene. J. Bacteriol. 2001, 183, 7213–7223. [Google Scholar] [CrossRef]
- Gerstel, U.; Park, C.; Römling, U. Complex regulation of csgD promoter activity by global regulatory proteins: Regulation of csgD expression. Mol. Microbiol. 2004, 49, 639–654. [Google Scholar] [CrossRef]
- White, A.P.; Gibson, D.L.; Grassl, G.A.; Kay, W.W.; Finlay, B.B.; Vallance, B.A.; Surette, M.G. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect. Immun. 2008, 76, 1048–1058. [Google Scholar] [CrossRef]
- Grantcharova, N.; Peters, V.; Monteiro, C.; Zakikhany, K.; Römling, U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J. Bacteriol. 2009, 192, 456–466. [Google Scholar] [CrossRef]
- DePas, W.H.; Hufnagel, D.A.; Lee, J.S.; Blanco, L.P.; Bernstein, H.C.; Fisher, S.T.; James, G.A.; Stewart, P.S.; Chapman, M.R. Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.D.; Wang, Y.; Shivak, D.J.; Wong, C.S.; Hoffman, L.J.L.; Lam, S.; Kröger, C.; Cameron, A.D.S.; Townsend, H.G.G.; Köster, W.; et al. Bistable expression of CsgD in Salmonella enterica serovar Typhimurium connects virulence to persistence. Infect. Immun. 2015, 83, 2312–2326. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. Mmbr 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Cimdins, A.; Beske, T.; Römling, U. Detailed analysis of c-di-GMP mediated regulation of csgD expression in Salmonella typhimurium. Bmc Microbiol. 2017, 17, 27. [Google Scholar] [CrossRef]
- Solano, C.; García, B.; Latasa, C.; Toledo-Arana, A.; Zorraquino, V.; Valle, J.; Casals, J.; Pedroso, E.; Lasa, I. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc. Natl. Acad. Sci. USA 2009, 106, 7997–8002. [Google Scholar] [CrossRef]
- Dworkin, J.; Losick, R. Developmental Commitment in a Bacterium. Cell 2005, 121, 401–409. [Google Scholar] [CrossRef]
- Bjarnason, J.; Southward, C.M.; Surette, M.G. Genomic Profiling of Iron-Responsive Genes in Salmonella enterica Serovar Typhimurium by High-Throughput Screening of a Random Promoter Library. J. Bacteriol. 2003, 185, 4973–4982. [Google Scholar] [CrossRef]
- White, A.P.; Surette, M.G. Comparative Genetics of the rdar Morphotype in Salmonella. J. Bacteriol. 2006, 188, 8395–8406. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Maloy, S.R. Experimental Techniques in Bacterial Genetics; Jones & Bartlett Learning: Boston, MA, USA, 1990; p. 180. [Google Scholar]
- Collinson, S.K.; Emödy, L.; Müller, K.H.; Trust, T.J.; Kay, W.W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 1991, 173, 4773–4781. [Google Scholar] [CrossRef]
- Romling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Danese, P.N.; Silhavy, T.J. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 1998, 180, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Hunke, S.; Keller, R.; Müller, V.S. Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 2011, 326, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishihama, A. Characterization of Copper-Inducible Promoters Regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 2006, 70, 1688–1695. [Google Scholar] [CrossRef]
- Novick, A.; Weiner, M. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 1957, 43, 553–566. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Keen, J.E.; Elder, R.O. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microb. 2001, 67, 2367–2370. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Chapman, M.R. Curli Biogenesis and Function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef]
- Jackson, D.W.; Simecka, J.W.; Romeo, T. Catabolite Repression of Escherichia coli Biofilm Formation. J. Bacteriol. 2002, 184, 3406–3410. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Veening, J.-W.; Smits, W.K.; Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef]
- Veening, J.-W.; Stewart, E.J.; Berngruber, T.W.; Taddei, F.; Kuipers, O.P.; Hamoen, L.W. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. USA 2008, 105, 4393–4398. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Hengge, R. Stress responses go three dimensional—The spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 2014, 16, 1455–1471. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Soo, J.; Rao, C.; Leung, A.S.; Ngai, D.H.-M.; Ensminger, A.W.; Navarre, W.W. Silencing by H-NS potentiated the evolution of Salmonella. Plos Pathog. 2014, 10, e1004500. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M. Salmonella persistence and transmission strategies. Curr. Opin. Microbiol. 2012, 15, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.J.; Benitez, J.A. Vibrio cholerae Biofilms and Cholera Pathogenesis. PloS Neglect. Trop. Dis. 2016, 10, e0004330. [Google Scholar] [CrossRef]
- Monds, R.D.; O’Toole, G.A. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2009, 17, 73–87. [Google Scholar] [CrossRef]
- Kragh, K.N.; Hutchison, J.B.; Melaugh, G.; Rodesney, C.; Roberts, A.E.L.; Irie, Y.; Jensen, P.Ø.; Diggle, S.P.; Allen, R.J.; Gordon, V.; et al. Role of Multicellular Aggregates in Biofilm Formation. MBio 2016, 7, e00237. [Google Scholar] [CrossRef]
- Bokranz, W.; Wang, X.; Tschäpe, H.; Römling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef]
- Ogasawara, H.; Yamada, K.; Kori, A.; Yamamoto, K.; Ishihama, A. Regulation of the Escherichia coli csgD promoter: Interplay between five transcription factors. Microbiol. 2010, 156, 2470–2483. [Google Scholar] [CrossRef]
- Brown, P.K.; Dozois, C.M.; Nickerson, C.A.; Zuppardo, A.; Terlonge, J.; Curtiss, R. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium: Regulation of curli and aggregation by mlrA. Mol. Microbiol. 2001, 41, 349–363. [Google Scholar] [CrossRef]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Szczesny, M.; Beloin, C.; Ghigo, J.-M. Increased Osmolarity in Biofilm Triggers RcsB-Dependent Lipid A Palmitoylation in Escherichia coli. MBio 2018, 9, e01415-18. [Google Scholar] [CrossRef] [PubMed]
- Kimkes, T.E.P.; Heinemann, M. Reassessing the role of the Escherichia coli CpxAR system in sensing surface contact. PLoS ONE 2018, 13, e0207181. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.L. Everything old is new again: An update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 2013, 1843, 1529–1541. [Google Scholar] [CrossRef]
- Bremer, E.; Krämer, R. Responses of Microorganisms to Osmotic Stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef]
- Grinberg, M.; Orevi, T.; Steinberg, S.; Kashtan, N. Bacterial survival in microscopic surface wetness. Elife 2019, 8, e48508. [Google Scholar] [CrossRef]
- Solopova, A.; van Gestel, J.; Weissing, F.J.; Bachmann, H.; Teusink, B.; Kok, J.; Kuipers, O.P. Bet-hedging during bacterial diauxic shift. Proc. Natl. Acad. Sci. USA 2014, 111, 7427–7432. [Google Scholar] [CrossRef]
- Paytubi, S.; Cansado, C.; Madrid, C.; Balsalobre, C. Nutrient Composition Promotes Switching between Pellicle and Bottom Biofilm in Salmonella. Front. Microbiol. 2017, 8, 2160. [Google Scholar] [CrossRef]
- Sokaribo, A.; Novakovski, B.A.A.; Cotelesage, J.; White, A.P.; Sanders, D.; Goldie, H. Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants. Biochim. Et Biophys. Acta Gen. Subj. 2020, 1864, 129517. [Google Scholar] [CrossRef]
- Lehninger, A.L.; Nelson, D.L.; Cox, M.M. Lehninger principles of biochemistry, 4th ed.; Worth Publishers: New York, NY, USA, 2000. [Google Scholar]
- Chirwa, N.T.; Herrington, M.B. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 2003, 149, 525–535. [Google Scholar] [CrossRef]
- Malcova, M.; Karasova, D.; Rychlik, I. aroA and aroD mutations influence biofilm formation in Salmonella Enteritidis. Fems Microbiol. Lett. 2008, 291, 44–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hamilton, S.; Bongaerts, R.J.M.; Mulholland, F.; Cochrane, B.; Porter, J.; Lucchini, S.; Lappin-Scott, H.M.; Hinton, J.C.D. The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genom. 2009, 10, 599. [Google Scholar] [CrossRef] [PubMed]
- Nolan, R. Amino Acids and Growth Factors in Vitamin-Free Casamino Acids. Mycologia 1971, 63, 1231–1234. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Durfee, T.; Cabrera, J.; Zhao, K.; Jin, D.; Blattner, F. Global Transcriptional Programs Reveal a Carbon Source Foraging Strategy by Escherichia coli. J. Biol. Chem. 2005, 280, 15921–15927. [Google Scholar] [CrossRef] [PubMed]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Hufnagel, D.A.; Evans, M.L.; Greene, S.E.; Pinkner, J.S.; Hultgren, S.J.; Chapman, M.R. The Catabolite Repressor Protein-Cyclic AMP Complex Regulates csgD and Biofilm Formation in Uropathogenic Escherichia coli. J. Bacteriol. 2016, 198, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli Fibers Are Highly Conserved between Salmonella typhimurium and Escherichia coli with Respect to Operon Structure and Regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef]
- Bren, A.; Park, J.O.; Towbin, B.D.; Dekel, E.; Rabinowitz, J.D.; Alon, U. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 2016, 6, 24834. [Google Scholar] [CrossRef]
- Miller, A.L.; Pasternak, J.A.; Medeiros, N.J.; Nicastro, L.K.; Tursi, S.A.; Hansen, E.G.; Krochak, R.; Sokaribo, A.S.; MacKenzie, K.D.; Palmer, M.B.; et al. In vivo synthesis of bacterial amyloid curli contributes to joint inflammation during S. typhimurium infection. Plos Pathog. 2020, 16. [Google Scholar] [CrossRef]
- González, J.F.; Tucker, L.; Fitch, J.; Wetzel, A.; White, P.; Gunn, J.S. Human Bile-Mediated Regulation of Salmonella Curli Fimbriae. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Simm, R.; Lusch, A.; Kader, A.; Andersson, M.; Romling, U. Role of EAL-Containing Proteins in Multicellular Behavior of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2007, 189, 3613–3623. [Google Scholar] [CrossRef] [PubMed]
- Kader, A.; Simm, R.; Gerstel, U.; Morr, M.; Romling, U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006, 60, 602–616. [Google Scholar] [CrossRef] [PubMed]
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]

| Strains or Plasmids | Genotype | Reference |
|---|---|---|
| Strains | ||
| Salmonella enterica subsp. enterica serovar Typhimurium | ||
| 14028 | Wild-type strain | ATCC |
| 14028 ∆cpxR | Deletion of cpxR | This study |
| 14028 ∆iraP | Deletion of iraP | This study |
| 14028 ∆rpoS | Deletion of rpoS | [12] |
| Plasmids | ||
| pCS26, pU220 | Bacterial luciferase | [47] |
| pCS26-stm1987::luxCDABE | stm1987 promoter | This study |
| pU220-cpxP::luxCDABE | cpxP promoter | This study |
| pU220-csgD::luxCDABE | csgDEFG promoter | [12] |
| pCS26-csgB::luxCDABE | csgBAC promoter | [12] |
| pCS26-adrA::luxCDABE | adrA promoter | [12] |
| pBR322 | ||
| pBR322/stm1987 | stm1987^14028 | This study |
| pBR322/yhjH | yhjH^14028 | This study |
| pACYC184 | ||
| pACYC/rpoS | rpoS^14028 | [48] |
| Primer | Sequence (5′–3′) a | Purpose |
|---|---|---|
| cpxRko sense | AAGATGCGCGCGGTTAAACTTCCTATCATGAAGCGGAAACCATCAGATAGGTGTAGGCTGGAGCTGCTTC | To amplify cat gene product from pKD3 to generate ∆cpxR strain by lambda red recombination |
| cpxRko antisense | CCTGTTAGTTGATGATGACCGAGAGCTGACTTCCCTGTTAAAAGAGCTCCCCTCCTTAGTTCCTATTCCG | |
| cpxR ver F | CCAGCATTAGCACCAGCGCC | To confirm the deletion of cpxR from S. typhimurium 14028 |
| cpxR ver R | TCTGCCTCGGAGGTACGTAAACA | |
| cpxR1 | GCCCTCGAGGTAACTTTGCGCATCGCTTG | To amplify the cpxR and cpxP promoter regions from S. typhimurium 14028 |
| cpxR2 | GCCGGATCCTTCATTGTTTACGTACCTCCG | |
| iraPko sense | GGCAGTGGTTCTTCATAGTGATAACGTCACCCTGGAACTAATAAGGAAATGTGTAGGCTGTAGCTGCTTC | To amplify cat gene product from pKD3 to generate ∆iraP strain by lambda red recombination |
| iraPko antisense | TGTTATTTCATAAAAGTAACGTTATAACAACTGTGTTGTTTTAAATACGACCTCCTTAGTTCCTATTCCG | |
| iraPko-detect1 iraPko-detect2 | CAAAAAGCGAAAGGCCAATA TAGCACCATCCTTTTGTCAG | To confirm the deletion of iraP from S. typhimurium 14028 |
| STM14_2408for1 | GATCCTCGAGAAATTCGCGGTGTTTCGCAC | To amplify the stm1987 promoter region from S. typhimurium 14028 |
| STM14_2408rev2 | GATCGGATCCCTAACAGTGTTTCGTGCGGC | |
| STM1987forEco | GATCGAATTCAAACGGTGTTTCGCAC | To amplify stm1987 with native promoter region from S. typhimurium 14028 |
| STM1987revAatII | GATCGACGTCGGACTATTTCTTTTCCCGCT | |
| yhjHforEco | GATCGAATTCTTGACAAGTTTCGGGGGCTG | To amplify yhjH with native promoter region from S. typhimurium 14028 |
| yhjHrevAatll | GATCGACGTCGTATTACGGGAACAGTCTGG | |
| pZE05 | CCAGCTGGCAATTCCGA | Used to verify promoter fusions to luxCDABE |
| pZE06 | AATCATCACTTTCGGGAA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokaribo, A.S.; Hansen, E.G.; McCarthy, M.; Desin, T.S.; Waldner, L.L.; MacKenzie, K.D.; Mutwiri, G., Jr.; Herman, N.J.; Herman, D.J.; Wang, Y.; et al. Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms. Microorganisms 2020, 8, 964. https://doi.org/10.3390/microorganisms8070964
Sokaribo AS, Hansen EG, McCarthy M, Desin TS, Waldner LL, MacKenzie KD, Mutwiri G Jr., Herman NJ, Herman DJ, Wang Y, et al. Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms. Microorganisms. 2020; 8(7):964. https://doi.org/10.3390/microorganisms8070964
Chicago/Turabian StyleSokaribo, Akosiererem S., Elizabeth G. Hansen, Madeline McCarthy, Taseen S. Desin, Landon L. Waldner, Keith D. MacKenzie, George Mutwiri, Jr., Nancy J. Herman, Dakoda J. Herman, Yejun Wang, and et al. 2020. "Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms" Microorganisms 8, no. 7: 964. https://doi.org/10.3390/microorganisms8070964
APA StyleSokaribo, A. S., Hansen, E. G., McCarthy, M., Desin, T. S., Waldner, L. L., MacKenzie, K. D., Mutwiri, G., Jr., Herman, N. J., Herman, D. J., Wang, Y., & White, A. P. (2020). Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms. Microorganisms, 8(7), 964. https://doi.org/10.3390/microorganisms8070964





