Genomic and Proteomic Characterization of the Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Strain CCUG 73778: A Virulent, Nosocomial Outbreak Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Growth Conditions
2.2. DNA Extraction
2.3. DNA Sequencing
2.4. Genome Assembly
2.5. Whole Genome Sequence Quality Control and Annotation
2.6. Genome Sequence Analysis
2.7. Plasmid Sequence Characterization
2.8. Protein Expression Determination
2.9. Data Availability
3. Results and Discussion
3.1. Sequencing and Assembly Comparison
3.2. Antibiotic Resistance Factors
3.3. Virulence Related Elements
3.4. Toxin–Antitoxin Systems
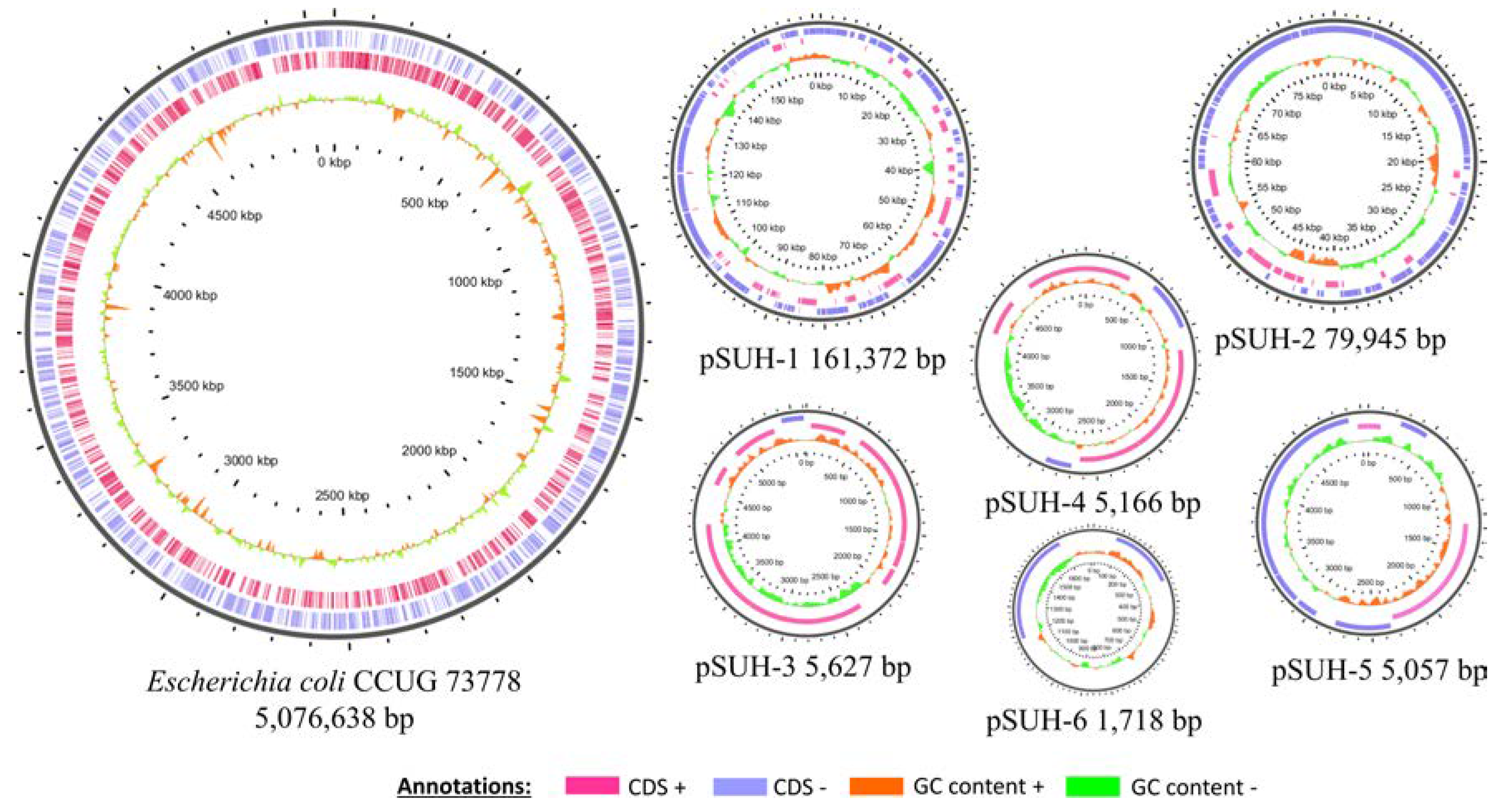
3.5. Plasmid Sequence Characterization
3.6. Protein Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ena, J.; Arjona, F.; Martínez-Peinado, C.; del mar López-Perezagua, M.; Amador, C. Epidemiology of urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Urology 2006, 68, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Baquero, F.; Cantón, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008, 13, 19051. [Google Scholar]
- Peirano, G.; Pitout, J.D. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: The worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 2010, 35, 316–321. [Google Scholar] [CrossRef]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Turner, P.J. Extended-spectrum beta-lactamases. Clin. Infect. Dis. 2005, 41, S273–S275. [Google Scholar] [CrossRef]
- Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: www.genome.gov/sequencingcostsdata (accessed on 10 June 2020).
- Mardis, E.R. A decade’s perspective on DNA sequencing technology. Nature 2011, 470, 198–203. [Google Scholar] [CrossRef]
- Sabat, A.J.; Hermelijn, S.M.; Akkerboom, V.; Juliana, A.; Degener, J.E.; Grundmann, H.; Friedrich, A.W. Complete-genome sequencing elucidates outbreak dynamics of CA-MRSA USA300 (ST8-spa t008) in an academic hospital of Paramaribo, Republic of Suriname. Sci. Rep. 2017, 7, 41050. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, S.; Beka, L.; Graf, J.; Klassen, J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genom. 2019, 20, 23. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Miao, M.; Yan, J.; Wang, M.; Tang, Y.-W.; Kreiswirth, B.N.; Zhang, X.; Chen, L.; Du, H. Expression characteristics of the plasmid-borne mcr-1 colistin resistance gene. Oncotarget 2017, 8, 107596–107602. [Google Scholar] [CrossRef] [PubMed]
- Karami, N.; Helldal, L.; Welinder-Olsson, C.; Åhrén, C.; Moore, E.R. Sub-typing of extended-spectrum-β-lactamase- producing isolates from a nosocomial outbreak: Application of a 10-loci generic Escherichia coli multi- locus variable number tandem repeat analysis. PLoS ONE 2013, 8, e83030. [Google Scholar] [CrossRef]
- Müller, V.; Karami, N.; Nyberg, L.K.; Pichler, C.; Torche Pedreschi, P.C.; Quaderi, S.; Fritzsche, J.; Ambjörnsson, T.; Åhrén, C.; Westerlund, F. Rapid tracing of resistance plasmids in a nosocomial outbreak using optical DNA mapping. ACS Infect. Dis. 2016, 2, 322–328. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Salvà-Serra, F.; Svensson-Stadler, L.; Busquets, A.; Jaén-Luchoro, D.; Karlsson, R.; Moore, E.R.; Gomila, M. A protocol for extraction and purification of high-quality and quantity bacterial DNA applicable for genome sequencing: A modified version of the Marmur procedure. Protoc. Exch. 2018. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. Available online: https://github.com/najoshi/sickle (accessed on 10 June 2020).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.-Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Karlsson, R.; Thorsell, A.; Gomila, M.; Salvà-Serra, F.; Jakobsson, H.E.; Gonzales-Siles, L.; Jaén-Luchoro, D.; Skovbjerg, S.; Fuchs, J.; Karlsson, A.; et al. Discovery of species-unique peptide biomarkers of bacterial pathogens by tandem mass spectrometry-based proteotyping. Mol. Cell. Proteom. 2020, 19, 518–528. [Google Scholar] [CrossRef]
- Karlsson, R.; Gonzales-Siles, L.; Gomila, M.; Busquets, A.; Salvà-Serra, F.; Jaén-Luchoro, D.; Jakobsson, H.E.; Karlsson, A.; Boulund, F.; Kristiansson, E.; et al. Proteotyping bacteria: Characterization, differentiation and identification of pneumococcus and other species within the Mitis Group of the genus Streptococcus by tandem mass spectrometry proteomics. PLoS ONE 2018, 13, e0208804. [Google Scholar] [CrossRef] [PubMed]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of oxford nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Takahata, S.; Ida, T.; Hiraishi, T.; Sakakibara, S.; Maebashi, K.; Terada, S.; Muratani, T.; Matsumoto, T.; Nakahama, C.; Tomono, K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003, 51, 1109–1117. [Google Scholar] [CrossRef]
- Bahrani, F.K.; Buckles, E.L.; Lockatell, C.V.; Hebel, J.R.; Johnson, D.E.; Tang, C.M.; Donnenberg, M.S. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 2002, 45, 1079–1093. [Google Scholar] [CrossRef]
- Kallenius, G.; Mollby, R.; Svenson, S.B.; Helin, I.; Hultberg, H.; Cedergren, B.; Winberg, J. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet 1981, 2, 1369–1372. [Google Scholar] [CrossRef]
- Rick, P.D.; Silver, R.P. Enterobacterial common antigen and capsular polysaccharides. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; ASM Press: Washington, DC, USA, 1996; pp. 104–122. [Google Scholar]
- Welch, R.A.; Burland, V.; Plunkett, G.; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.-R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef]
- Lloyd, A.L.; Rasko, D.A.; Mobley, H.L.T. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 2007, 189, 3532–3546. [Google Scholar] [CrossRef]
- Rendón, M.A.; Saldaña, Z.; Erdem, A.L.; Monteiro-Neto, V.; Vázquez, A.; Kaper, J.B.; Puente, J.L.; Girón, J.A. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA 2007, 104, 10637–10642. [Google Scholar] [CrossRef]
- Nasr, A.; Olsén, A.; Sjöbring, U.; Müller-Esterl, W.; Björck, L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 1996, 20, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [CrossRef] [PubMed]
- Xicohtencatl-Cortes, J.; Monteiro-Neto, V.; Saldaña, Z.; Ledesma, M.A.; Puente, J.L.; Girón, J.A. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 2009, 191, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, S.-H.; Wass, C.A.; Stins, M.F.; Kim, K.S. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 1999, 67, 4751–4756. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.; Nies, D.H.; Franke, S. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 2001, 147, 965–972. [Google Scholar]
- Grass, G.; Rensing, C. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 2001, 183, 2145–2147. [Google Scholar] [CrossRef]
- Phung, L.T.; Silver, S.; Gupta, A.; Taylor, D.E. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 2001, 147, 3393–3402. [Google Scholar]
- Janakiraman, A.; Slauch, J.M. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 2000, 35, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Heras, B.; Shouldice, S.R.; Totsika, M.; Scanlon, M.J.; Schembri, M.A.; Martin, J.L. DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 2009, 7, 215–225. [Google Scholar] [CrossRef]
- Bjur, E.; Eriksson-Ygberg, S.; Aslund, F.; Rhen, M. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 2006, 74, 5140–5151. [Google Scholar] [CrossRef]
- Robbins, J.B.; McCracken, G.H.J.; Gotschlich, E.C.; Orskov, F.; Orskov, I.; Hanson, L.A. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. N. Engl. J. Med. 1974, 290, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.G.S. Neonatal Sepsis and Meningitis; Hall Medical Publishers: Boston, MA, USA, 1985. [Google Scholar]
- Silver, R.P.; Vimr, E.R. Polysialic acid capsule of Escherichia coli K1. In The Bacteria, Molecular Basis of Bacterial Pathogenesis; Academic Press: Cambridge, MA, USA, 1990; pp. 39–60. [Google Scholar]
- Engelberg-Kulka, H.; Glaser, G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 1999, 53, 43–70. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000, 182, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Engelberg-Kulka, H.; Amitai, S.; Kolodkin-Gal, I.; Hazan, R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006, 2, e135. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef]
- Sala, A.; Calderon, V.; Bordes, P.; Genevaux, P. TAC from Mycobacterium tuberculosis: A paradigm for stress-responsive toxin-antitoxin systems controlled by SecB-like chaperones. Cell Stress Chaperones 2013, 18, 129–135. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef]
- Christensen, S.K.; Mikkelsen, M.; Pedersen, K.; Gerdes, K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 2001, 98, 14328–14333. [Google Scholar] [CrossRef]
- Tsuchimoto, S.; Ohtsubo, H.; Ohtsubo, E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 1988, 170, 1461–1466. [Google Scholar] [CrossRef]
- Wozniak, R.A.F.; Waldor, M.K. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Garcillán-Barcia, M.P.; Francia, M.V.; de La Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Francia, M.V.; Varsaki, A.; Garcillán-Barcia, M.P.; Latorre, A.; Drainas, C.; de la Cruz, F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004, 28, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M.; Sanchez, Z.K.; Kimbara, K. Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 2015, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Ilangovan, A.; Connery, S.; Waksman, G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 2015, 23, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Kwong, S.M.; Murphy, R.J.T.; Yui Eto, K.; Price, K.J.; Nguyen, Q.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Firth, N. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elem. 2016, 6, e1208317. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wyres, K.L.; Wick, R.R.; Judd, L.M.; Fostervold, A.; Holt, K.E.; Löhr, I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019, 74, 1218–1222. [Google Scholar] [CrossRef]
- Boyd, D.A.; Tyler, S.; Christianson, S.; McGeer, A.; Muller, M.P.; Willey, B.M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey, M.R. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef]
- Goodacre, N.F.; Gerloff, D.L.; Uetz, P. Protein domains of unknown function are essential in bacteria. mBio 2014, 5, e00744-13. [Google Scholar] [CrossRef]
| Replicon | System/Gene | Drug Class | Resistance Mechanism |
|---|---|---|---|
| Chromosome | acrD | aminoglycoside antibiotic | antibiotic efflux |
| mdtNOP | acridine dye; nucleoside antibiotic | antibiotic efflux | |
| mdtABC-tolC | aminocoumarin antibiotic | antibiotic efflux | |
| mdtG | fosfomycin | antibiotic efflux | |
| mdtEF-tolC | multidrug | antibiotic efflux | |
| mdtH | fluoroquinolone antibiotic | antibiotic efflux | |
| patA | fluoroquinolone antibiotic | antibiotic efflux | |
| emrAB-tolC | fluoroquinolone antibiotic | antibiotic efflux | |
| emrE | macrolide antibiotic | antibiotic efflux | |
| acrAB-tolC | multidrug | antibiotic efflux | |
| acrEF-tolC | multidrug | antibiotic efflux | |
| emrKY-tolC | tetracycline antibiotic | antibiotic efflux | |
| mdfA | multidrug | antibiotic efflux | |
| msbA | nitroimidazole antibiotic | antibiotic efflux | |
| msrB | macrolide antibiotic; streptogramin antibiotic | antibiotic efflux | |
| bacA | nitroimidazole antibiotic | antibiotic efflux | |
| yojL | peptide antibiotic | antibiotic efflux | |
| glpT (mutation E350Q) | fosfomycin | antibiotic target alteration | |
| uhpT (mutation E448K) | fosfomycin | antibiotic target alteration | |
| pmrEFC and arnA (pmrL) | peptide antibiotic | antibiotic target alteration | |
| ampC | cephalosporin; penam | antibiotic inactivation | |
| kdpE, cpxA, baeRS, soxRS, gadX, gadW, evgSA, CRP, H-HS, mdtE, emrR, acrS, ptsI | /// | regulators | |
| pSUH1 | aadA5 | aminoglycoside antibiotic | antibiotic inactivation |
| blaTEM-1 | cephalosporin; monobactam; penam; penem | antibiotic inactivation | |
| dfrA17 | diaminopyrimidine antibiotic | antibiotic target replacement | |
| mphA, Mrx | macrolide antibiotic | antibiotic inactivation | |
| vgaC | streptogramin antibiotic; pleuromutilin antibiotic | antibiotic efflux | |
| sul1 | sulfonamide antibiotic | antibiotic target replacement | |
| pSUH2 | AAC(3)-IIc | aminoglycoside antibiotic | antibiotic inactivation |
| blaCTX-M-15 | cephalosporin | antibiotic inactivation | |
| blaOXA-1 | cephalosporin; penam | antibiotic inactivation | |
| AAC(6′)-Ib-cr | fluoroquinolone antibiotic; aminoglycoside antibiotic | antibiotic inactivation | |
| catB3 | phenicol antibiotic | antibiotic inactivation |
| Contig | Genes Detected | Function (PATRIC) |
|---|---|---|
| Chromosome | ecpRABCDE, | Adherence |
| csgBAC and csgDEFG | Adherence | |
| fimABCDEFGHI | Adherence, invasion, virulence | |
| papAIX, papCDFK | Adherence | |
| ppdD | Invasion, adhesion, biofilm, cell motility | |
| eptC | Invasion | |
| aslA | Invasion | |
| cusCFBA and cusRS | Invasion | |
| kpsCDEFSTM | Invasion, antiphagocytosis | |
| dsbAB | Virulence | |
| sitABCD | Virulence | |
| ompF | Virulence | |
| wecE | Virulence | |
| assT | Virulence | |
| trxA | Modulate host immune response |
| Toxin Family | Number | Cellular Process |
|---|---|---|
| COG2929/SplT | 1 | translation |
| Fic | 3 | translation |
| GNAT | 1 | translation |
| HipA | 2 | translation |
| MazF | 3 | translation |
| PIN | 2 | translation |
| RelE | 7 | translation |
| YeeV | 3 | cytoskeleton |
| MosT | 1 | protection of ICE |
| PemK | 2 | plasmid maintenance |
| unclear | 2 | ND * |
| null | 1 | ND * |
| Plasmid | Size (bp) | Replicon Types | Relaxase Type | MPF Type | OriT | Predicted Mobility |
|---|---|---|---|---|---|---|
| pSUH-1 | 161,372 | IncFIA, IncFIB | MOBF | MPFF | Yes | Conjugative |
| pSUH-2 | 79,945 | IncFIIA, IncFII | MOBF | MPFF | Yes | Conjugative |
| pSUH-3 | 5627 | ColRNAI rep cluster 1857 | MOBP | - | Yes | Mobilizable |
| pSUH-4 | 5166 | ColRNAI rep cluster 1291 | MOBQ | - | ND * | Mobilizable |
| pSUH-5 | 5057 | ColRNAI rep cluster 1857 | MOBP | - | Yes | Mobilizable |
| pSUH-6 | 1718 | - | - | - | Yes | Mobilizable |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaén-Luchoro, D.; Busquets, A.; Karlsson, R.; Salvà-Serra, F.; Åhrén, C.; Karami, N.; Moore, E.R.B. Genomic and Proteomic Characterization of the Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Strain CCUG 73778: A Virulent, Nosocomial Outbreak Strain. Microorganisms 2020, 8, 893. https://doi.org/10.3390/microorganisms8060893
Jaén-Luchoro D, Busquets A, Karlsson R, Salvà-Serra F, Åhrén C, Karami N, Moore ERB. Genomic and Proteomic Characterization of the Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Strain CCUG 73778: A Virulent, Nosocomial Outbreak Strain. Microorganisms. 2020; 8(6):893. https://doi.org/10.3390/microorganisms8060893
Chicago/Turabian StyleJaén-Luchoro, Daniel, Antonio Busquets, Roger Karlsson, Francisco Salvà-Serra, Christina Åhrén, Nahid Karami, and Edward R. B. Moore. 2020. "Genomic and Proteomic Characterization of the Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Strain CCUG 73778: A Virulent, Nosocomial Outbreak Strain" Microorganisms 8, no. 6: 893. https://doi.org/10.3390/microorganisms8060893
APA StyleJaén-Luchoro, D., Busquets, A., Karlsson, R., Salvà-Serra, F., Åhrén, C., Karami, N., & Moore, E. R. B. (2020). Genomic and Proteomic Characterization of the Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Strain CCUG 73778: A Virulent, Nosocomial Outbreak Strain. Microorganisms, 8(6), 893. https://doi.org/10.3390/microorganisms8060893






