Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. Isolation and Screening of Phosphate Solubilizing Bacteria on Plates
2.3. Quantitative Assay of Phosphate Solubilization in Liquid Medium
2.4. DNA Amplification and Sequences Analysis
2.5. In Vitro Screening for PGP Activities
2.5.1. Indole-3-Acetic Acid (IAA) Production Assay
2.5.2. Bacterial Salinity and Heat Stress Monitoring
2.5.3. Biofilm Formation Assay
2.5.4. Siderophores Production Assay
2.5.5. Ammonia Production Assay
2.5.6. HCN Production Assay
2.5.7. Extracellular Enzymes Production
2.6. In Vivo Assessment of Selected PSB Strains
2.6.1. Seed Germination Assay
2.6.2. In Vivo Experiment under Saline Conditions
Plant Harvest and Phenotypes Monitoring
Plant Nutrient and Ionic Analysis
2.7. Statistical Analysis
3. Results
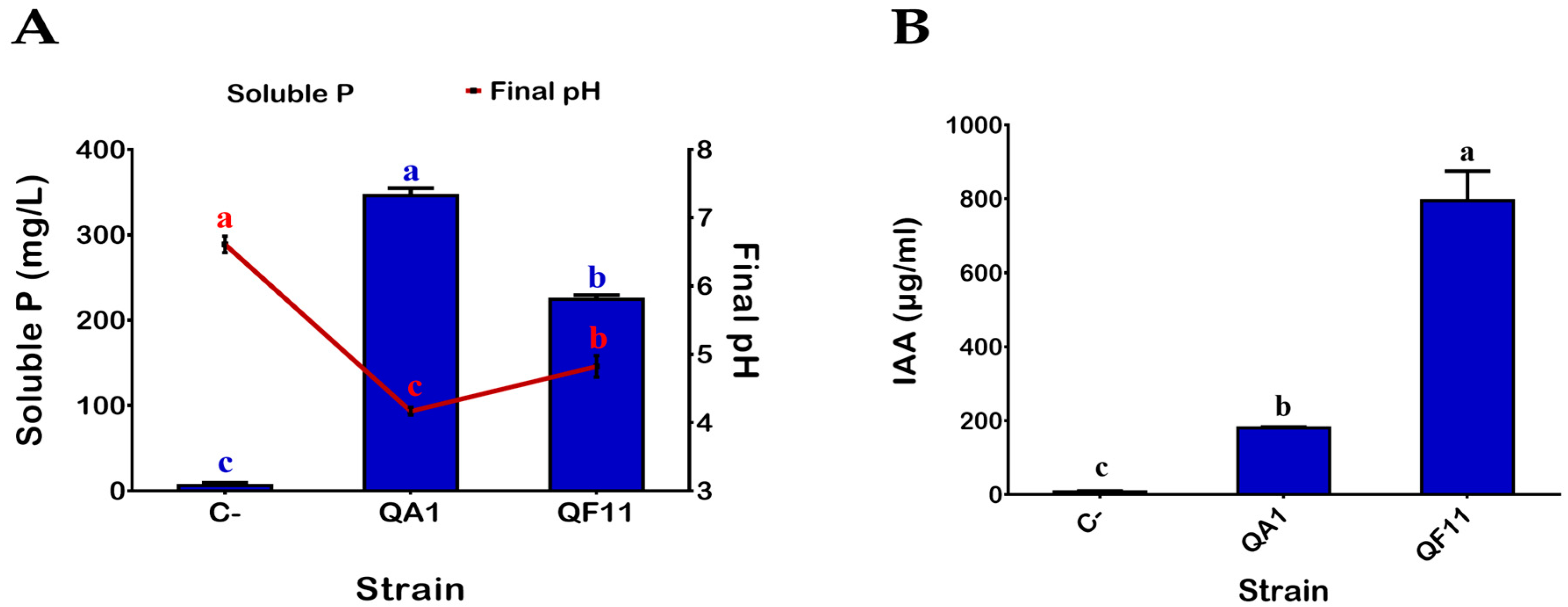
3.1. QA1 and QF11 Are Strains Exhibiting High Phosphate Solubilization Activities
3.2. QF11 Strain Overproduces Indole-3-Acetic Acid (IAA)
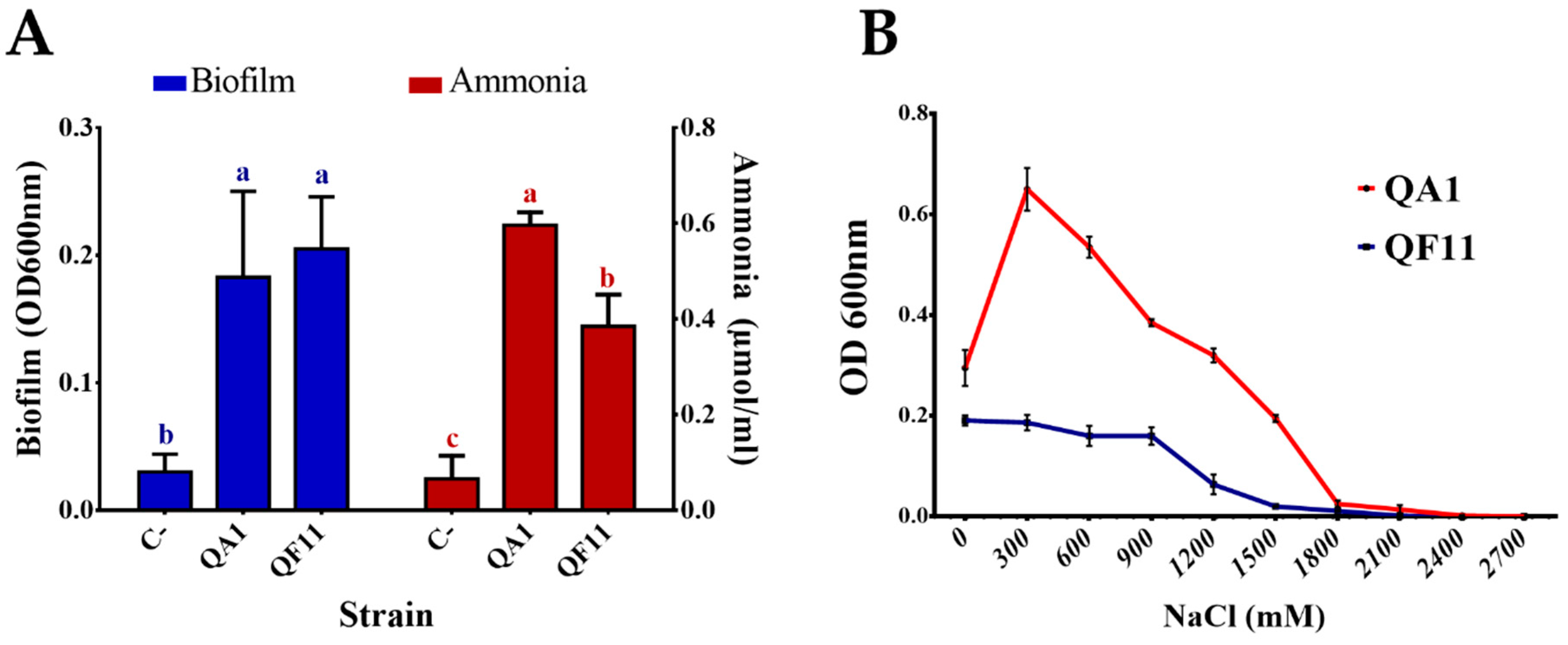
3.3. Strain QA1 Tolerates High Salt Concentrations
3.4. QA1 Strain Overproduces Siderophores And QF11 Enhances Biofilm Formation
3.5. Strain QA1 Overproduces Ammonia and Hydrogen Cyanide (HCN)
3.6. Strain QA1 Overproduces Extracellular Enzymes
3.7. QA1 and QF11 Belong to the Genus of Bacillus Licheniformis and Enterobacter Asburiae, Respectively
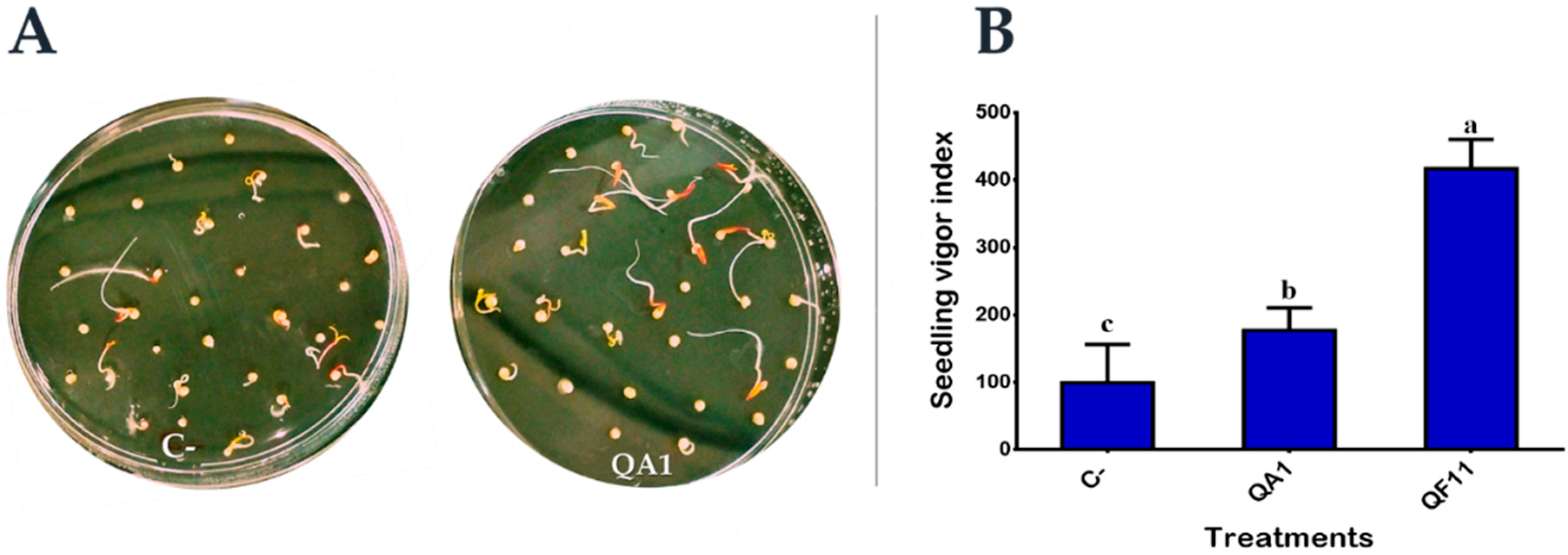
3.8. Strain QF11 Increases Seed Germination
3.9. B. licheniformis QA1 Improves Shoot Biomass while E. asburiae QF11 Enhances Root Development
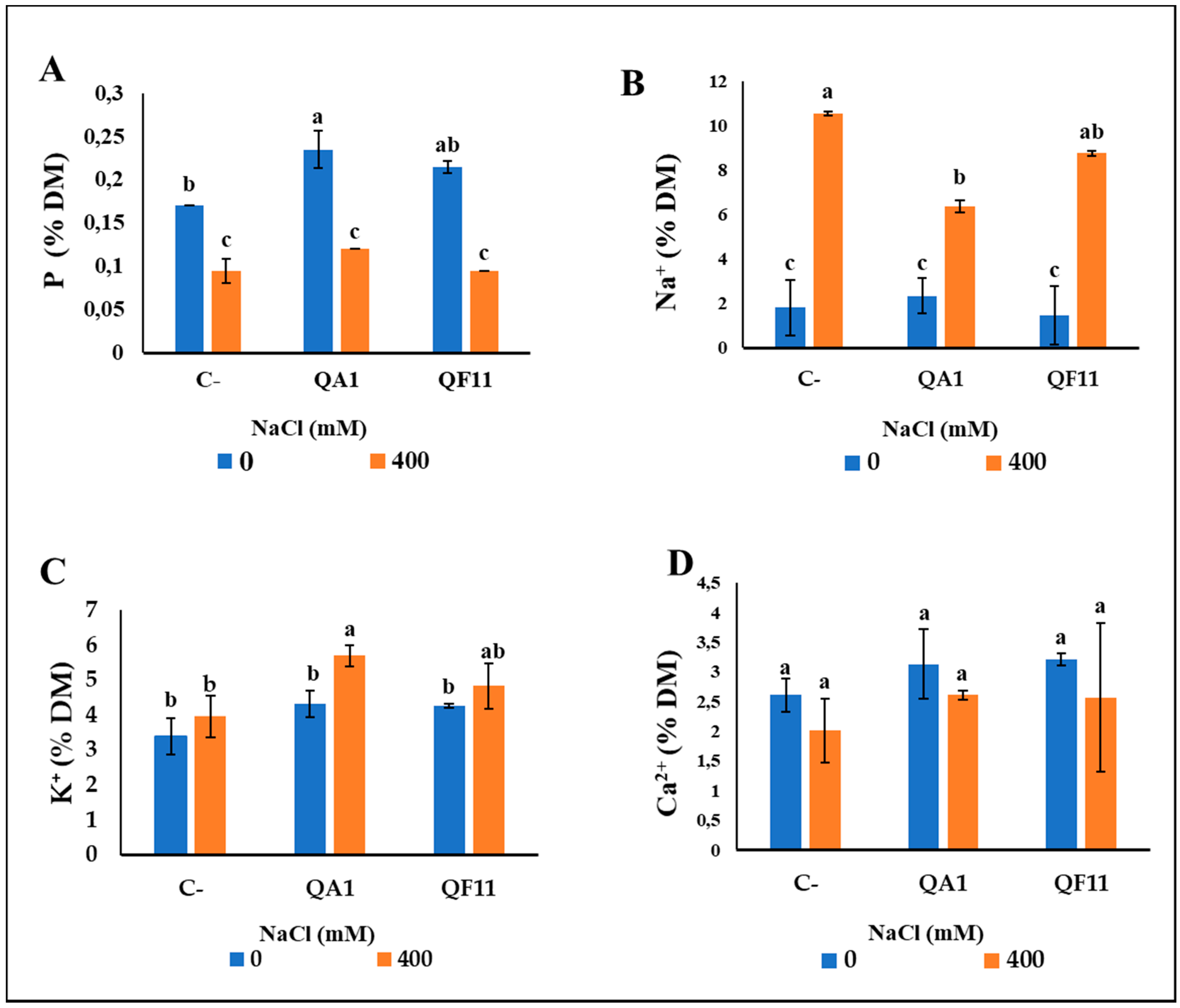
3.10. B. licheniformis QA1 Strain Increases Leaf CCI, P and K+ and Decreases Na+ Uptake
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pitzschke, A. Developmental peculiarities and seed-borne endophytes in quinoa: Omnipresent, robust bacilli contribute to plant fitness. Front. Microbiol. 2016, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014, 32, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Wei, Y.; Zhao, Y.; Wang, H.; Lu, Q.; Cao, Z.; Cui, H.; Zhu, L.; Wei, Z. An optimized regulating method for composting phosphorus fractions transformation based on biochar addition and phosphate-solubilizing bacteria inoculation. Bioresour Technol 2016, 221, 139–146. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Wani, P.A. Role of phosphate-solubilizing microorganisms in sustainable agriculture — A review. Agron. Sustain. Dev. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 587. [Google Scholar] [CrossRef]
- Saghir Khan, M.; Zaidi, A.; Ahmad, E. Mechanism of Phosphate Solubilization and Physiological Functions of Phosphate-Solubilizing Microorganisms. Phosphate Solubilizing Microorg. Princ. Appl. Microphos. Technol. 2014, 31–62. [Google Scholar] [CrossRef]
- Bhat, N.A.; Riar, A.; Ramesh, A.; Iqbal, S.; Sharma, M.P.; Sharma, S.K.; Bhullar, G.S. Soil Biological Activity Contributing to Phosphorus Availability in Vertisols under Long-Term Organic and Conventional Agricultural Management. Front. Plant Sci. 2017, 8, 1523. [Google Scholar] [CrossRef]
- Zhu, J.; Li, M.; Whelan, M. Phosphorus activators contribute to legacy phosphorus availability in agricultural soils: A review. Sci. Total Environ. 2018, 612, 522–537. [Google Scholar] [CrossRef]
- Panhwar, Q.A.; Naher, U.A.; Shamshuddin, J.; Othman, R.; Latif, M.A.; Ismail, M.R. Biochemical and molecular characterization of potential phosphate-solubilizing bacteria in acid sulfate soils and their beneficial effects on rice growth. PLoS ONE 2014, 9, e97241. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, S.; Shahid, I.; Mehnaz, S.; Malik, K. Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum L.). Microbiol. Res. 2017, 205. [Google Scholar] [CrossRef] [PubMed]
- Yin, R. Phosphate-solubilizing microbes in non-irrigated soils in China. Soils 1988, 20, 243–246. [Google Scholar]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Al-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Ruan, C.-J.; da Silva, J.A.T.; Mopper, S.; Qin, P.; Lutts, S. Halophyte improvement for a salinized world. Crit. Rev. Plant Sci. 2010, 29, 329–359. [Google Scholar] [CrossRef]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene Expression Profiling of Plants under Salt Stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Vega-Galvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martinez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Nowak, V.; Du, J.; Charrondiere, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hirich, A.; Choukr-Allah, R.; Jacobsen, S.-E. Quinoa in Morocco – Effect of Sowing Dates on Development and Yield. J. Agron. Crop Sci. 2014, 200. [Google Scholar] [CrossRef]
- Liu, F.-P.; Liu, H.-Q.; Zhou, H.-L.; Dong, Z.-G.; Bai, X.-H.; Bai, P.; Qiao, J.-J. Isolation and characterization of phosphate-solubilizing bacteria from betel nut (Areca catechu) and their effects on plant growth and phosphorus mobilization in tropical soils. Biol. Fertil. Soils 2014, 50, 927–937. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Kumar Johri, J.; Surange, S.; Shekhar Nautiyal, C. Occurrence of Salt, pH, and Temperature-tolerant, Phosphate-solubilizing Bacteria in Alkaline Soils. Curr. Microbiol. 1999, 39, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Kryachko, Y.; Semler, D.; Vogrinetz, J.; Lemke, M.; Irvine, R.; Davidson, J.; Links, M.G.; McCarthy, E.L.; Haug, B.; Hemmingsen, S.M. Analyses of 16S rRNA and cpn60 gene sequences provide complementary information about potentially useful and harmful oil field microbiota. Int. Biodeterior. Biodegrad. 2017, 123, 320–327. [Google Scholar] [CrossRef]
- Oliver, F. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acid. Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.; Lindow, S.E. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl. Environ. Microbiol. 2005, 71, 2365–2371. [Google Scholar] [CrossRef] [PubMed]
- Latif Khan, A.; Ahmed Halo, B.; Elyassi, A.; Ali, S.; Al-Hosni, K.; Hussain, J.; Al-Harrasi, A.; Lee, I.-J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solarium lycopersicum. Electron. J. Biotechnol. 2016, 19, 58–64. [Google Scholar] [CrossRef]
- Biswas, J.; Banerjee, A.; Rai, M.; Naidu, R.; Biswas, B.; Vithanage, M.; Chandra Dash, M.; Sarkar, S.; Meers, E. Potential application of selected metal resistant phosphate solubilizing bacteria isolated from the gut of earthworm (Metaphire posthuma) in plant growth promotion. Geoderma 2018, 330, 117–124. [Google Scholar] [CrossRef]
- Patel, K.S.; Naik, J.H.; Chaudhari, S.; Amaresan, N. Characterization of culturable bacteria isolated from hot springs for plant growth promoting traits and effect on tomato (Lycopersicon esculentum) seedling. Comptes Rendus Biol. 2017, 340, 244–249. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.; Baddour, L.; Barrett, F.; Melton, D.; Beachey, E. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual, 3rd ed.; Benjamin-Cummings Pub Co (B.c.P): New York, NY, USA, 1992. [Google Scholar]
- Chrouqi, L.; Lahcen, O.; Jadrane, I.; Koussa, T.; Alfeddy, M.N. Screening of soil rhizobacteria isolated from wheat plants grown in the Marrakech region (Morocco, North Africa) for plant growth promoting activities. JMES 2017, 8, 3382–3390. [Google Scholar]
- Lorck, H. Production of hydrocyanic acid by bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Kavitha, T.; Nelson, R.; Jesi, S.J. Screening of rhizobacteria for plant growth promoting traits and antifungal activity against charcoal rot pathogen Macrophomina phaseolina. Int. J. Pharma Bio. Sci. 2013, 4. [Google Scholar]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururani, M.A.; Chun, S.C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Suleman, M.; Yasmin, S.; Rasul, M.; Yahya, M.; Atta, B.M.; Mirza, M.S. Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 2018, 13, e0204408. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Mannan Akanda, A.; Prova, A.; Islam, T.; Hossain, M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Akhtar, S.S.; Iqbal, S.; Amjad, M.; Naveed, M.; Zahir, Z.A.; Jacobsen, S.-E. Enhancing salt tolerance in quinoa by halotolerant bacterial inoculation. Funct. Plant Biol. 2016, 43, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Zandi, P.; Ali, S.; Mehmood, A.; Shahid, M.A.; Yang, J. Corrigendum: Impact of Salicylic acid and PGPR on the Drought Tolerance and Phytoremediation potential of Helianthus annus. Front. Microbiol. 2019, 10, 2222. [Google Scholar] [CrossRef]
- Petiole. Available online: https://web.archive.org/web/20200106144735/http://petioleapp.com/ (accessed on 1 June 2020).
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.; Alexander, M. Lysis of soil fungi by bacteria. Can. J. Microbiol. 2011, 9, 169–177. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Del Rio, T.G.; Jones, C.D.; Tringe, S.G. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- Barka, E.A.; Nowak, J.; Clément, C. Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl. Environ. Microbiol. 2006, 72, 7246–7252. [Google Scholar] [CrossRef] [PubMed]
- Sukweenadhi, J.; Kim, Y.-J.; Choi, E.-S.; Koh, S.-C.; Lee, S.-W.; Kim, Y.-J.; Yang, D.C. Paenibacillus yonginensis DCY84T induces changes in Arabidopsis thaliana gene expression against aluminum, drought, and salt stress. Microbiol. Res. 2015, 172, 7–15. [Google Scholar] [CrossRef]
- Mishra, A.; Morang, P.; Deka, M.; Kumar, S.N.; Kumar, B.D. Plant growth-promoting rhizobacterial strain-mediated induced systemic resistance in tea (Camellia sinensis (L.) O. Kuntze) through defense-related enzymes against brown root rot and charcoal stump rot. Appl. Biochem. Biotechnol. 2014, 174, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Huang, X.-F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil 2016, 401, 259–272. [Google Scholar] [CrossRef]
- Scholz, R.; Molohon, K.; Nachtigall, J.; Vater, J.; Markley, A.; Süssmuth, R.D.; Mitchell, D.A.; Borriss, R. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2011, 193, 215–224. [Google Scholar]
- Seema, R.; Asrar, I.; Amir, K. Bacterial diversity in wheat rhizosphere and their characterization. Adv. Appl. Sci. Res. 2011, 2, 351–356. [Google Scholar]
- Sridevi, M.; Mahalakshmi, M.; Ranjan, A. Isolation and characterization of phosphate-solubilizing bacterial species from different crop fields of Salem, Tamil Nadu, India. Int. J. Nutr. Pharmacol. Neurol. Dis. 2013, 3, 29. [Google Scholar] [CrossRef]
- Stephen, J.; Shabanamol, S.; Rishad, K.; Jisha, M. Growth enhancement of rice (Oryza sativa) by phosphate solubilizing Gluconacetobacter sp.(MTCC 8368) and Burkholderia sp.(MTCC 8369) under greenhouse conditions. 3 Biotech 2015, 5, 831–837. [Google Scholar] [CrossRef]
- Ortuño, N.; Castillo, J.A.; Claros, M.; Navia, O.; Angulo, M.; Barja, D.; Gutiérrez, C.; Angulo, V. Enhancing the sustainability of quinoa production and soil resilience by using bioproducts made with native microorganisms. Agronomy 2013, 3, 732–746. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Ali, S.; Ali, B.; Farooq, M.A.; Gill, R.A. Priming-induced antioxidative responses in two wheat cultivars under saline stress. Acta Physiol. Plant. 2015, 37, 153. [Google Scholar] [CrossRef]
- Perrig, D.; Boiero, M.; Masciarelli, O.; Penna, C.; Ruiz, O.; Cassán, F.; Luna, M. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl. Microbiol. Biotechnol. 2007, 75, 1143–1150. [Google Scholar] [CrossRef]
- Lai, W.-A.; Rekha, P.; Arun, A.; Young, C.-C. Effect of mineral fertilizer, pig manure, and Azospirillum rugosum on growth and nutrient contents of Lactuca sativa L. Biol. Fertil. Soils 2008, 45, 155–164. [Google Scholar] [CrossRef]
- Richardson, A.E.; Barea, J.-M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Sapre, S.; Sharma, A.; Tiwari, S. Alleviation of mercury toxicity in wheat by the interaction of mercury-tolerant plant growth-promoting rhizobacteria. J. Plant Growth Regul. 2016, 35, 1000–1012. [Google Scholar] [CrossRef]
- Whipps, J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001, 52, 487–511. [Google Scholar] [CrossRef]
- Tailor, A.J.; Joshi, B.H. Characterization and optimization of siderophore production from Pseudomonas fluorescens strain isolated from sugarcane rhizosphere. J. Environ. Res. Dev. 2012, 6, 688–694. [Google Scholar]
- Jetiyanon, K. Multiple Mechanisms of Enterobacter asburiae strain RS83 for Plant Growth Enhancement. Songklanakarin J. Sci. Technol. 2015, 37, 29–36. [Google Scholar]
- Marchese, A.; Orhan, I.E.; Daglia, M.; Barbieri, R.; Di Lorenzo, A.; Nabavi, S.F.; Gortzi, O.; Izadi, M.; Nabavi, S.M. Antibacterial and antifungal activities of thymol: A brief review of the literature. Food Chem. 2016, 210, 402–414. [Google Scholar] [CrossRef]
- Behar, A.; Yuval, B.; Jurkevitch, E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 2005, 14, 2637–2643. [Google Scholar] [CrossRef] [PubMed]
- Neilson, A.; Sparell, L. Acetylene reduction (nitrogen fixation) by Enterobacteriaceae isolated from paper mill process waters. Appl. Environ. Microbiol. 1976, 32, 197–205. [Google Scholar] [CrossRef]
- Walsh, U.F.; Morrissey, J.P.; O’Gara, F. Pseudomonas for biocontrol of phytopathogens: From functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 2001, 12, 289–295. [Google Scholar] [CrossRef]
- Agbodjato, N.A.; Noumavo, P.A.; Baba-Moussa, F.; Salami, H.A.; Sina, H.; Sèzan, A.; Bankolé, H.; Adjanohoun, A.; Baba-Moussa, L. Characterization of potential plant growth promoting rhizobacteria isolated from Maize (Zea mays L.) in central and Northern Benin (West Africa). Appl. Environ. Soil Sci. 2015, 2015. [Google Scholar] [CrossRef]
- Xun, F.; Xie, B.; Liu, S.; Guo, C. Effect of plant growth-promoting bacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) inoculation on oats in saline-alkali soil contaminated by petroleum to enhance phytoremediation. Environ. Sci. Pollut. Res. 2015, 22, 598–608. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Gong, B.; Li, X.; VandenLangenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-l-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Cuin, T.A.; Tian, Y.; Betts, S.A.; Chalmandrier, R.; Shabala, S. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Funct. Plant Biol. 2009, 36, 1110–1119. [Google Scholar] [CrossRef]
- Adolf, V.I.; Shabala, S.; Andersen, M.N.; Razzaghi, F.; Jacobsen, S.-E. Varietal differences of quinoa’s tolerance to saline conditions. Plant Soil 2012, 357, 117–129. [Google Scholar] [CrossRef]
- Razzaghi, F.; Jacobsen, S.-E.; Jensen, C.R.; Andersen, M.N. Ionic and photosynthetic homeostasis in quinoa challenged by salinity and drought–mechanisms of tolerance. Funct. Plant Biol. 2015, 42, 136–148. [Google Scholar] [CrossRef]
- Khalid, A.; Athar, H.-U.-R.; Zafar, Z.U.; Akram, A.; Hussain, K.; Manzoor, H.; Al-Qurainy, F.; Ashraf, M. Photosynthetic capacity of canola (Brassica napus L.) plants as affected by glycinebetaine under salt stress. J. Appl. Bot. Food Qual. 2015, 88, 78–86. [Google Scholar] [CrossRef]
- Qurashi, A.W.; Sabri, A.N. Osmolyte accumulation in moderately halophilic bacteria improves salt tolerance of chickpea. Pak. J. Bot. 2013, 45, 1011–1016. [Google Scholar]
- Zhao, H.; Yan, H.; Zhou, S.; Xue, Y.; Zhang, C.; Dong, X.; Cui, Q.; Zhang, Y.; Zhang, B.; Zhang, Z. The growth promotion of mung bean (Phaseolus radiatus) by Enterobacter asburiae HPP16 in acidic soils. Afr. J. Biotechnol. 2011, 10, 13802–13814. [Google Scholar] [CrossRef]
- Kayasth, M.; Kumar, V.; Gera, R. Exploring the potential of PGPR strain Bacillus licheniformis to be developed as multifunctional biofertilizer. Cent. Eur. J. Biol. 2013, 2, 12–17. [Google Scholar]
- Went, F.W.; Thimann, K.V. Phytohormones. Phytohormones; The Macmillan Company: New York, NY, USA, 1937. [Google Scholar]
- Höflich, G.; Wiehe, W.; Kühn, G. Plant growth stimulation by inoculation with symbiotic and associative rhizosphere microorganisms. Experientia 1994, 50, 897–905. [Google Scholar] [CrossRef]
- Asim, M.; Aslam, M.; Bano, A.; Munir, M.; Majeed, A.; Abbas, S.H. Role of phytohormones in root nodulation and yield of soybean under salt stress. Am. J. Res. Commun. 2013, 1, 191–208. [Google Scholar]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Tahir, M.; Mirza, M.S.; Zaheer, A.; Dimitrov, M.R.; Smidt, H.; Hameed, S. Isolation and identification of phosphate solubilizer Azospirillum, Bacillus and Enterobacter strains by 16SrRNA sequence analysis and their effect on growth of wheat (Triticum aestivum L.). Aust. J. Crop Sci. 2013, 7, 1284–1292. [Google Scholar]
- Ashraf, M.; Hasnain, S.; Berge, O.; Mahmood, T. Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 2004, 40, 157–162. [Google Scholar] [CrossRef]
- Gupta, G.; Snehi, S.K.; Singh, V. Role of PGPR in biofilm formations and its importance in plant health. Biofilms Plant Soil Health 2017, 27. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. A halotolerant bacterium Bacillus licheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Front. Plant Sci. 2016, 7, 1890. [Google Scholar] [CrossRef]


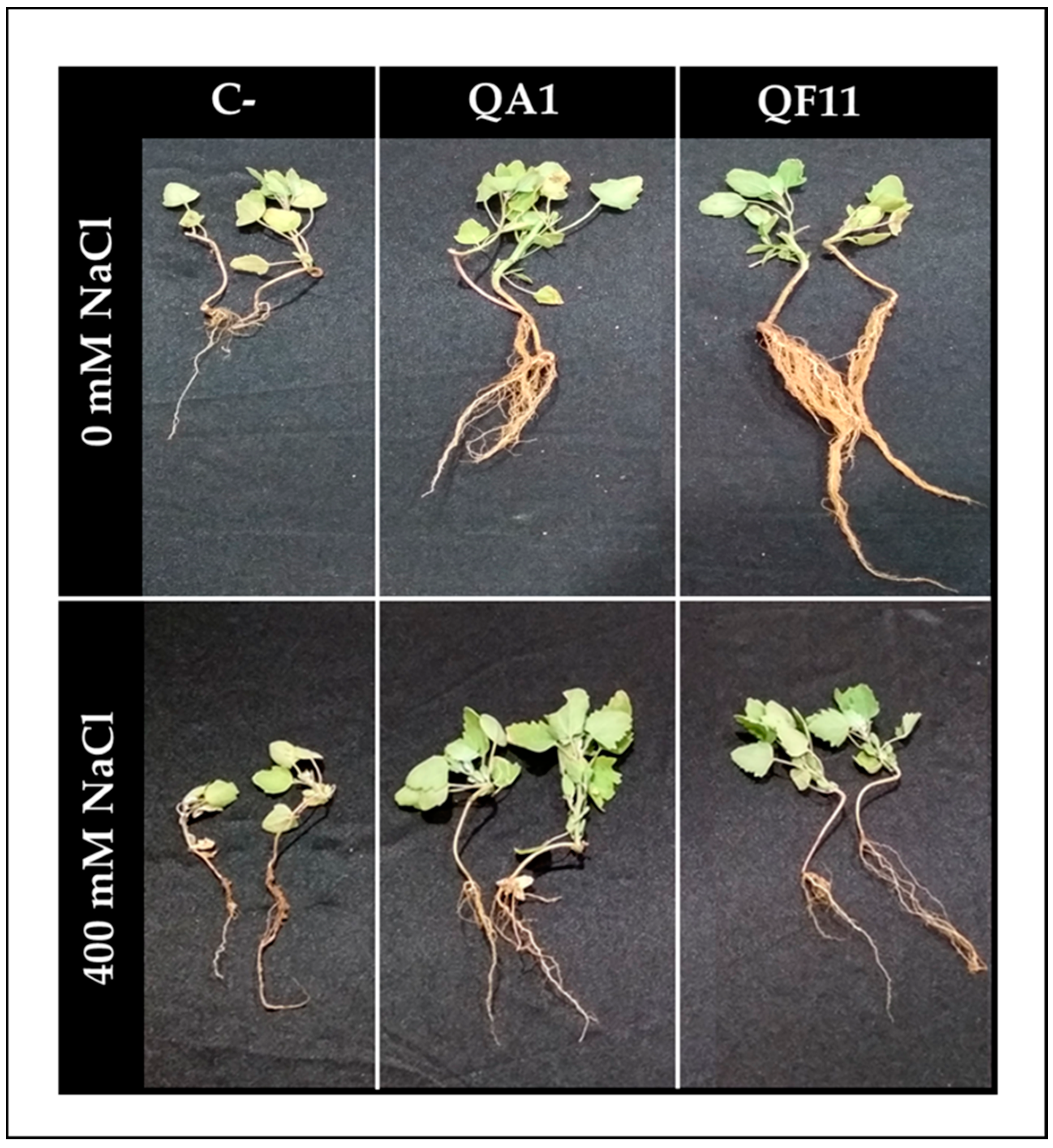

| 0 mM NaCl | 400 mM NaCl | ||
|---|---|---|---|
| Symbol | Treatment | Symbol | Treatment |
| C- | Seeds and plants treated with sterilized distilled water (Negative control) | C- | Seeds and plants treated with sterilized distilled water (Negative control) |
| QA1 | Seeds and plants treated with QA1 strain | QA1 | Seeds and plants treated with QA1 strain |
| QF11 | Seeds and plants treated with QF11 strain | QF11 | Seeds and plants treated with QF11 strain |
| Bacterial Strain | Bacterial Treatment | QA1 | QF11 |
|---|---|---|---|
| Extreme properties | NaCl tolerance on plates | 2 M | <0.9 M |
| Temperature tolerance | 55 °C | 37 °C | |
| Siderophore production | +++ | ++ | |
| HCN production | +++ | + | |
| Extracellular enzymes (Halo Colony diameter) | Protease Cellulase | 1.07 ± 0.08 4.21 ± 0.24 | − − |
| Parameter | Incubation Time | C- | QF11 | QA1 |
|---|---|---|---|---|
| Germination rate (%) | 24 h | 16.6 ± 6.65 c | 67.3 ± 12.5 a | 43.2 ± 6.7 b |
| 48 h | 58.6 ± 5.13 c | 77.6 ± 6.8 a | 67.7 ± 5.09 b | |
| Total length (cm) | 1.7 ± 0.55 c | 5.3 ± 0.32 a | 2.56 ± 0.94 bc | |
| Fresh weight (mg) | 36 ± 10.5 c | 50 ± 10.8 a | 41.3 ± 3.51 bc | |
| Dry weight (mg) | 6.6 ± 1.52 b | 11.6 ± 2.51 a | 7.3 ± 1.52 ab | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahdi, I.; Fahsi, N.; Hafidi, M.; Allaoui, A.; Biskri, L. Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd. Microorganisms 2020, 8, 948. https://doi.org/10.3390/microorganisms8060948
Mahdi I, Fahsi N, Hafidi M, Allaoui A, Biskri L. Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd. Microorganisms. 2020; 8(6):948. https://doi.org/10.3390/microorganisms8060948
Chicago/Turabian StyleMahdi, Ismail, Nidal Fahsi, Mohamed Hafidi, Abdelmounaaim Allaoui, and Latefa Biskri. 2020. "Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd" Microorganisms 8, no. 6: 948. https://doi.org/10.3390/microorganisms8060948
APA StyleMahdi, I., Fahsi, N., Hafidi, M., Allaoui, A., & Biskri, L. (2020). Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd. Microorganisms, 8(6), 948. https://doi.org/10.3390/microorganisms8060948







