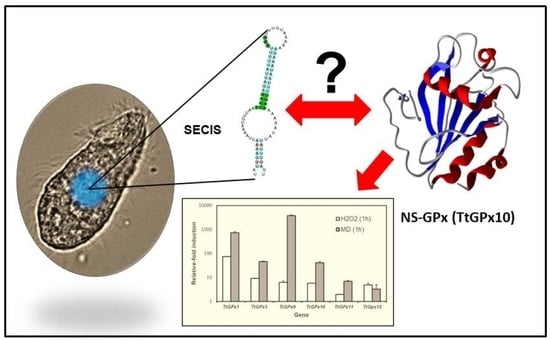
Tetrahymena Glutathione Peroxidase Family: A Comparative Analysis of These Antioxidant Enzymes and Differential Gene Expression to Metals and Oxidizing Agents
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Stress Treatments
2.3. Total RNA Isolation and cDNA Synthesis
2.4. Quantitative Real Time RT-PCR
2.5. Bioinformatics Analysis
3. Results
3.1. The GPx Family of T. thermophila Comprises 12 Isoforms
3.2. A Ciliate GPx Comparative Analysis
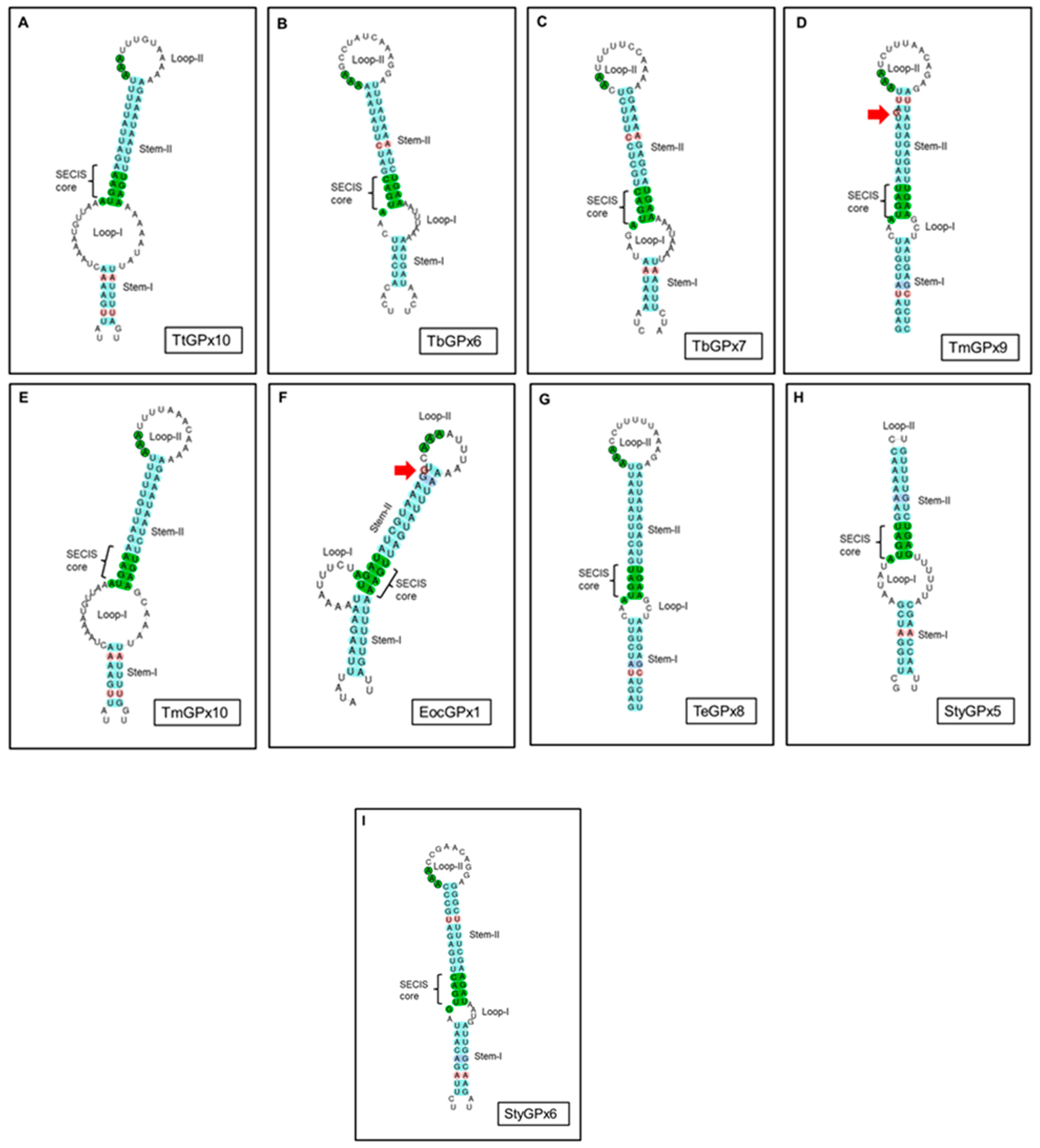
3.3. SECIS Elements
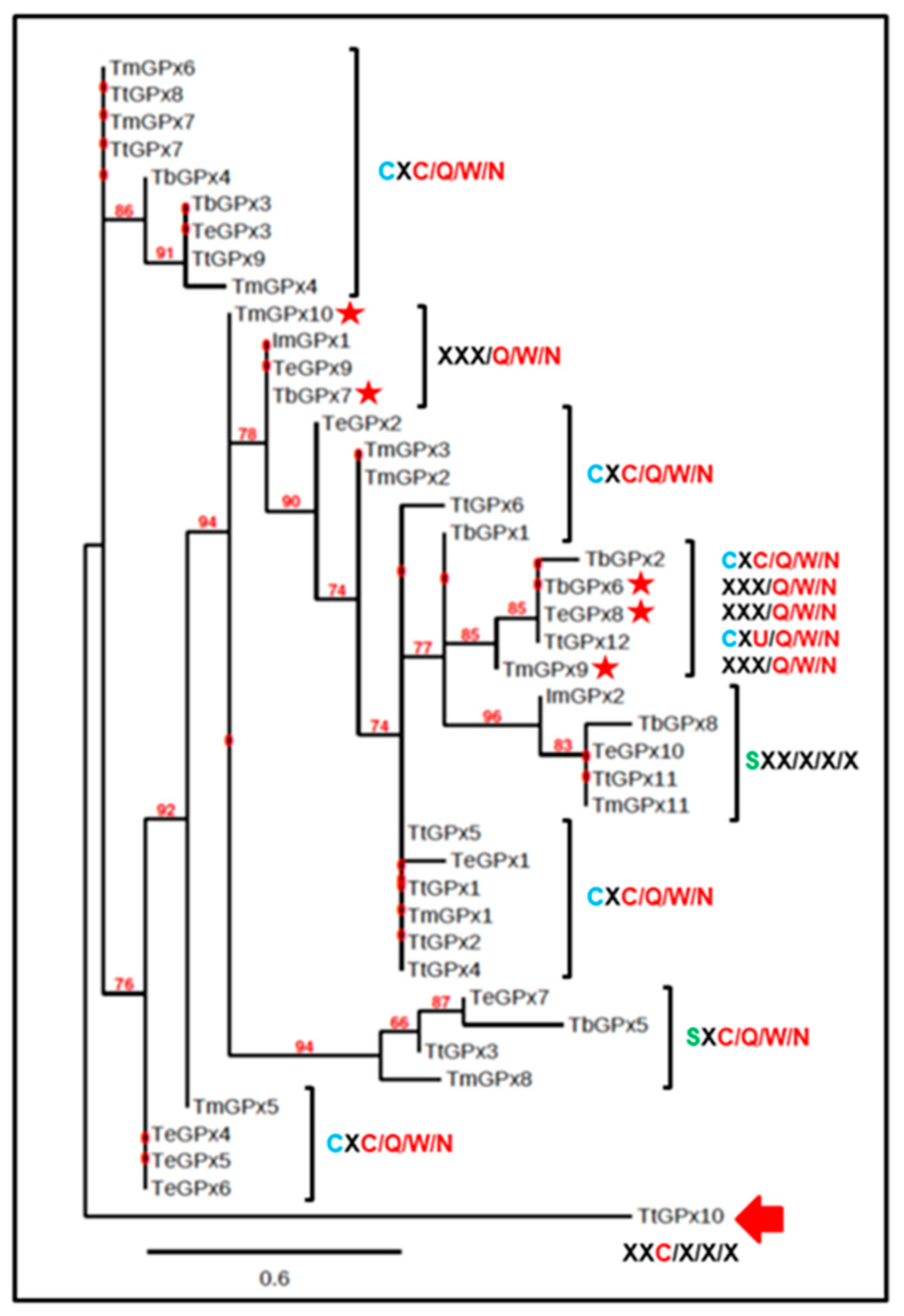
3.4. Phylogenetic Relationships
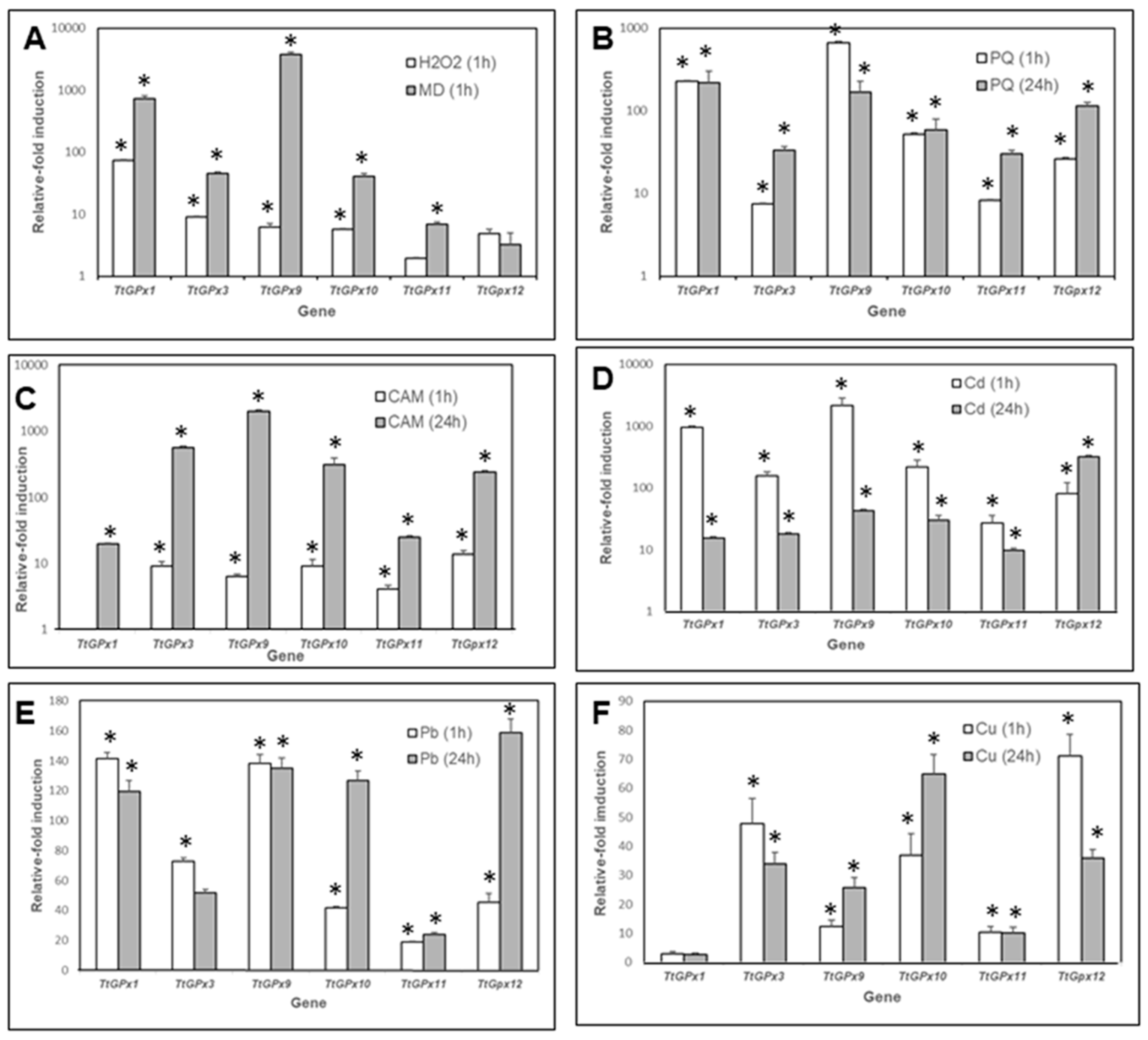
3.5. TtGPx Paralog Gene Expression Patterns under Different Stressful Conditions
4. Discussion
4.1. Ciliate GPx
4.2. Presence of Sec in Ciliate GPx
4.3. The Mysterious Presence or Absence of SECIS Elements in Some Ciliate GPx
4.4. Phylogenetic Considerations
- It has been suggested [11,43] that Sec was probably present in the last universal common ancestor (LUCA), and has been preserved in organisms present in all three domains of the tree of life. Although ciliates also have genes encoding selenoproteins, within the genus Tetrahymena, it seems that the GPx family had a common ancestor which lost the Sec in its catalytic tetrad, replacing it with Cys (C/Q/W/N). A change from Cys to Sec would arise later and in a single isoform (TtGPx12) of the species T. thermophila. As indicated by Lobanov et al., 2007 [11], the Cys/Sec substitution can be in both directions, and both Cys → Sec or Sec → Cys changes are evolutionarily possible. When the latter occurs, the presence of a SECIS element does not have any evolutionary advantage, and tends to be lost. However, it seems that among some ciliates, this process has not happened, since supposedly the change from Sec to Cys has taken place without losing the SECIS element. Moreover, the emergence of Sec in ciliate GPx into more recent phylogenetic classes, such as Hypotrichea (Monoeuplotes crassus), could be explained as a subsequent minority acquisition among ciliate GPx, as is also the case with TtGPx12.
- Among ciliates, including T. thermophila, an unusual feature is to use an alternative genetic code, in which the UAG and UAA stop codons code for glutamine, leaving only one codon (UGA) as a usable stop codon. However, T. thermophila presumably uses a Sec-tRNA, with a UCA anticodon, to incorporate Sec into its selenoproteins. Therefore, we can conclude that UGA is the only stop codon which is also translated to Sec, which makes T. thermophila the first known organism to use all 64 codons for the incorporation of amino acids in protein biosynthesis, as previously reported [49]. The fact of using a single type of stop codon (UGA), which is also used to code Sec, has probably exerted a strong selective pressure for the massive loss of this amino acid and its replacement by Cys in most ciliate GPx, despite the lower catalytic efficiency involved in replacing Sec with Cys [40].
- The loss of the complete GPx catalytic tetrad appears to be an apomorphic character, arising from ancestors with the complete catalytic tetrad and present in one member of each studied Tetrahymena species and even in a related subclass (I. multifiliis). Similarly, the later emergence of Sec in a single Tetrahymena species defines it as an apomorphic character.
- From the phylograms obtained, it can be inferred that many of the ciliate GPx isoforms, which are present in each of the species, were produced by gene duplication, as occurs in many other ciliate gene families [49,50]. The fact that similar GPx isoforms appear both within and between related ciliate species supports the idea of extensive gene duplication throughout the evolution of these microorganisms.
- T. thermophila, a freshwater microorganism, has only five selenoproteins, and the rest of the ciliates probably have a similar or even lower number (with NS-GPx in almost all of them). This number of selenoproteins is closer to the seleproteomes of terrestrial organisms, such as Dictyostelium discoideum (with five selenoproteins), Plasmodium falciparum (with four) or Drosophila melanogaster (with three), which have smaller selenoproteomes than those living in aquatic ecosystems, such as the freshwater microalgae Chlamydomonas reinnhardii (with 10 selenoproteins) or the marine microalgae Ostreococcus tauri (with 26). Therefore, the ciliate selenoproteomes, at least the known ones, do not confirm the established hypothesis associating large selenoproteomes with aquatic life [11].
4.5. TtGPx Gene Expression Patterns
5. Conclusions
- Ciliated protozoa GPx are mostly NS-GPx (with Cys in their catalytic tetrads), and a minority are of the Sec-GPx type.
- Many ciliate GPx could be considered as PHGPx, because they have Cys instead of Sec as part of their catalytic center, and probably use Trx rather than GSH as a reducing substrate.
- None of the nine ciliate GPx, where SECIS elements are detected, has Sec in their sequence, so they could be considered evolutionary relics that have remained in the genome after losing Sec or replacing it with Cys.
- Although T. thermophila has in its genome the main elements for the incorporation of Sec into its selenoproteins, in the isoform TtGPx12, which is a Sec-GPx, no SECIS elements are detected, after scanning with the SECISearch3 program.
- The loss of the complete GPx catalytic tetrad appears to be an apomorphic character, arising from ancestors with the complete catalytic tetrad, and is present in one member of each studied Tetrahymena species and even in a related subclass.
- Extensive genetic duplication appears to have occurred throughout the evolution of ciliate GPx, as similar paralogs exist both within and between related ciliate species.
- The size of the T. thermophila selenoproteome, and probably those from other ciliates, is closer to those present in terrestrial organisms, despite the fact that this ciliate has an aquatic life.
- Differential gene expression patterns exist between all selected T. thermophila GPx genes, depending on the nature of the stressor and the time of exposure. Regarding the metal treatments, both Cd and Pb could have a greater affinity for the -SH groups of the Cys residues than for the -SeH groups of the Sec, which could explain the faster response of NS-GPx-coding genes with respect to those coding Sec-GPx.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Staerck, C.; Gastebois, A.; Vandeputte, P.; Calenda, A.; Larcher, G.; Gillmann, L.; Papon, N.; Bouchara, J.P.; Fleury, M.J.J. Microbial antioxidant defense enzymes. Microb. Pathog. 2017, 110, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Al-Assai, S.; Malik, A.; Bakiu, R.; Santovito, G.; Menz, I.; Schuller, K. Characterization of the peroxiredoxin 1 subfamily from Tetrahymena thermophila. Cell. Mol. Life Sci. 2019. [Google Scholar] [CrossRef]
- Rhee, S.G.; Kil, I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017, 86, 749–775. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Speckmann, B.; Klotz, L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016, 595, 113–119. [Google Scholar] [CrossRef]
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Selenium and redox signaling. Arch. Biochem. Biophys. 2017, 617, 48–59. [Google Scholar] [CrossRef]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef]
- Passaia, G.; Margis-Pinheiro, M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015, 234, 22–26. [Google Scholar] [CrossRef]
- Fu, L.; Wang, X.; Eyal, Y.; She, Y.; Donald, L.J.; Standing, K.G.; Benhayyim, G. A Selenoprotein in the plant kingdom. Mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii gluththione peroxidase. J. Biol. Chem. 2002, 277, 25983–25991. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Fomenko, D.E.; Zhang, Y.; Sengupta, A.; Hatfield, D.L.; Gladyshev, V.N. Evolutionary dynanics of eukaryotic selenoproteomes: Large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007, 8, R198. [Google Scholar] [CrossRef] [PubMed]
- Toppo, S.; Flohé, L.; Ursini, F.; Vanin, S.; Maiorino, M. Catalytic mechanisms and specificities of glutathione peroxidases: Variations of a basic scheme. Biochim. Biophys. Acta 2009, 1790, 1486–1500. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.A.; Cai, G.B.; Kim, S.H.; Zo, Y.G.; Kong, Y. Modular evolution of glutathione peroxidase genes in association with different biochemical properties of their encoded proteins in invertebrate animals. BMC Evol. Biol. 2009, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Bang, M.A.; Lee, S.; Chae, H.Z.; Kim, K. Distinct functional roles of peroxiredoxin isozymes and glutathione peroxidase from fission yeast, Schizosaccharomyces pombe. BMB Rep. 2010, 43, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Dayer, R.; Fisher, B.B.; Eggen, R.I.L.; Lemaire, S.D. The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics 2008, 179, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Pey, A.; Zamoum, T.; Christen, R.; Merle, P.L.; Furla, P. Characterization of glutathione peroxidase diversity in the symbiotic sea anemone Anemonia viridis. Biochimie 2017, 132, 94–101. [Google Scholar] [CrossRef]
- Schlecker, T.; Comini, M.A.; Melchers, J.; Ruppert, T.; Krauth-Siegel, L. Catalytic mechanism of the glutathione peroxidase-type tryparedoxin peroxidase of Trypanosoma brucei. Biochem. J. 2007, 405, 445–454. [Google Scholar] [CrossRef]
- Miramón, P.; Dunker, C.; Kasper, L.; Jacobsen, I.D.; Barz, D.; Kurzai, O.; Hube, B. A family of glutathione peroxidases contributes to oxidative stress resistance in Candida albicans. Med. Mycol. 2014, 52, 223–239. [Google Scholar] [CrossRef]
- Machado-Silva, A.; Cerqueira, P.G.; Grazielle-Silva, V.; Gadelha, F.R.; Peloso, E.F.; Teixeira, S.M.; Machado, C.R. How Trypanosoma cruzi deals with oxidative stress: Antioxidant defence and DNA repair pathways. Mutat. Res. Rev. Mutat. Res. 2016, 767, 8–22. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Toppo, S.; Vanin, S.; Bosello, V.; Tosaltto, C.E. Evolutionary and structural insights into multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid. Redox Signal. 2008, 10, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Sztajer, H.; Gamain, B.; Aumann, K.D.; Slomianny, C.; Becker, K.; Brigelius-Flohé, R.; Flohé, L. The putative glutathione peroxidase gene of Plasmodium falciparum codes for a thioredoxin peroxidase. J. Biol. Chem. 2001, 276, 7397–7403. [Google Scholar] [CrossRef]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta 2009, 1790, 1424–1428. [Google Scholar] [CrossRef]
- Chapple, C.E.; Guigó, R.; Krol, A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics 2009, 25, 674–675. [Google Scholar] [CrossRef][Green Version]
- Bock, A.; Rother, M.; Leibundgut, M.; Ban, N. Selenium metabolism in prokaryotes. In Selenium: Its Molecular Biology and Role in Human Health, 2nd ed.; Hatfield, D.L., Berry, M.J., Gladyshev, V.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 9–28. [Google Scholar]
- Wilting, R.; Schorling, S.; Persson, B.C.; Böck, A. Selenoprotein synthesis in archaea: Identification of an mRNA of Methanococcus jannaschii probably directing selenocysteine insertion. J. Mol. Biol. 1997, 266, 637–641. [Google Scholar] [CrossRef][Green Version]
- Lu, J.; Holmgren, A. Selenoproteins. J. Biol. Chem. 2009, 284, 723–727. [Google Scholar] [CrossRef]
- Gallego, A.; Martín-González, A.; Ortega, R.; Gutiérrez, J.C. Flow cytometry assessment of cytotoxicity and reactive oxygen species generation by single and binary mixtures of cadmium, zinc and copper on populations of the ciliated protozoan Tetrahymena thermophila. Chemosphere 2007, 68, 647–661. [Google Scholar] [CrossRef]
- Amaro, F.; del Pilar de Lucas, M.; Martín-González, A.; Gutiérrez, J.C. Two new members of the Tetrahymena multi-stress-inducible metallothionein family: Characterization and expression analysis of T. rostrata Cd/Cu metallothionein genes. Gene 2008, 423, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Martin-González, A.; Cubas, L.; Ortega, R.; Amaro, F.; Rodríguez-Martin, D.; Gutiérrez, J.C. High resistance of Tetrahymena thermophila to paraquat: Mitochondrial alterations, oxidative stress and antioxidant genes expression. Chemosphere 2016, 144, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M.; Lobanov, A.V.; Guigo, R.; Gladyshev, V.N. SECISearch3 and Seblastian: New tools for prediction of SECIS elements and selenoproteins. Nucl. Acids Res. 2013, 41. [Google Scholar] [CrossRef]
- Imai, H.; Nakagawa, Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Rad. Biol. Med. 2003, 3, 145–169. [Google Scholar] [CrossRef]
- Coyne, R.S.; Thiagarajan, M.; Jones, K.M.; Wortman, J.R.; Tallon, L.J.; Haas, B.J.; Cassidy-Hanley, D.M.; Wiley, E.A.; Smith, J.J.; Collins, K.; et al. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genom. 2008, 9, 562. [Google Scholar] [CrossRef]
- Xu, X.M.; Carlson, B.A.; Mix, H.; Zhang, Y.; Saira, K.; Glass, R.S.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007, 5, e4. [Google Scholar] [CrossRef]
- Tosatto, S.C.E.; Bosello, V.; Fogolari, F.; Mauri, P.; Roveri, A.; Toppo, S.; Flohé, L.; Ursini, F.; Maiorino, M. The catalytic site of glutathione peroxidases. Antioxid. Redox Signal. 2008, 10, 1515–1525. [Google Scholar] [CrossRef]
- Allmang, C.; Wurth, L.; Krol, A. The selenium to selenoproteinpathway in eukaryotes: More nolecular partners than anticipated. Biochim. Biophys. Acta 2009, 1790, 1415–1423. [Google Scholar] [CrossRef]
- Shetty, S.P.; Sturts, R.; Vetick, M.; Copeland, P.R. Processive incorporation of multiple selenocysteine residues is driven by a novel feature of the selenocysteine insertion sequence. J. Biol. Chem. 2018, 293, 19377–19386. [Google Scholar] [CrossRef]
- Mix, H.; Lobanov, A.V.; Gladyshev, V.N. SECIS elements in the coding regions of selenoprotein transcripts are functional in higher eukaryotes. Nucl. Acids Res. 2007, 35, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M.; Lobanov, A.V.; Manta, B.; Santesmasses, D.; Bofill, A.; Guigó, R.; Gabaldón, T.; Gladyshev, V.N. Lokiarchaeota marks the transition between the Archaeal and Eukaryotic selenocysteine encoding systems. Mol. Biol. Evol. 2016, 33, 2441–2453. [Google Scholar] [CrossRef] [PubMed]
- Allmang, C.; Krol, A. SECIS RNAs and K-turn binding proteins. A survey of evolutionary conserved RNA and protein motifs. In Selenium, Its Molecular Biology and Role in Human Health, 2nd ed.; Hatfield, D.L., Marla, J., Berry, M.J., Gladyshev, V.N., Eds.; Springer US: Boston, MA, USA, 2006; Volume 5, pp. 51–61. [Google Scholar]
- Hirosawa-Takamori, M.; Ossipov, D.; Novoselov, S.V.; Turanov, A.A.; Zhang, Y.; Gladyshev, V.N.; Krol, A.; Vorbrüggen, G.; Jäckle, H. A novel stem loop control element-dependent UGA read-through system without translational selenocysteine incorporation in Drosophila. FASEB J. 2009, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Shrimali, R.K.; Lobanov, A.V.; Xu, X.M.; Rao, M.; Carlson, B.A.; Mahadeo, D.C.; Parent, C.A.; Gladyshev, V.N.; Hatfield, D.L. Selenocysteine tRNA identification in the model organisms Dictyostelium discoideum and Tetrahymena thermophila. Biochem. Biophys. Res. Commun. 2005, 329, 147–151. [Google Scholar] [CrossRef]
- Simonovic, M.; Puppala, A.K. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins. Biochem. Biophys. Acta 2018, 1862, 2463–2472. [Google Scholar] [CrossRef]
- Bubenik, J.L.; Driscoll, D.M. Altered RNA binding activity underlies abnormal thyroid hormone metabolism linked to a mutation in selenocysteine insertion sequence-binding protein 2. J. Biol. Chem. 2007, 282, 34653–34662. [Google Scholar] [CrossRef]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, e286. [Google Scholar] [CrossRef]
- De Francisco, P.; Melgar, L.M.; Díaz, S.; Martín-González, A.; Gutiérrez, J.C. The Tetrahymena metallothionein gene family: Twenty-one new cDNAs, molecular characterization, phylogenetic study and comparative analysis of the gene expression under different abiotic stressors. BMC Genom. 2016, 17, 1–23. [Google Scholar] [CrossRef]
- Miao, W.; Xiong, J.; Bowen, J.; Wang, W.; Liu, Y.F.; Braguinets, O.; Grigull, J.; Pearlman, R.E.; Orias, E.; Gorovsky, M.A. Microarray Analyses of Gene Expression during the Tetrahymena thermophila Life Cycle. PLoS ONE 2009, 4, e4429. [Google Scholar] [CrossRef]
- Osorio, H.; Carvalho, E.; del Valle, M.; Günther Sillero, M.A.; Moradas-Ferreira, P.; Sillero, A. H2O2, but not menadione, provokes a decrease in the ATP and an increase in the inosine levels in Saccharomyces cerevisiae. An experimental and theoretical approach. Eur. J. Biochem. 2003, 270, 1578–1589. [Google Scholar] [CrossRef]
- Grant, C.M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001, 39, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Dawes, I.W.; Roe, J.H. Adaptive response of Schizosaccharomyces pombe to hydrogen peroxide and menadione. Microbiology 1995, 141, 3127–3132. [Google Scholar] [CrossRef]
- Arenas, F.A.; Díaz, W.A.; Leal, C.A.; Pérez-Donoso, J.M.; Imlay, J.A.; Vásquez, C.C. The Escherichia coli btuE gene, encodes a glutathione peroxidase that is induced under oxidative stress conditions. Biochem. Biophys. Res. Commun. 2010, 398, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Djukic, M.M.; Jovanovic, M.D.; Ninkovic, M.; Stevanovic, I.; Ilic, K.; Curcic, M.; Vekic, J. Protective role of glutathione reductase in paraquat induced neurotoxicity. Chem. Biol. Interact. 2012, 199, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Feinstein, S.I.; Wang, Y.; Dodia, C.; Fisher, D.; Yu, K.; Ho, Y.S.; Fisher, A.B. Comparison of glutathione peroxidase 1 and peroxiredoxin 6 in protection against oxidative stress in the mouse lung. Free Radic. Biol. Med. 2010, 49, 1172–1181. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, D.S.; Lee, G.H.; Kim, K.W.; Kim, H.R.; Chae, H.J. Apoptosis Induced by Manganese on Neuronal SK-N-MC Cell Line: Endoplasmic Reticulum (ER) Stress and Mitochondria Dysfunction. Environ. Health Toxicol. 2011, 26, e2011017. [Google Scholar] [CrossRef]
- Hegedüs, A.; Erdei, S.; Horváth, G. Comparative studies of H2O2 detoxifying enzymes in green and greening barley seedlings under cadmium stress. Plant Sci. 2001, 160, 1085–1093. [Google Scholar] [CrossRef]
- Messaoudi, I.; Barhoumi, S.; Saïd, K.; Kerken, A. Study on the sensitivity to cadmium of marine fish Salaria basilisca (Pisces: Blennidae). J. Environ. Sci. 2009, 21, 1620–1624. [Google Scholar] [CrossRef]
- Ramos, J.; Matamoros, M.A.; Naya, L.; James, E.K.; Rouhier, N.; Sato, S.; Tabata, S.; Becana, M. The glutathione peroxidase gene family of Lotus japonicus: Characterization of genomic clones, expression analyses and immunolocalization in legumes. New Phytol. 2009, 181, 103–114. [Google Scholar] [CrossRef]
- Cirillo, T.; Cocchieri, R.; Fasano, E.; Lucisano, A.; Tafuri, S.; Ferrante, M.C.; Carpenè, E.; Andreani, G.; Isani, G. Cadmium accumulation and antioxidant responses in Sparus aurata exposed to waterborne cadmium. Arch. Environ. Contam. Toxicol. 2012, 62, 118–126. [Google Scholar] [CrossRef]
- Ahamed, M.; Siddiqui, M.K. Environmental lead toxicity and nutritional factors. Clin. Nutr. 2007, 26, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Upasani, C.D.; Khera, A.; Balaraman, R. Effect of lead with vitamin E, C, or spirulina on malondialdehyde, conjugated dienes and hydroperoxides in rats. Indian J. Exp. Biol. 2001, 39, 70–74. [Google Scholar] [PubMed]
- Yagi, K.; Komura, S.; Kojima, H.; Sun, Q.; Nagata, N.; Ohishi, N.; Nishikimi, M. Expression of human phospholipid hydroperoxide glutathione peroxidase gene for protection of host cells from lipid hydroperoxide-mediated injury. Biochem. Biophys. Res. Commun. 1996, 219, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Templeton, D.M.; O’brien, P.J. Mitochondrial involvement in genetically determined transition metal toxicity II. Copper toxicity. Chem. Biol. Interact. 2006, 163, 77–85. [Google Scholar] [CrossRef]
- Nagalakshmi, N.; Prasad, M.N. Responses of glutathione cycle enzymes and glutathione metabolism to copper stress in Scenedesmus bijugatus. Plant Sci. 2001, 160, 291–299. [Google Scholar] [CrossRef]
| CILIATES | |||||
|---|---|---|---|---|---|
| Name | AccessionNumber (1) | MW (KDa) | Introns | SECIS | Sec |
| TtGPx1 | TTHERM_00630010 | 19.9 | 0 | - | - |
| TtGPx2 | TTHERM_00895660 | 24.5 | 0 | - | - |
| TtGPx3 | TTHERM_01099010 | 15.5 | 1 | - | - |
| TtGPx4 | TTHERM_00895640 | 20.0 | 0 | - | - |
| TtGPx5 | TTHERM_00895650 | 20.0 | 0 | - | - |
| TtGPx6 | TTHERM_00895630 | 20.0 | 0 | - | - |
| TtGPx7 | TTHERM_00046110 | 21.9 | 0 | - | - |
| TtGPx8 | TTHERM_00046090 | 22.0 | 0 | - | - |
| TtGPx9 | TTHERM_00141170 | 24.5 | 0 | - | - |
| TtGPx10 | TTHERM_000141169 | 15.5 | 2 | + | - |
| TtGPx11 | TTHERM_00661720 | 21.0 | 0 | - | - |
| TtGPx12 | TTHERM_000279829 | 20.9 | 1 | - | + |
| TbGPx1 | E19_04307.1 | 20.1 | 0 | - | - |
| TbGPx2 | EI9_08583.1 | 21.2 | 1 | - | - |
| TbGPx3 | EI9_04856.1 | 21.6 | 0 | - | - |
| TbGPx4 | EI9_04855.1 | 21.5 | 0 | - | - |
| TbGPx5 | EI9_19376.1 | 19.8 | 1 | - | - |
| TbGPx6 | EI9_08932.1 | 18.4 | 1 | + | - |
| TbGPx7 | EI9_04857.1 | 19.6 | 3 | + | - |
| TbGPx8 | EI9_17207.1 | 21.2 | 0 | - | - |
| TeGPx1 | E17_10142.1 | 19.9 | 0 | - | - |
| TeGPx2 | E17_10229.1 | 20.0 | 0 | - | - |
| TeGPx3 | E17_10737.1 | 21.4 | 0 | - | - |
| TeGPx4 | E17_05831.1 | 22.1 | 0 | - | - |
| TeGPx5 | E17_05830.1 | 22.0 | 0 | - | - |
| TeGPx6 | E17_05832.1 | 22.1 | 0 | - | - |
| TeGPx7 | E17_20047.1 | 19.4 | 1 | - | - |
| TeGPx8 | E17_12195.1 | 25.5 | 4 | + | - |
| TeGPx9 (2) | E17_10736.1 | 1 | - | - | |
| E17_10736.2 | 16.0 | 2 | |||
| TeGPx10 | E17_11408.1 | 21.0 | 0 | - | - |
| TmGPx1 | EIA_10427.1 | 19.9 | 0 | - | - |
| TmGPx2 | EIA_05417.1 | 19.9 | 0 | - | - |
| TmGPx3 | EIA_05416.1 | 21.8 | 1 | - | - |
| TmGPx4 | EIA_04640.1 | 21.5 | 0 | - | - |
| TmGPx5 | EIA_01773.1 | 22.1 | 0 | - | - |
| TmGPx6 | EIA_01775.1 | 22.0 | 0 | - | - |
| TmGPx7 | EIA_01770.1 | 22.0 | 0 | - | - |
| TmGPx8 | EIA_20673.1 | 19.4 | 1 | - | - |
| TmGPx9 | EIA_08958.1 | 17.1 | 1 | + | - |
| TmGPx10 | EIA_04639.1 | 19.7 | 3 | + | - |
| TmGPx11 | EIA_07345.1 | 21.0 | 0 | - | - |
| ImGPx1 | IMG5_104650.1 | 16.3 | 2 | - | - |
| ImGPx2 | IMG5_178250.1 | 24.7 | 1 | - | - |
| PtGPx1 | GSPATT00033641001 | 18.3 | 0 | - | - |
| PtGPx2 | GSPATT00011474001 | 20.8 | 2 | - | - |
| PtGPx3 | GSPATT00004297001 | 20.7 | 2 | - | - |
| PtGPx4 | GSPATT00002394001 | 21.3 | 2 | - | - |
| PtGPx5 | GSPATT00011189001 | 20.8 | 2 | - | - |
| OxyGPx1 | OXYTRI_14679 | 24.4 | 3 | - | - |
| OxyGPx2 | OXYTRI_14831 | 13.8 | 0 | - | - |
| OxyGPx3 | OXYTRI_00243 | 19.7 | 0 | - | - |
| OxyGPx4 | OXYTRI_11899 | 21.0 | 0 | - | - |
| OxyGPx5 | OXYTRI_09235 | 37.2 | 0 | - | - |
| OxyGPx6 | OXYTRI_00914 | 20.8 | 0 | - | - |
| OxyGPx7 | OXYTRI_24397 | 19.9 | 1 | - | - |
| OxyGPx8 | OXYTRI_17994 | 19.3 | 0 | - | - |
| OxyGPx9 | OXYTRI_19181 | 20.2 | 1 | - | - |
| OxyGPx10 | Contig6997.0.g51 | 25.9 | 2 | - | - |
| StyGPx1 | Contig8855.g9459 | 15.5 | 0 | - | - |
| StyGPx2 | Contig13525.g14435 | 19.0 | 0 | - | - |
| StyGPx3 | Contig14711.g15669 | 23.3 | 0 | - | - |
| StyGPx4 | Contig17005.g18115 | 19.0 | 0 | - | - |
| StyGPx5 | Contig18173.g19318 | 122.7 | 8 | + | - |
| StyGPx6 | Contig18431.g19575 | 19.8 | 1 | + | - |
| StyGPx7 | Contig18781.g19927 | 17.6 | 1 | - | - |
| StyGPx8 | Contig19413.g20586 | 21.3 | 0 | - | - |
| StyGPx9 | Contig19790.g20991 | 20.5 | 1 | - | - |
| EvGPx1 | MSTRG.6189 | 16.8 | 1 | ? | - |
| EvGPx2 | MSTRG.7263 | 22.1 | 0 | ? | - |
| SteGPx1 | SteCoe_16643 | 19.8 | ? | - | + |
| SteGPx2 | SteCoe_21264 | 18.4 | ? | - | + |
| SteGPx3 | SteCoe_26857 | 21.5 | 0 | - | + |
| SteGPx4 | SteCoe_40903 | 18.7 | ? | - | + |
| SteGPx5 | SteCoe_40904 | 19.0 | ? | - | + |
| SteGPx6 | SteCoe_40905 | 20.0 | ? | - | + |
| SteGPx7 | SteCoe_6726 | 20.2 | ? | - | + |
| McGPx1 | ACL81236.1 | 21.3 | 0 | - | + |
| McGPx2 | AFR60589.1 | 21.2 | 0 | - | + |
| McGPx3 | ACL81237.1 | 21.2 | 0 | - | + |
| PcpGPx1 | PPERSA_00007440 | 17.1 | 2 | ? | - |
| PcpGPx2 | PPERSA_00008250 | 25.7 | 3 | ? | - |
| PcpGPx3 | PPERSA_00077430 | 13.3 | 2 | ? | - |
| PcpGPx4 | PPERSA_00077470 | 20.2 | 4 | ? | - |
| PcpGPx5 | PPERSA_00073590 | 26.2 | 4 | ? | - |
| PcpGPx6 | PPERSA_00077460 | 14.5 | 3 | ? | - |
| PcpGPx7 | PPERSA_00040530 | 27.3 | 2 | ? | - |
| EocGPx1 | Contig11975.g1890 | 21.4 | 0 | + | - |
| EocGPx2 | Contig13463.g3154 | 21.1 | 0 | - | - |
| EocGPx3 | Contig23176.g11961 | 45.1 | 4 | - | - |
| EocGPx4 | Contig32168.g20842 | 21.1 | 0 | - | - |
| EocGPx5 | Contig8754.g27852 | 22.7 | 1 | - | - |
| Other selected organisms | |||||
| CrGPx5 | AAB66330.1 | 18.0 | 4 | - | - |
| CrGPx1 | AAL14348.1 | 21.4 | 0 | + | + |
| TcGPx1 | TcCL_NomESM05066 | 17.4 | 0 | ? | - |
| PfGPx1 | PF3D7_1212000 | 23.9 | 2 | ? | - |
| ScGPx1 | EDV09454.1 | 18.6 | 0 | - | - |
| AtGPx6 | AEE83029.1 | 25.5 | 0 | - | - |
| DmGPx1 | NP_728869.1 | 26.2 | 0 | - | - |
| HsGPx1 | NP_000572.2 | 22.0 | 0 | + | + |
| HsGPx5 | NP_001500.1 | 25.2 | 0 | - | - |
| HsGPx4 | AAH32695.3 | 22.1 | 0 | + | + |
| Catalytic Tetrad (1) | Ciliate GPx |
|---|---|
| CXC/Q/W/N | TtGPx1, 2, 4, 5, 6, 7, 8, 9 |
| TbGPx1, 2, 3, 4 | |
| TeGPx1, 2, 3, 4, 5, 6 | |
| TmGPx1, 2, 3, 4, 5, 6, 7 | |
| XXC/Q/W/N | OxyGPx6 |
| CSC/Q/W/N | PtGPx2, 3, 4, 5 |
| SXC/Q/W/N | TtGPx3 TbGPx5 TeGPx7 TmGPx8 StyGPx2, 3, 4 OxyGPx3, 5, 8 EvGPx2 EocGPx1, 2, 4 |
| CXU/Q/W/N | TtGPx12 |
| SXU/Q/W/N | SteGPx1, 4, 5, 7 McGPx1, 2, 3 |
| SXU/X/W/N | SteGPx2 |
| CXU/X/X/N | SteGPx3, 6 |
| SXX/Q/W/N | TmGPx10 StyGPx9 OxyGPx7 |
| SXS/X/X/N | OxyGPx4 |
| XXX/Q/X/N | EvGPx1 |
| XXX/Q/W/N | TbGPx7 TeGPx8, 9 TmGPx9 ImGPx1 StyGPx1, 6, 7 OxyGPx9 PcpGPx4, 6 |
| CXX/Q/W/N | TbGPx6 |
| SXC/X/X/X | PcpGPx3 |
| SXC/X/W/N | PcpGPx1 |
| SXX/Q/X/N | StyGPx8 |
| XXX/X/W/N | OxyGPx2 |
| Treatments | TtGPx Genes |
|---|---|
| H2O2 (1 h) | TtGPx1 >> TtGPx3 > TtGPx9 > TtGPx10 > TtGPx12 > TtGPx11 |
| MD (1 h) | TtGPx9 >> TtGPx1 > TtGPx3 > TtGPx10 > TtGPx11 > TtGPx12 |
| PQ (1 h) | TtGPx9 >> TtGPx1 > TtGPx10 > TtGPx12 > TtGPx11 > TtGPx3 |
| PQ (24 h) | TtGPx1 > TtGPx9 > TtGPx12 > TtGPx10 > TtGPx3 > TtGPx11 |
| CAM (1 h) | TtGPx12 > TtGPx3 > TtGPx10 > TtGPx9 > TtGPx11 > TtGPx1 |
| CAM (24 h) | TtGPx9 > TtGPx3 > TtGPx10 > TtGPx12 > TtGPx11 > TtGPx1 |
| Cd (1 h) | TtGPx9 >> TtGPx1 > TtGPx10 > TtGPx3 > TtGPx12 > TtGPx11 |
| Cd (24 h) | TtGPx12 >> TtGPx9 > TtGPx10 > TtGPx3 > TtGPx1 > TtGPx11 |
| Pb (1 h) | TtGPx1 > TtGPx9 > TtGPx3 > TtGPx12 > TtGPx10 > TtGPx11 |
| Pb (24 h) | TtGPx12 > TtGPx9 > TtGPx10 > TtGPx1 > TtGPx3 > TtGPx11 |
| Cu (1 h) | TtGPx12 > TtGPx3 > TtGPx10 > TtGPx9 > TtGPx11 > TtGPx1 |
| Cu (24 h) | TtGPx10 > TtGPx12 > TtGPx3 > TtGPx9 > TtGPx11 > TtGPx1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cubas-Gaona, L.L.; de Francisco, P.; Martín-González, A.; Gutiérrez, J.C. Tetrahymena Glutathione Peroxidase Family: A Comparative Analysis of These Antioxidant Enzymes and Differential Gene Expression to Metals and Oxidizing Agents. Microorganisms 2020, 8, 1008. https://doi.org/10.3390/microorganisms8071008
Cubas-Gaona LL, de Francisco P, Martín-González A, Gutiérrez JC. Tetrahymena Glutathione Peroxidase Family: A Comparative Analysis of These Antioxidant Enzymes and Differential Gene Expression to Metals and Oxidizing Agents. Microorganisms. 2020; 8(7):1008. https://doi.org/10.3390/microorganisms8071008
Chicago/Turabian StyleCubas-Gaona, Liliana L., Patricia de Francisco, Ana Martín-González, and Juan Carlos Gutiérrez. 2020. "Tetrahymena Glutathione Peroxidase Family: A Comparative Analysis of These Antioxidant Enzymes and Differential Gene Expression to Metals and Oxidizing Agents" Microorganisms 8, no. 7: 1008. https://doi.org/10.3390/microorganisms8071008
APA StyleCubas-Gaona, L. L., de Francisco, P., Martín-González, A., & Gutiérrez, J. C. (2020). Tetrahymena Glutathione Peroxidase Family: A Comparative Analysis of These Antioxidant Enzymes and Differential Gene Expression to Metals and Oxidizing Agents. Microorganisms, 8(7), 1008. https://doi.org/10.3390/microorganisms8071008