Uncovered Microbial Diversity in Antarctic Cryptoendolithic Communities Sampling Three Representative Locations of the Victoria Land
Abstract
1. Introduction
2. Materials and Methods
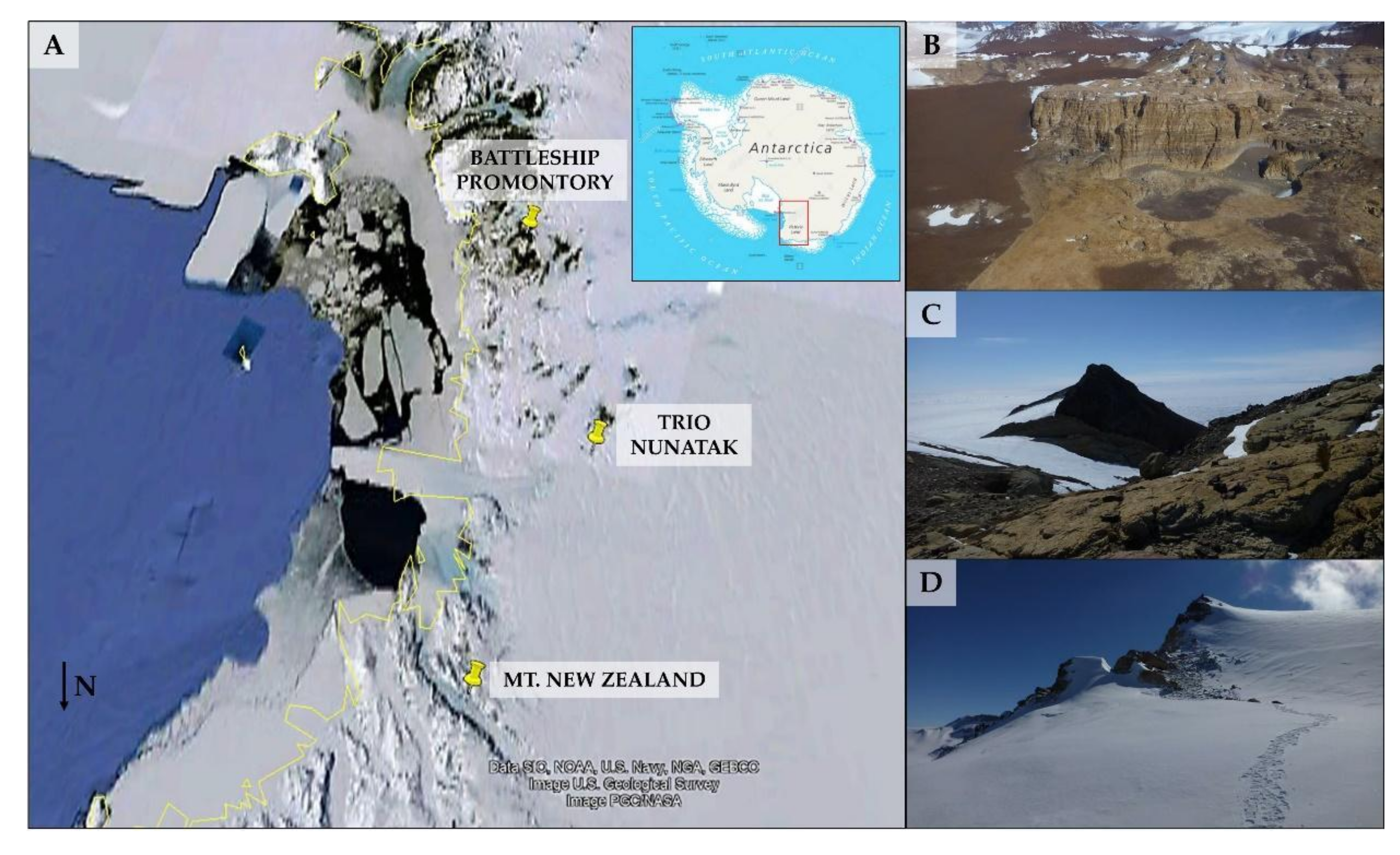
2.1. Study Area
2.2. Metagenomics DNA Extraction and Amplicon Sequencing
2.3. Raw Data Processing
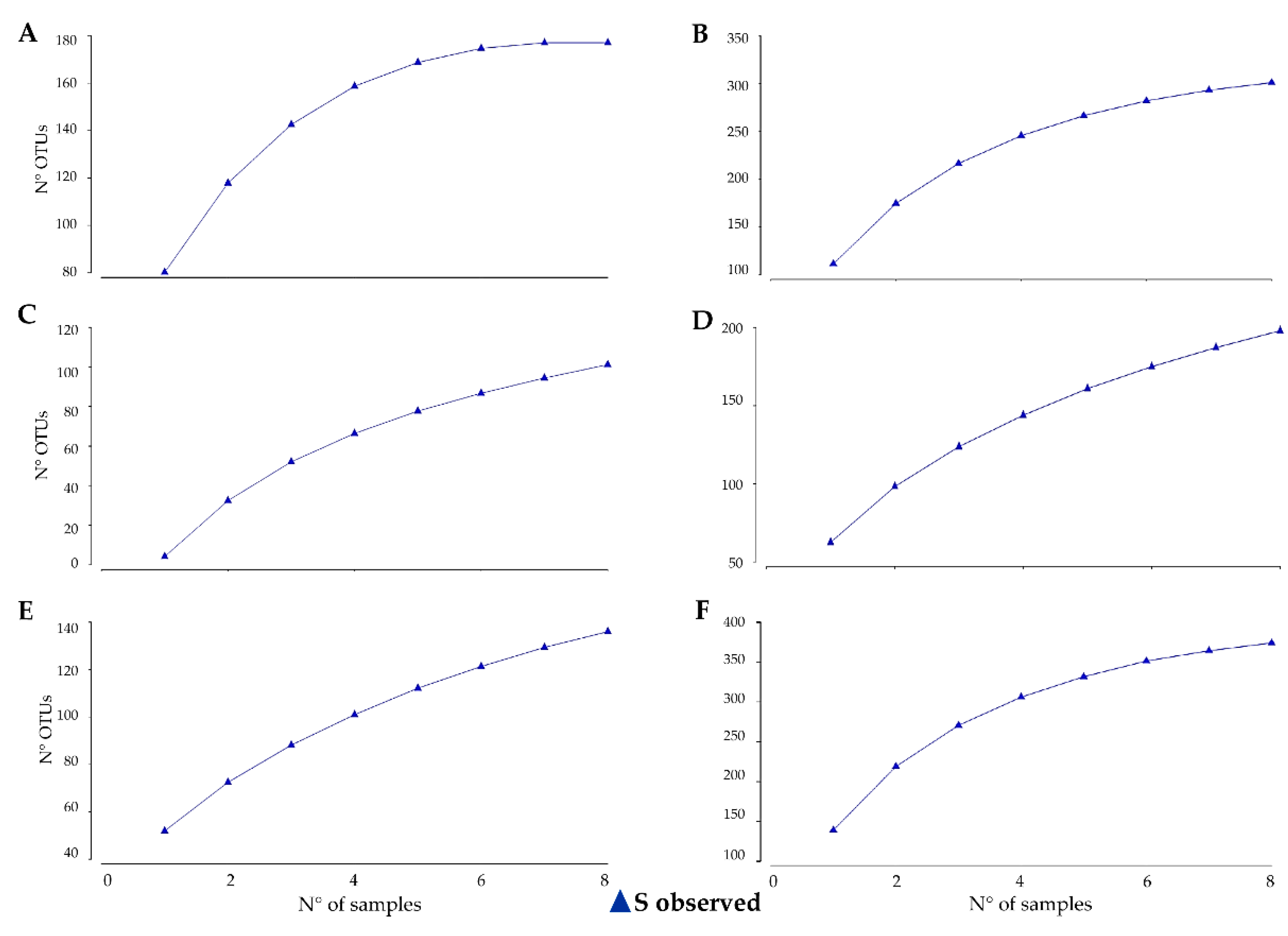
2.4. Accumulation and Extrapolation
2.5. Downstream Analysis
3. Results
3.1. Estimating and Determining Sampling Effort Across Sampling Areas
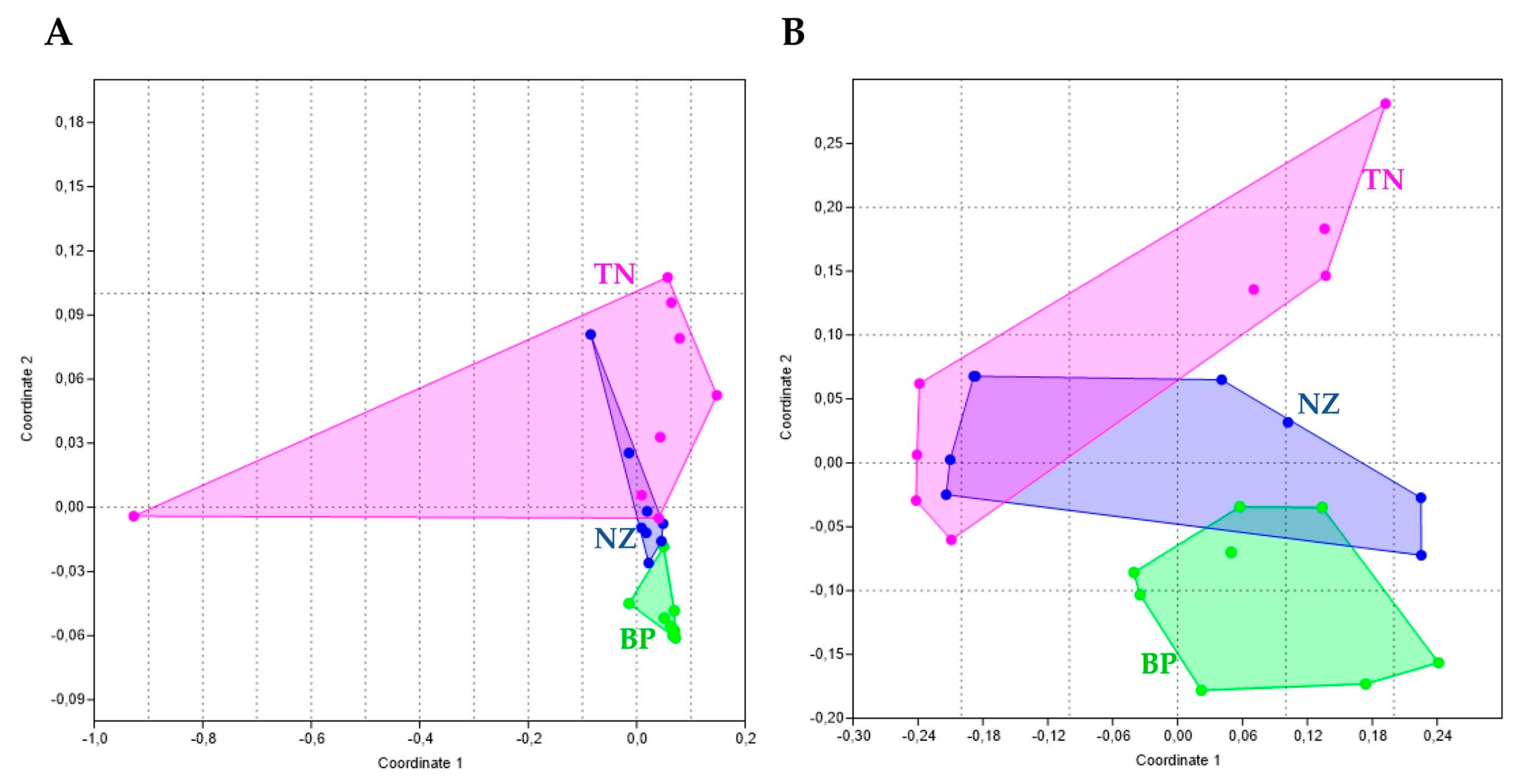
3.2. Differences in Community’ Composition by Site
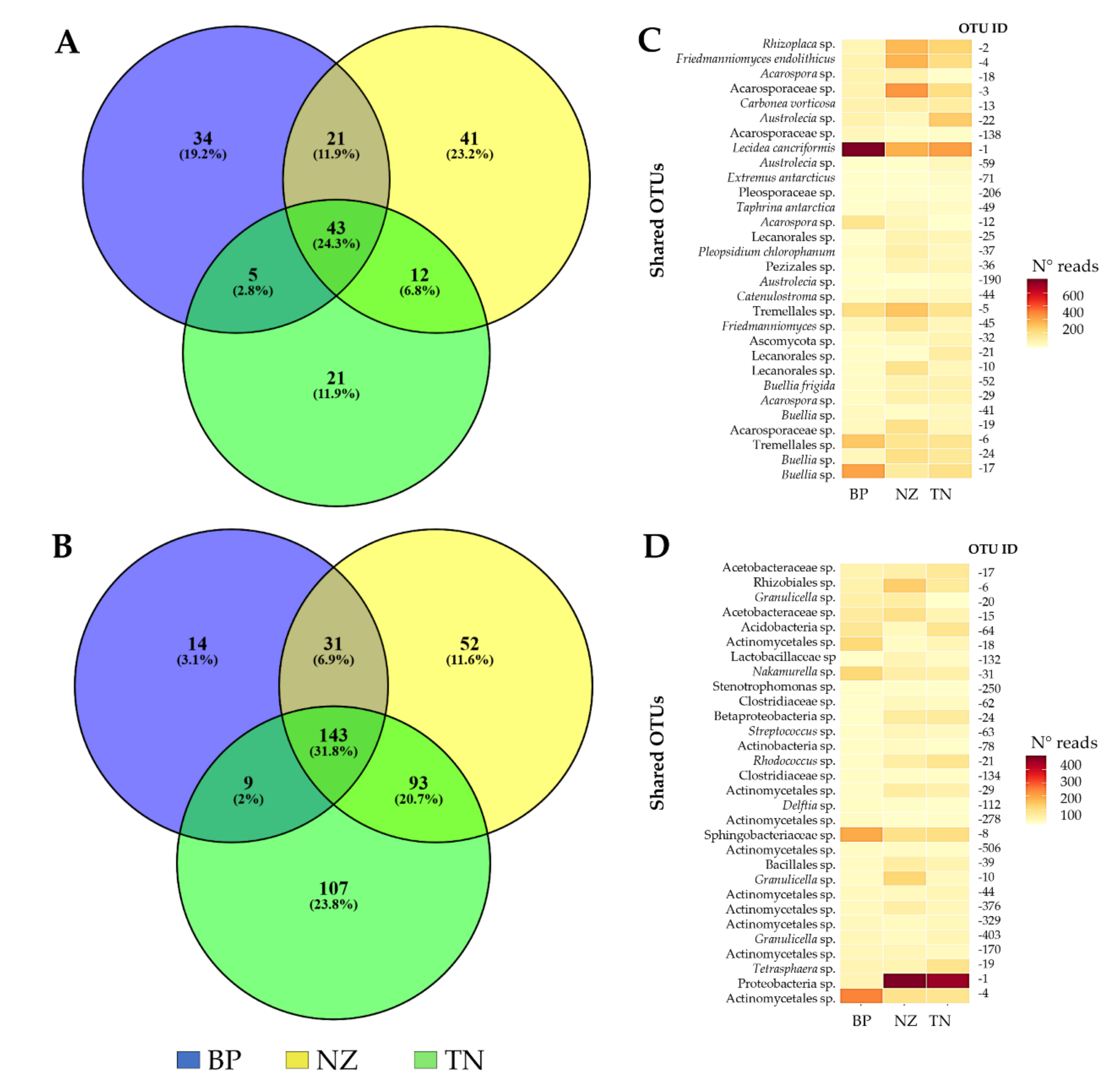
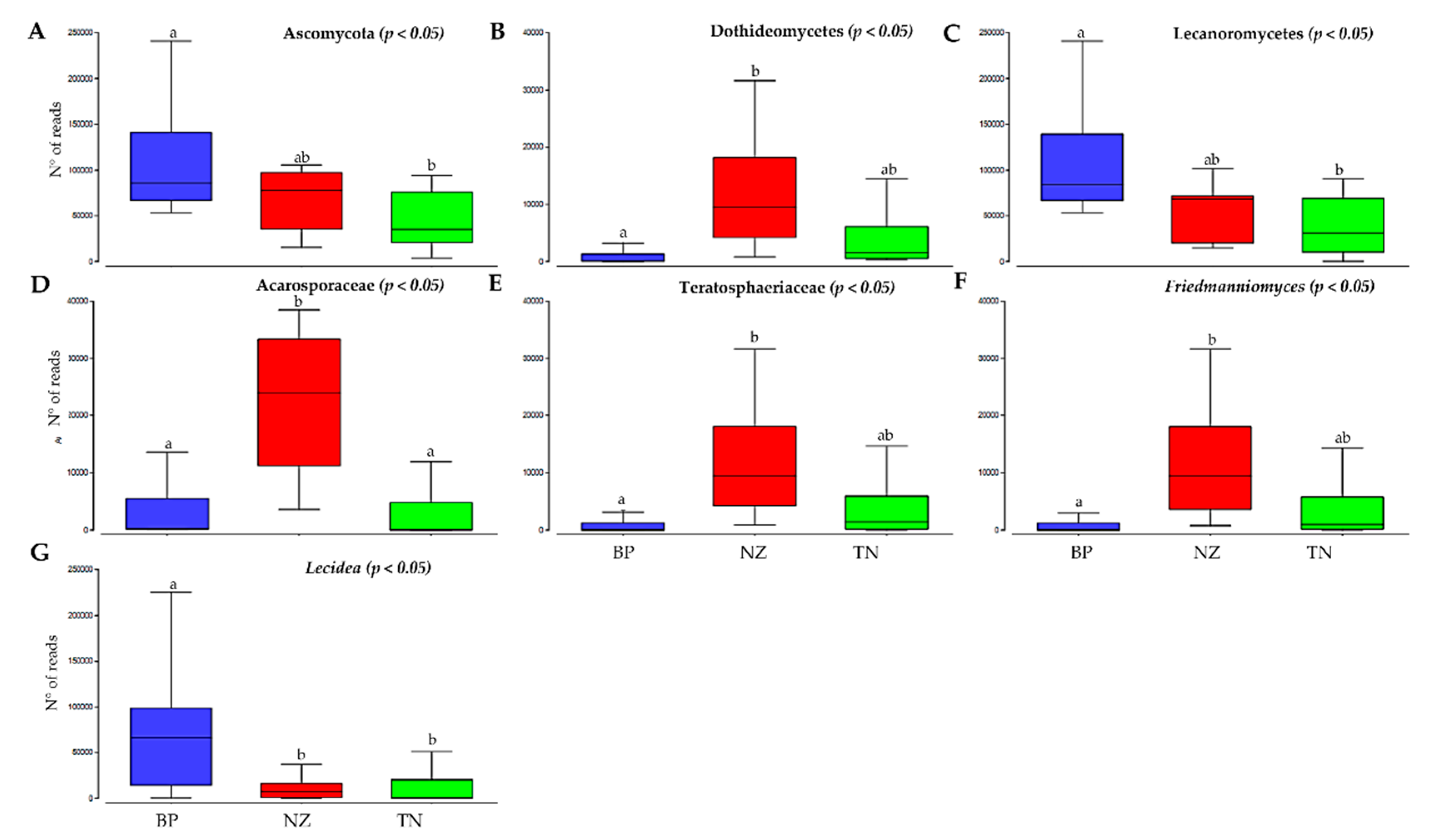
3.3. Differences in Alpha Diversity and Taxonomic Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gorbushina, A. Life on the rocks. Environ. Microbiol. 2007, 9, 1613–1631. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.J.; Spear, J.R.; Pace, N.R. Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 2005, 434, 1011–1014. [Google Scholar] [CrossRef] [PubMed]
- Friedmann, E.I.; Ocampo, R. Endolithic blue-green algae in dry valleys-primary producers in Antarctic desert ecosystem. Science 1976, 193, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Friedmann, E.I. Endolithic microorganisms in the Antarctic cold desert. Science 1982, 215, 1045–1053. [Google Scholar] [CrossRef]
- Bell, R.A. Cryptoendolithic algae of hot semiarid lands and deserts. J. Phycol. 1993, 29, 133–139. [Google Scholar] [CrossRef]
- Wierzchos, J.; de los Ríos, A.; Ascaso, C. Microorganisms in desert rocks: The edge of life on Earth. Int. Microbiol. 2012, 15, 173–183. [Google Scholar]
- Wierzchos, J.; DiRuggiero, J.; Vítek, P.; Artieda, O.; Souza-Egipsy, V.; Škaloud, P.; Tisza, T.; Davila, A.F.; Vílchez, C.; Garbayo, I.; et al. Adaptation strategies of endolithic chlorophototrophs to survive the hyperarid and extreme solar radiation environment of the Atacama Desert. Front. Microbiol. 2015, 6, 934. [Google Scholar] [CrossRef]
- Cowan, D.; Tow, L. Endangered Antarctic environments. Annu. Rev. Microbiol. 2004, 58, 649–690. [Google Scholar] [CrossRef]
- Cary, S.C.; McDonald, I.R.; Barrett, J.E.; Cowan, D.A. On the rocks: The microbiology of Antarctic dry valley soils. Nat. Rev. Microbiol. 2010, 8, 129–138. [Google Scholar] [CrossRef]
- Cowan, D.A.; Makhalanyane, T.P.; Dennis, P.G.; Hopkins, D.W. Microbial ecology and biogeochemistry of continental Antarctic soils. Front. Microbiol. 2014, 5, 154. [Google Scholar] [CrossRef]
- Nienow, J.A.; Friedmann, E.I. Terrestrial lithophytic (rock) communities. In Antarctic Microbiology; Friedmann, E.I., Ed.; Wiley-Liss: New York, NY, USA, 1993; pp. 343–412. [Google Scholar]
- Zucconi, L.; Onofri, S.; Cecchini, C.; Isola, D.; Ripa, C.; Fenice, M.; Madonna, S.; Reboleiro-Rivas, P.; Selbmann, L. Mapping the lithic colonization at the boundaries of life in Northern Victoria Land, Antarctica. Polar. Biol. 2016, 39, 91–102. [Google Scholar] [CrossRef]
- Selbmann, L.; Onofri, S.; Coleine, C.; Buzzini, P.; Canini, F.; Zucconi, L. Effect of environmental parameters on biodiversity of the fungal component in the lithic Antarctic communities. Extremophiles 2017, 21, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, J.R.; Goebel, B.M.; Friedmann, E.; Pace, N.R. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2003, 69, 3858–3867. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.T.; Lacap-Bugler, D.C.; Lau, M.C.; Caruso, T.; Rao, S.; de Los Ríos, A.; Archer, S.K.; Chiu, J.M.Y.; Higgins, C.; Van Nostrand, J.D.; et al. Taxonomic and functional diversity of soil and hypolithic microbial communities in Miers Valley, McMurdo Dry Valleys. Antarct. Front. Microbiol. 2016, 7, 1642. [Google Scholar] [CrossRef]
- Archer, S.D.; de los Ríos, A.; Lee, K.C.; Niederberger, T.S.; Cary, S.C.; Coyne, K.J.; Douglas, S.; Lacap-Bugler, D.C.; Pointing, S.B. Endolithic microbial diversity in sandstone and granite from the McMurdo Dry Valleys. Antarctica. Polar Biol. 2017, 40, 997–1006. [Google Scholar] [CrossRef]
- Coleine, C.; Stajich, J.E.; Zucconi, L.; Onofri, S.; Pombubpa, N.; Egidi, E.; Franks, A.; Buzzini, P.; Selbmann, L. Antarctic cryptoendolithic fungal communities are highly adapted and dominated by Lecanoromycetes and Dothideomycetes. Frontiers Microbiol. 2018, 9, 1392. [Google Scholar] [CrossRef]
- Coleine, C.; Zucconi, L.; Onofri, S.; Pombubpa, N.; Stajich, J.E.; Selbmann, L. Sun exposure shapes functional grouping of fungi cryptoendolithic Antarctic communities. Life 2018, 8, 19. [Google Scholar] [CrossRef]
- Coleine, C.; Stajich, J.E.; Pombubpa, N.; Zucconi, L.; Onofri, S.; Canini, F.; Selbmann, L. Altitude and fungal diversity influence the structure of Antarctic cryptoendolithic Bacteria communities. Environ. Microbiol. Rep. 2019, 11, 718–726. [Google Scholar] [CrossRef]
- Coleine, C.; Stajich, J.E.; Pombubpa, N.; Zucconi, L.; Onofri, S.; Selbmann, L. Sampling strategies to assess microbial diversity of Antarctic cryptoendolithic communities. Polar Biol. 2020, 43, 225–235. [Google Scholar] [CrossRef]
- Bradshaw, M.A. Paleoenvironmental interpretations and systematics of Devonian trace fossils from the Taylor Group (lower Beacon Supergroup), Antarctica. New Zealand J. Geol. Geophys. 1981, 24, 615–652. [Google Scholar] [CrossRef]
- Casnedi, R.; Di Giulio, A. Sedimentology of the Section Peak Formation (Jurassic), northern Victoria Land, Antarctica. Spec. Publ. Int. Assoc. Sedimentol. 1999, 28, 435–448. [Google Scholar]
- Smith, D.P.; Peay, K.G. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE 2014, 9, 90234. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Jusino, M.A.; Banik, M.T.; Lindner, D.L. Non-biological synthetic spike-in controls and the AMPtk software pipeline improve mycobiome data. PeerJ 2018, 6, e4925. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Kõljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J.; et al. Fungal community analysis by high-throughput sequencing of amplified markers—A user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef]
- Colwell, R.K.; Coddington, J.A. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1994, 345, 101–118. [Google Scholar]
- Moreno, C.E.; Halffter, G. Assessing the completeness of bat biodiversity inventories using species accumulation curves. J. Appl. Ecol. 2000, 37, 149–158. [Google Scholar] [CrossRef]
- Palmer, M.W. The estimation of species richness by extrapolation. Ecology 1990, 71, 1195–1198. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Entsminger, G.L. Swap and fill algorithms in null model analysis: Rethinking the knight’s tour. Oecologia 2001, 129, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Pielou, E.C. Mathematical Ecology; No. 574.50151; Wiley: Hoboken, NJ, USA, 1977. [Google Scholar]
- Colwell, R.K.; Chao, A.; Gotelli, N.J.; Lin, S.Y.; Mao, C.X.; Chazdon, R.L.; Longino, J.T. Models and estimators linking individual-based and sample-based rarefaction, extrapolation, and comparison of assemblages. J. Plant. Ecol. 2012, 5, 3–21. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST-Palaeontological Statistics. 2001. Available online: www.uv.es/~pardomv/pe/2001_1/past/pastprog/past.pdf (accessed on 23 June 2020).
- Anderson, M.J.; Walsh, D.C.I. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Eco.l Monogr. 2013, 83, 557574. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1963. [Google Scholar]
- Ludwig, J.A. Statistical Ecology: A Primer in Methods and Computing; John Wiley & Sons: New York, NY, USA, 1988. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Pombubpa, N.; Zucconi, L.; Onofri, S.; Stajich, J.E.; Selbmann, L. Endolithic Fungal Species Markers for Harshest Conditions in the McMurdo Dry Valleys, Antarctica. Life 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; Turchetti, B.; Yurkov, A.; Cecchini, C.; Zucconi, L.; Isola, D.; Buzzini, P.; Onofri, S. Description of Taphrina antarctica f.a. sp. nov., a new anamorphic ascomycetous yeast species associated with Antarctic endolithic microbial communities and transfer of four Lalaria species in the genus Taphrina. Extremophiles 2014, 18, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Saul, D.J.; Aislabie, J.M.; Brown, C.E.; Harris, L.; Foght, J.M. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 2005, 53, 141–155. [Google Scholar] [CrossRef]
- Aislabie, J.M.; Chhour, K.L.; Saul, D.J.; Miyauchi, S.; Ayton, R.; Paetzold, R.F.; Balks, M.R. Dominant bacteria in soils of Marble point and Wright Valley, Victoria Land, Antarctica. Soil Biol. Biochem. 2006, 38, 3041–3056. [Google Scholar] [CrossRef]
- Christner, B.C.; Kvitko, B.H.; Reeve, J.N. Molecular identification of bacteria and eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 2003, 7, 177–183. [Google Scholar] [CrossRef]
- Hirsch, P.; Hoffmann, B.; Gallikowski, C.C.; Mevs, U.; Siebert, J.; Sittig, M. Diversity and Identification of Heterotrophs from Antarctic Rocks of the McMurdo Dry Valleys (Ross Desert). Polarforschung 1988, 58, 261–269. [Google Scholar]
- Hirsch, P.; Mevs, U.; Kroppenstedt, R.M.; Schumann, P.; Stackebrandt, E. Cryptoendolithic actinomycetes from Antarctic sandstone rock samples: Micromonospora endolithica sp. nov. and two isolates related to Micromonospora coerulea Jensen 1932. Syst. App. Microbiol. 2004, 27, 166. [Google Scholar] [CrossRef] [PubMed]
- Choe, Y.H.; Kim, M.; Woo, J.; Lee, M.J.; Lee, J.I.; Lee, E.J.; Lee, Y.K. Comparing rock-inhabiting microbial communities in different rock types from a high arctic polar desert. FEMS Microbiol. Ecol. 2018, 94, fiy070. [Google Scholar] [CrossRef] [PubMed]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus radiodurans: Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef]
- Ferreira, A.C.; Nobre, M.F.; Rainey, F.A.; Silva, M.T.; Wait, R.; Burghardt, J.; Chung, A.P.; Da Costa, M.S. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int. J. Syst. Evol. Micr. 1997, 47, 939–947. [Google Scholar] [CrossRef]
- Makarova, K.S.; Omelchenko, M.V.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A. Deinococcus geothermalis: The pool of extreme radiation resistance genes shrinks. PLoS ONE 2007, 2, e955. [Google Scholar] [CrossRef]
- Selbmann, L.; de Hoog, G.S.; Mazzaglia, A.; Friedmann, E.I.; Onofri, S. Fungi at the edge of life: Cryptoendolithic black fungi from Antarctic deserts. Stud. Mycol. 2005, 51, 1–32. [Google Scholar]
- Ametrano, C.G.; Muggia, L.; Grube, M. Extremotolerant Black Fungi from Rocks and Lichens. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Springer: Cham, Switzerland, 2019; pp. 119–143. [Google Scholar]
- Perini, L.; Gostinčar, C.; Anesio, A.M.; Williamson, C.; Tranter, M.; Gunde-Cimerman, N. Darkening of the Greenland Ice Sheet: Fungal abundance and diversity are associated with algal bloom. Front. Microbiol. 2019, 10, 557. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Sonjak, S.; Zalar, P.; Frisvad, J.C.; Diderichsen, B.; Plemenitaš, A. Extremophilic fungi in arctic ice: A relationship between adaptation to low temperature and water activity. Phys. Chem. Earth Parts 2003, 28, 1273–1278. [Google Scholar] [CrossRef]
- Chung, D.; Kim, H.; Choi, H.S. Fungi in salterns. J. Microbiol. 2019, 57, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Tesei, D.; Zakharova, K. Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol. 2012, 5, 453–462. [Google Scholar] [CrossRef]
- Coleine, C.; Masonjones, S.; Sterflinger, K.; Onofri, S.; Selbmann, L.; Stajich, J.E. Peculiar genomic traits in the stress-adapted cryptoendolithic endemic Antarctic fungus Friedmanniomyces endolithicus. Fungal Biol. 2020. [Google Scholar] [CrossRef] [PubMed]

| Site | Samples | N° of Species (Computed) | |
|---|---|---|---|
| Fungi | |||
| Trio Nunatak | 23 | 150 | |
| Mt New Zealand | 28 | 190 | |
| Bacteria | |||
| Battleship Promontory | 12 | 335 | |
| Trio Nunatak | 32 | 350 | |
| Mt New Zealand | 34 | 465 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coleine, C.; Pombubpa, N.; Zucconi, L.; Onofri, S.; Turchetti, B.; Buzzini, P.; Stajich, J.E.; Selbmann, L. Uncovered Microbial Diversity in Antarctic Cryptoendolithic Communities Sampling Three Representative Locations of the Victoria Land. Microorganisms 2020, 8, 942. https://doi.org/10.3390/microorganisms8060942
Coleine C, Pombubpa N, Zucconi L, Onofri S, Turchetti B, Buzzini P, Stajich JE, Selbmann L. Uncovered Microbial Diversity in Antarctic Cryptoendolithic Communities Sampling Three Representative Locations of the Victoria Land. Microorganisms. 2020; 8(6):942. https://doi.org/10.3390/microorganisms8060942
Chicago/Turabian StyleColeine, Claudia, Nuttapon Pombubpa, Laura Zucconi, Silvano Onofri, Benedetta Turchetti, Pietro Buzzini, Jason E. Stajich, and Laura Selbmann. 2020. "Uncovered Microbial Diversity in Antarctic Cryptoendolithic Communities Sampling Three Representative Locations of the Victoria Land" Microorganisms 8, no. 6: 942. https://doi.org/10.3390/microorganisms8060942
APA StyleColeine, C., Pombubpa, N., Zucconi, L., Onofri, S., Turchetti, B., Buzzini, P., Stajich, J. E., & Selbmann, L. (2020). Uncovered Microbial Diversity in Antarctic Cryptoendolithic Communities Sampling Three Representative Locations of the Victoria Land. Microorganisms, 8(6), 942. https://doi.org/10.3390/microorganisms8060942









