Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake, Northern China
Abstract
1. Introduction
2. Materials and Methods
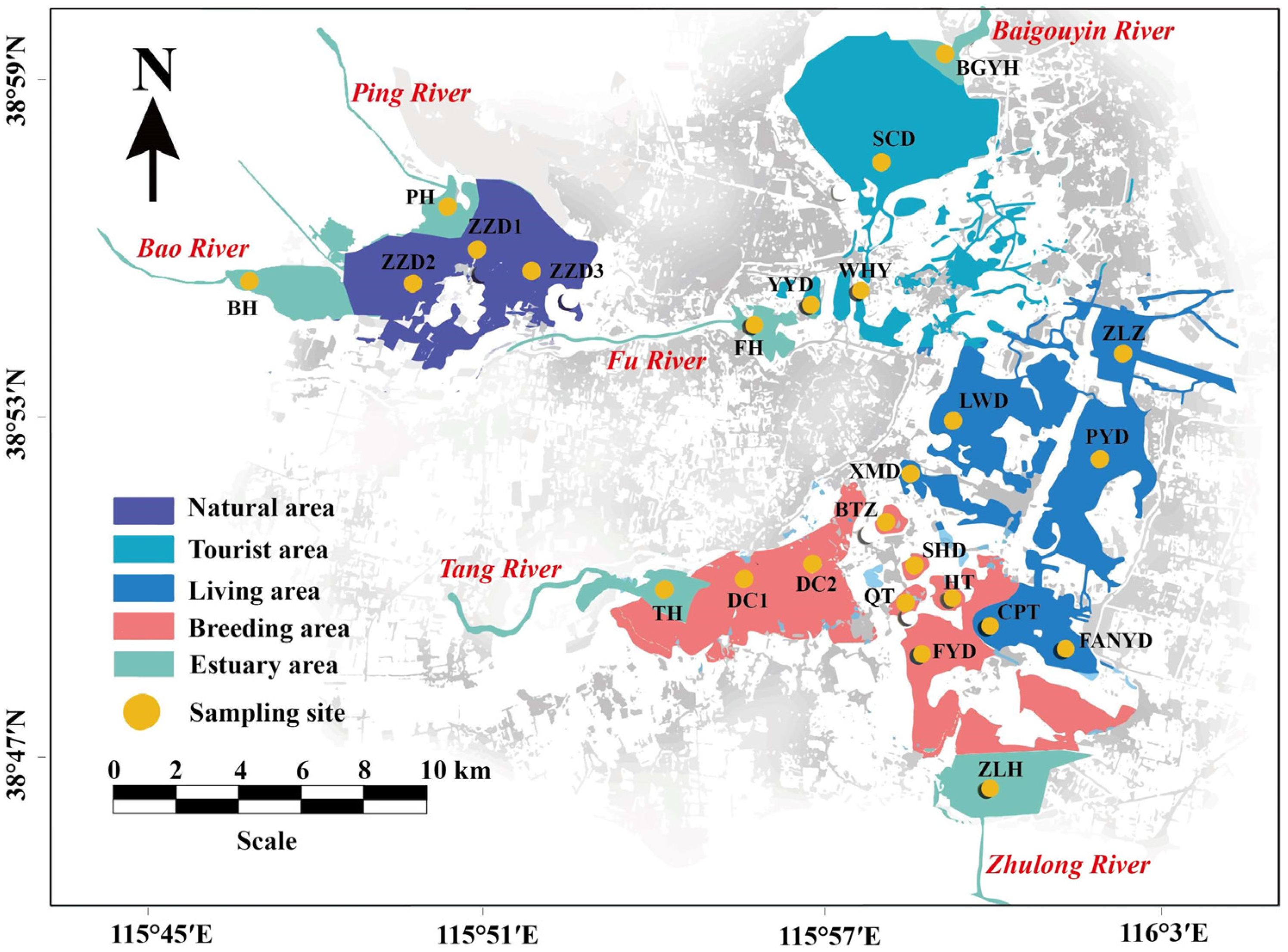
2.1. Research Area and Sample Collection
2.2. Measurement of Environmental Parameters
2.3. Spectral Characteristics of CDOM
2.4. DNA Extraction and PCR Amplification
2.5. Sequence Analysis
2.5.1. Water Bacterial Community Diversity
2.5.2. Key Environment Factor Analysis
2.5.3. Network Analysis
3. Results and Discussion
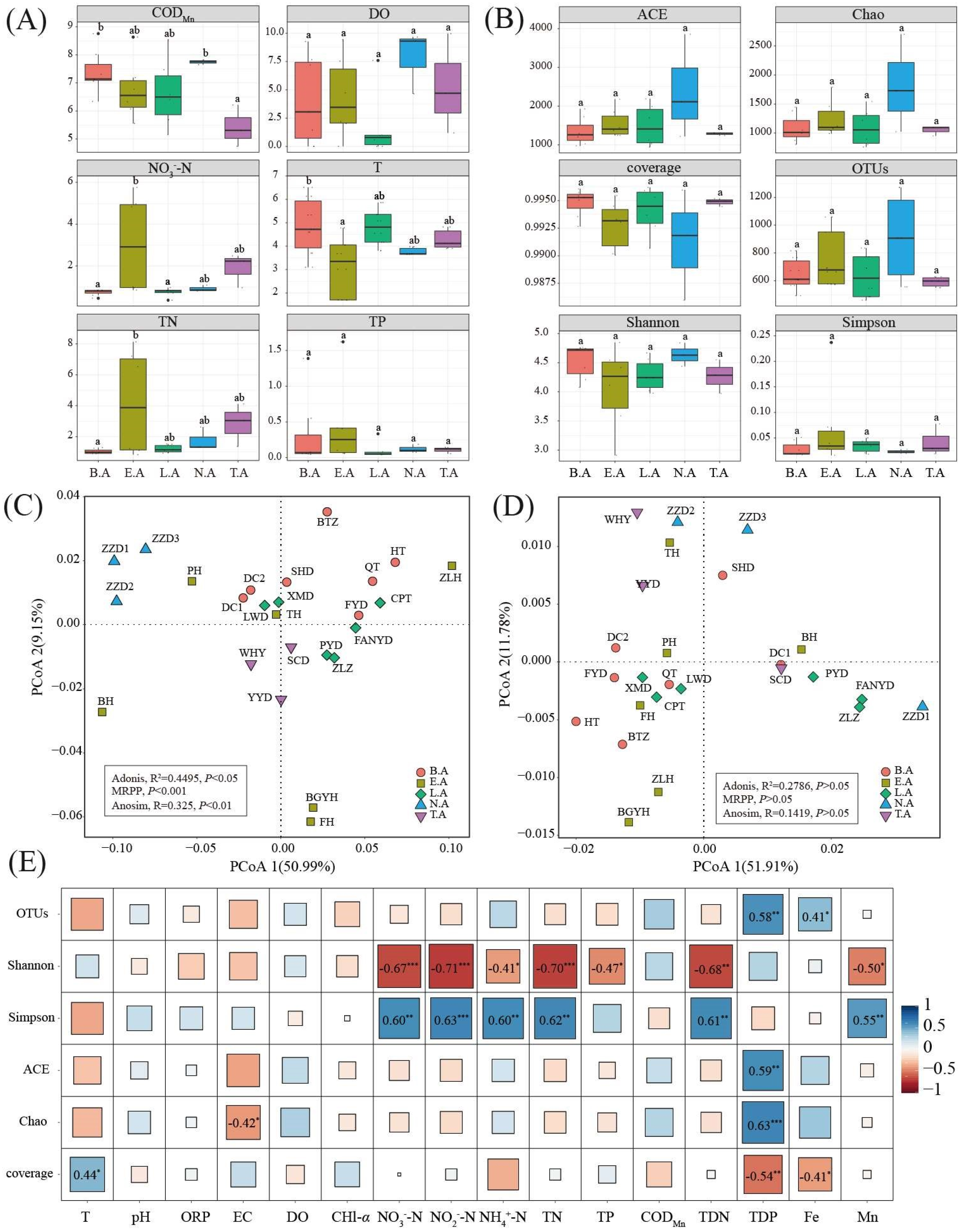
3.1. Spatial Differences in Environmental Parameters
3.2. Alpha Diversity of Bacterial Communities
3.3. Spatial Distribution of Bacterial Communities
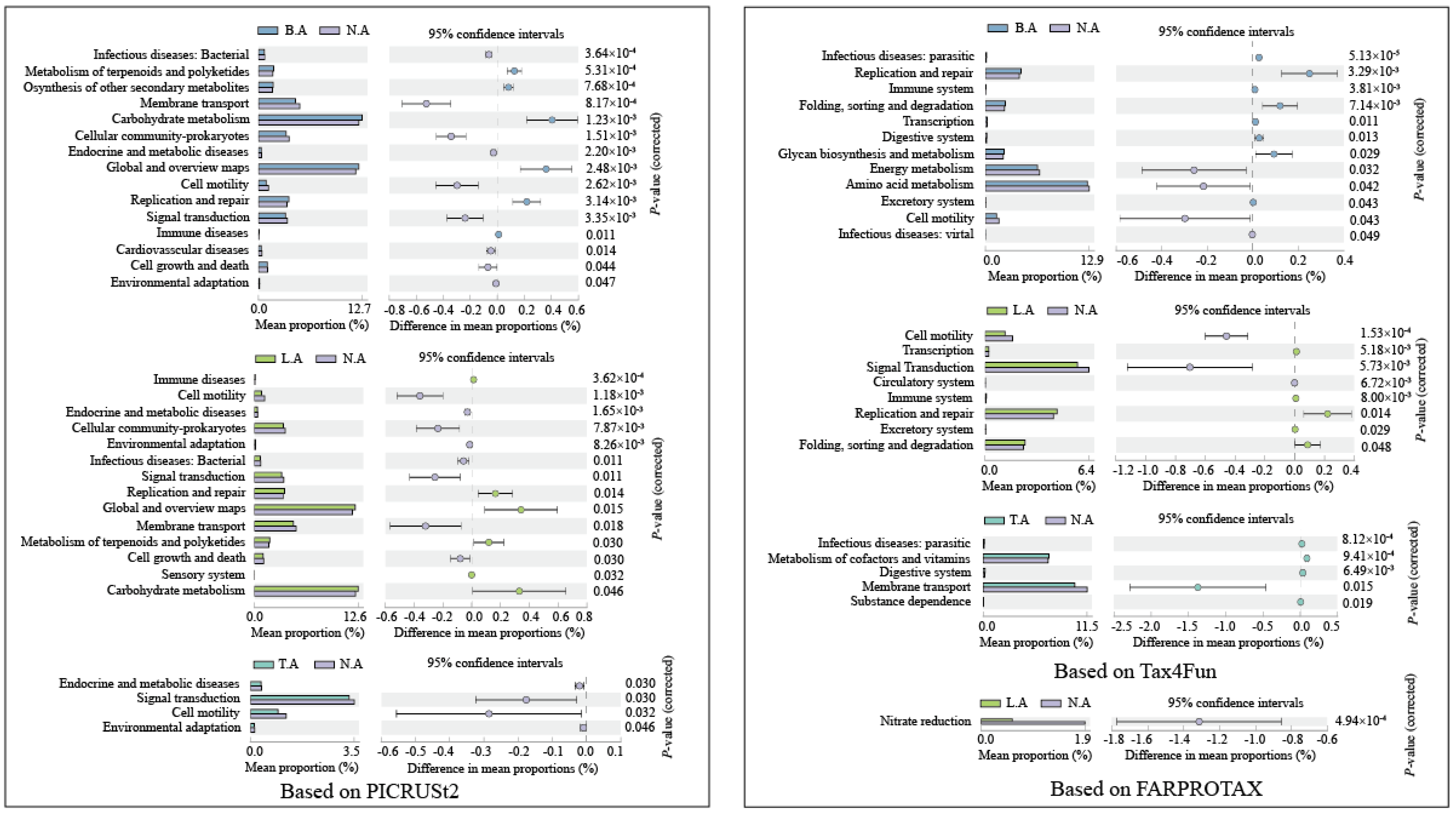
3.4. Comparison of Functional Properties
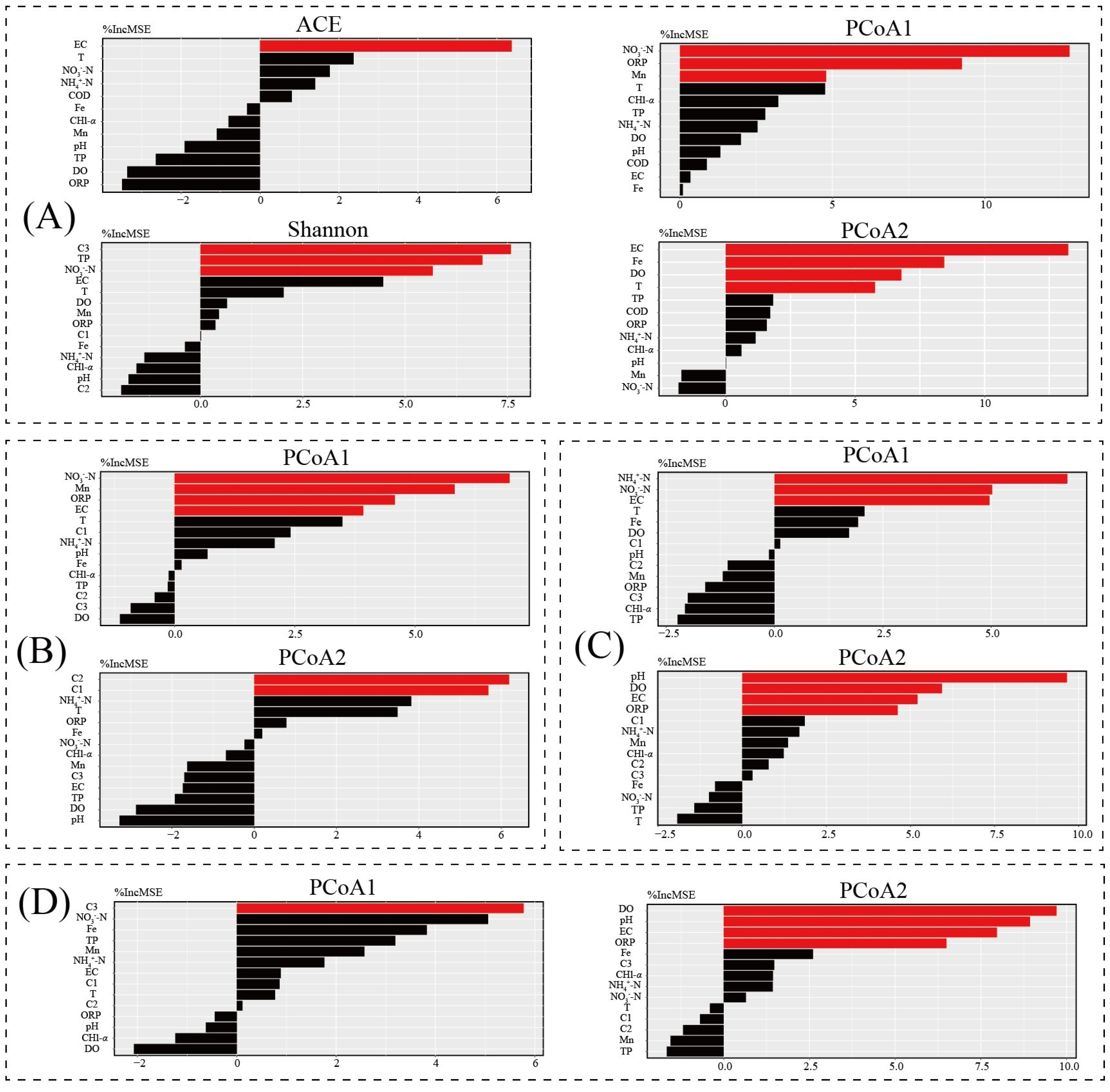
3.5. Key Environment Factor Analysis
3.6. CDOM Characteristics
3.6.1. EEM Spectroscopy Analysis
3.6.2. UV–Visible Absorption Spectroscopy Analysis
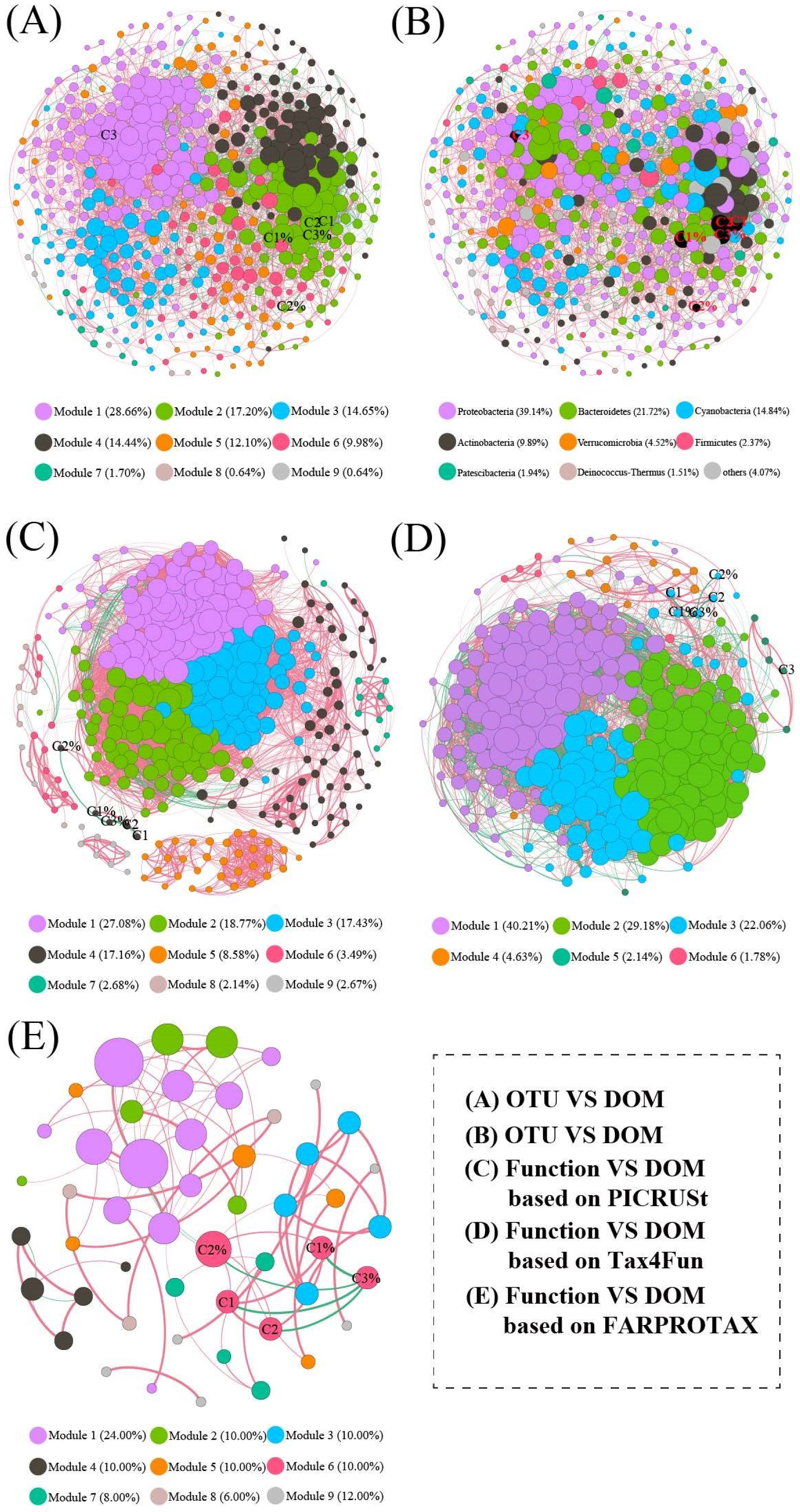
3.7. Co-Occurrence Network Analysis
3.8. Potential Drivers of the Water Bacterial Community and Its Functions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ávila, M.P.; Brandão, L.P.M.; Brighenti, L.S.; Tonetta, D.; Reis, M.P.; Stæhr, P.A.; Asmala, E.; Amado, A.M.; Barbosa, F.A.R.; Bezerra-Neto, J.F.; et al. Linking shifts in bacterial community with changes in dissolved organic matter pool in a tropical lake. Sci. Total Environ. 2019, 672, 990–1003. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, Y.; Jeppesen, E.; Wang, L.; Tan, H.; Zhang, J. Linking heterotrophic bacterioplankton community composition to the optical dynamics of dissolved organic matter in a large eutrophic Chinese lake. Sci. Total Environ. 2019, 679, 136–147. [Google Scholar] [CrossRef]
- Stedmon, C.A.; Markager, S. Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnol. Oceanogr. 2005, 50, 686–697. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, W.; Li, X.; Gao, G.; Jiang, J. Linking bacterial community shifts with changes in the dissolved organic matter pool in a eutrophic lake. Sci. Total Environ. 2020, 719, 137387. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; Yao, X.; Ding, W.; Zhang, Y.; Jeppesen, E.; Zhang, Y.; Podgorski, D.C.; Chen, C.; Ding, Y.; et al. Response of chromophoric dissolved organic matter dynamics to tidal oscillations and anthropogenic disturbances in a large subtropical estuary. Sci. Total Environ. 2019, 662, 769–778. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, K.; Guo, Z.; Li, X.; Chen, J.; Qi, Z.; Xu, S. Spatiotemporal variations of spectral characteristics of dissolved organic matter in river flowing into a key drinking water source in China. Sci. Total Environ. 2020, 700, 134360. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, Y.; Huang, T.; Liu, Y.; Fang, K.; Zhang, C. Microbial aerobic denitrification dominates nitrogen losses from reservoir ecosystem in the spring of Zhoucun reservoir. Sci. Total Environ. 2019, 651, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sun, Y.; Zhang, Y.; Huang, T.; Zhou, Z.; Li, Y.; Li, Z. Pollutant removal performance and microbial enhancement mechanism by water-lifting and aeration technology in a drinking water reservoir ecosystem. Sci. Total Environ. 2020, 709, 135848. [Google Scholar] [CrossRef]
- Wang, K.; Razzano, M.; Mou, X. Cyanobacterial blooms alter the relative importance of neutral and selective processes in assembling freshwater bacterioplankton community. Sci. Total Environ. 2020, 706, 135724. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, J.; Tong, T.; Li, B.; He, T.; Liu, Y.; Xie, S. Eutrophication influences methanotrophic activity, abundance and community structure in freshwater lakes. Sci. Total Environ. 2019, 662, 863–872. [Google Scholar] [CrossRef]
- Li, Y.; Xu, C.; Zhang, W.; Lin, L.; Wang, L.; Niu, L.; Zhang, H.; Wang, P.; Wang, C. Response of bacterial community in composition and function to the various DOM at river confluences in the urban area. Water Res. 2020, 169, 115293. [Google Scholar] [CrossRef]
- Han, Q.; Tong, R.; Sun, W.; Zhao, Y.; Yu, J.; Wang, G.; Shrestha, S.; Jin, Y. Anthropogenic influences on the water quality of the Baiyangdian Lake in North China over the last decade. Sci. Total Environ. 2020, 701, 134929. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, L.; Qin, S.; Cui, J.; Liu, Y. Quinolones antibiotics in the Baiyangdian Lake, China: Occurrence, distribution, predicted no-effect concentrations (PNECs) and ecological risks by three methods. Environ. Pollut. 2020, 256, 113458. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yan, J.; Wang, Y.; Zhang, B.-T.; Wang, H. Seasonal variation of aquatic macrophytes and its relationship with environmental factors in Baiyangdian Lake, China. Sci. Total Environ. 2020, 708, 135112. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence of antibiotics in water, sediments, aquatic plants, and animals from Baiyangdian Lake in North China. Chemosphere 2012, 89, 1307–1315. [Google Scholar] [CrossRef]
- Zhu, Y.; Jin, X.; Tang, W.; Meng, X.; Shan, B. Comprehensive analysis of nitrogen distributions and ammonia nitrogen release fluxes in the sediments of Baiyangdian Lake, China. J. Environ. Sci-China 2019, 76, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Chinese, N. Water and Wastewater Monitoring Methods; Chinese Environmental Science Publishing House: Beijing, China, 2002. (In Chinese) [Google Scholar]
- Huguet, A.; Vacher, L.; Relexans, S.; Saubusse, S.; Froidefond, J.-M.; Parlanti, E. Properties of fluorescent dissolved organic matter in the Gironde Estuary. Org. Geochem. 2009, 40, 706–719. [Google Scholar] [CrossRef]
- Ohno, T. Fluorescence inner-filtering correction for determining the humification index of dissolved organic matter. Environ. Sci. Technol. 2002, 36, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, M.; Qin, B.; Feng, S. Photochemical degradation of chromophoric-dissolved organic matter exposed to simulated UV-B and natural solar radiation. Hydrobiologia 2009, 627, 159–168. [Google Scholar] [CrossRef]
- Li, P.; Hur, J. Utilization of UV-Vis spectroscopy and related data analyses for dissolved organic matter (DOM) studies: A review. Crit. Rev. Environ. Sci. Technol. 2017, 47, 131–154. [Google Scholar] [CrossRef]
- Chen, Y.; Senesi, N.; Schnitzer, M. Information provided on humic substances by E4/E6 ratios 1. Soil Sci. Soc. Am. J. 1977, 41, 352–358. [Google Scholar] [CrossRef]
- Helms, J.R.; Stubbins, A.; Ritchie, J.D.; Minor, E.C.; Kieber, D.J.; Mopper, K. Absorption spectral slopes and slope ratios as indicators of molecular weight, source, and photobleaching of chromophoric dissolved organic matter. Limnol. Oceanogr. 2008, 53, 955–969. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, H.; Yang, J.R.; Liu, M.; Huang, B.; Yang, J. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 2018, 12, 2263. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef] [PubMed]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Zhou, S.; Huang, T.; Zhang, C.; Fang, K.; Xia, C.; Bai, S.; Zeng, M.; Qiu, X. Illumina MiSeq sequencing reveals the community composition of NirS-Type and NirK-Type denitrifiers in Zhoucun reservoir–a large shallow eutrophic reservoir in northern China. RSC Adv. 2016, 6, 91517–91528. [Google Scholar] [CrossRef]
- Zhou, S.; Huang, T.; Ngo, H.H.; Zhang, H.; Liu, F.; Zeng, M.; Shi, J.; Qiu, X. Nitrogen removal characteristics of indigenous aerobic denitrifiers and changes in the microbial community of a reservoir enclosure system via in situ oxygen enhancement using water lifting and aeration technology. Bioresour. Technol. 2016, 214, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Rui, J.; Li, J.; Dai, Y.; Bai, Y.; Heděnec, P.; Wang, J.; Zhang, S.; Pei, K.; Chi, L. Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol. Biochem. 2014, 79, 81–90. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.; Wang, J.; Du, N.; Li, Q.; Wei, G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Rohit, G.; Carolina Megumi, M.; Antonio, P.; Antonio, C.; Francisco, R.V. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol. Ecol. 2015, 23, 6073–6090. [Google Scholar]
- Hou, L.; Zhou, Q.; Wu, Q.; Gu, Q.; Sun, M.; Zhang, J. Spatiotemporal changes in bacterial community and microbial activity in a full-scale drinking water treatment plant. Sci. Total Environ. 2018, 625, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lu, W.; Liu, Y.; Ming, Z.; Liu, Y.; Meng, R.; Wang, H. Structure and diversity of bacterial communities in two large sanitary landfills in China as revealed by high-throughput sequencing (MiSeq). Waste Manag. 2017, 63, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-j.; Xi, B.-D.; Qiu, Z.-P.; He, X.-S.; Zhang, H.; Dang, Q.-L.; Zhao, X.-Y.; Li, D. Succession and diversity of microbial communities in landfills with depths and ages and its association with dissolved organic matter and heavy metals. Sci. Total Environ. 2019, 651, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Droppo, I.G.; Krishnappan, B.G.; Lawrence, J.R. Microbial interactions with naturally occurring hydrophobic sediments: Influence on sediment and associated contaminant mobility. Water Res. 2016, 92, 121–130. [Google Scholar] [CrossRef]
- Ou, D.; Li, H.; Li, W.; Wu, X.; Wang, Y.-q.; Liu, Y.-d. Salt-tolerance aerobic granular sludge: Formation and microbial community characteristics. Bioresour. Technol. 2018, 249, 132–138. [Google Scholar] [CrossRef]
- Reddy, B.; Pandey, J.; Dubey, S.K. Assessment of environmental gene tags linked with carbohydrate metabolism and chemolithotrophy associated microbial community in River Ganga. Gene 2019, 704, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Kube, M.; Teeling, H.; Richter, M.; Lombardot, T.; Allers, E.; Würdemann, C.A.; Quast, C.; Kuhl, H.; Knaust, F. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 2006, 8, 2201–2213. [Google Scholar] [CrossRef]
- Bittar, T.B.; Stubbins, A.; Vieira, A.A.H.; Mopper, K. Characterization and photodegradation of dissolved organic matter (DOM) from a tropical lake and its dominant primary producer, the cyanobacteria Microcystis aeruginosa. Mar. Chem. 2015, 177, 205–217. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, P.; Li, H.; Tian, Y.; Wang, S.; Song, Y.; Zeng, G.; Sun, C.; Tian, Z. Denitrification of landfill leachate under different hydraulic retention time in a two-stage anoxic/oxic combined membrane bioreactor process: Performances and bacterial community. Bioresour. Technol. 2018, 250, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, K.; Xia, Y.; Lau, F.T.; Tang, D.T.; Fung, W.C.; Fang, H.H.; Zhang, T. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl. Microbiol. Biot. 2014, 98, 5709–5718. [Google Scholar] [CrossRef] [PubMed]
- Rocker, D.; Brinkhoff, T.; Grüner, N.; Dogs, M.; Simon, M. Composition of humic acid-degrading estuarine and marine bacterial communities. FEMS Microbiol. Ecol. 2012, 80, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fu, W.; Yin, Y.; Chen, Z.; Qiu, R.; Simonnot, M.-O.; Wang, X. Adsorption-reduction removal of Cr(VI) by tobacco petiole pyrolytic biochar: Batch experiment, kinetic and mechanism studies. Bioresour. Technol. 2018, 268, 149–157. [Google Scholar] [CrossRef]
- Kong, X.-X.; Jiang, J.-L.; Qiao, B.; Liu, H.; Cheng, J.-S.; Yuan, Y.-J. The biodegradation of cefuroxime, cefotaxime and cefpirome by the synthetic consortium with probiotic Bacillus clausii and investigation of their potential biodegradation pathways. Sci. Total Environ. 2019, 651, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Consarnau, L.; Lindh, M.V.; Gasol, J.M.; Pinhassi, J. Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ. Microbiol. 2012, 14, 2361–2378. [Google Scholar] [CrossRef] [PubMed]
- Sarmento, H.; Morana, C.; Gasol, J.M. Bacterioplankton niche partitioning in the use of phytoplankton-derived dissolved organic carbon: Quantity is more important than quality. ISME J. 2016, 10, 2582–2592. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Identification of Novel Genes Involved in Long-Chain n-Alkane Degradation by Acinetobacter sp. Strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Eckert, E.M.; Salcher, M.M.; Posch, T.; Eugster, B.; Pernthaler, J. Rapid successions affect microbial N-acetyl-glucosamine uptake patterns during a lacustrine spring phytoplankton bloom. Environ. Microbiol. 2012, 14, 794–806. [Google Scholar] [CrossRef]
- Reichardt, W.; Gunn, B.; Colwell, R.R. Ecology and Taxonomy of Chitinoclastic Cytophaga and Related Chitin-Degrading Bacteria Isolated from an Estuary. Microb. Ecol. 1983, 9, 273–294. [Google Scholar] [CrossRef]
- Stewart, C.S.; Flint, H.J. Bacteroides (Fibrobacter) succinogenes, a cellulolytic anaerobic bacterium from the gastrointestinal tract. Appl. Microbiol. Biot. 1989, 30, 433–439. [Google Scholar] [CrossRef]
- Neis, E.P.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.R.; Stedmon, C.A.; Waite, T.D.; Ruiz, G.M. Distinguishing between terrestrial and autochthonous organic matter sources in marine environments using fluorescence spectroscopy. Mar. Chem. 2008, 108, 40–58. [Google Scholar] [CrossRef]
- Lavonen, E.; Kothawala, D.; Tranvik, L.; Gonsior, M.; Schmitt-Kopplin, P.; Köhler, S. Tracking changes in the optical properties and molecular composition of dissolved organic matter during drinking water production. Water Res. 2015, 85, 286–294. [Google Scholar] [CrossRef] [PubMed]

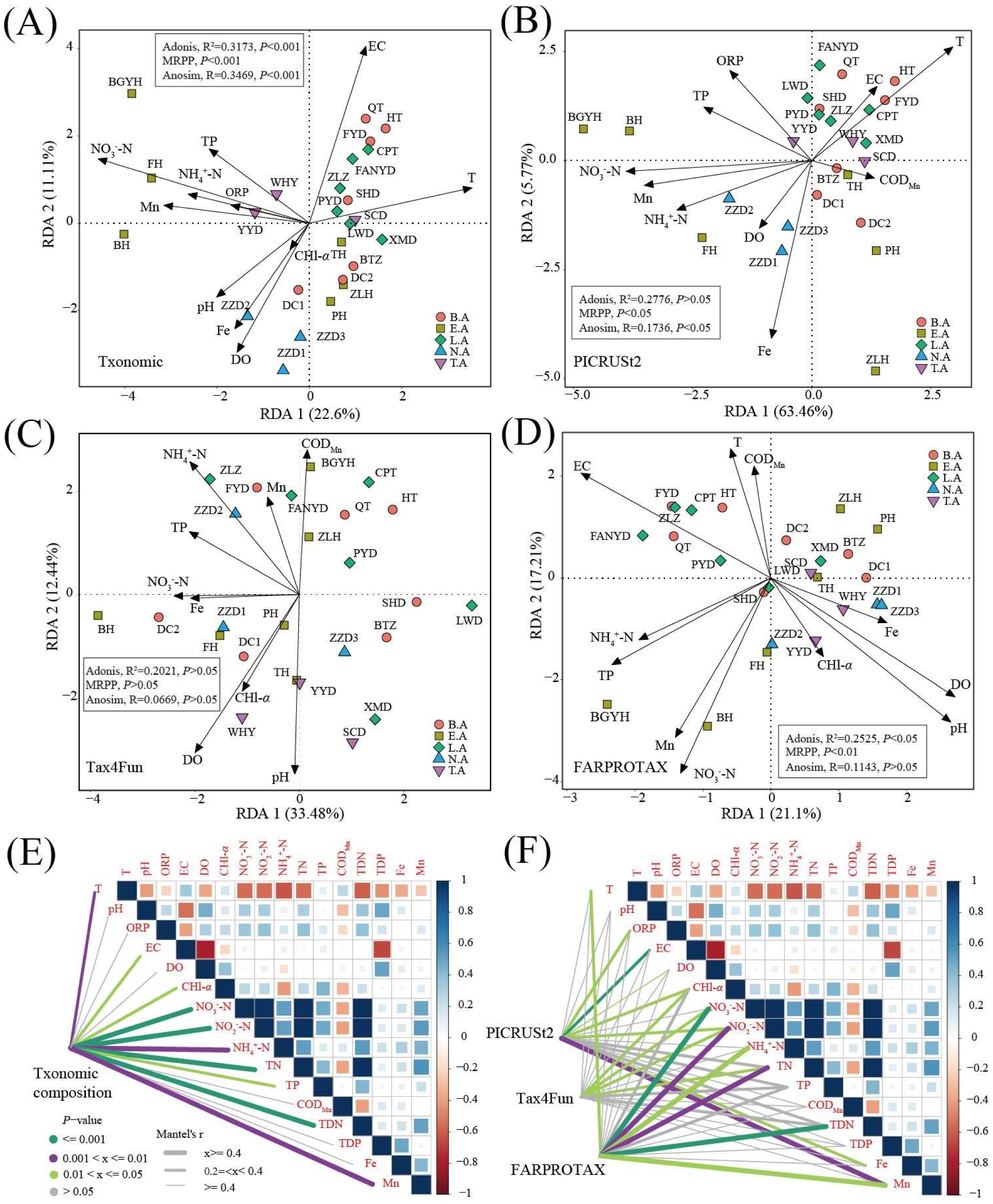

| VIF | RDA1 | RDA2 | R2 | p | ||
|---|---|---|---|---|---|---|
| Taxonomic composition (F = 2.156, p <0.001) | T | 9.66 | 0.98 | 0.21 | 0.49 | 0.003 |
| pH | 1.92 | −0.78 | −0.63 | 0.26 | 0.039 | |
| ORP | 2.09 | −0.97 | 0.23 | 0.12 | 0.227 | |
| EC | 4.2 | 0.29 | 0.96 | 0.65 | 0.001 | |
| DO | 6.61 | −0.48 | −0.88 | 0.41 | 0.004 | |
| CHl-α | 3.3 | −0.59 | −0.81 | 0.02 | 0.785 | |
| NO3−-N | 6.48 | −0.95 | 0.3 | 0.87 | 0.001 | |
| NH4+-N | 4.39 | −0.97 | 0.24 | 0.28 | 0.036 | |
| TP | 2.01 | −0.80 | 0.61 | 0.29 | 0.026 | |
| Fe | 1.68 | −0.57 | −0.82 | 0.31 | 0.019 | |
| Mn | 1.98 | −0.99 | 0.13 | 0.38 | 0.013 | |
| Functional composition based on PICRUSt2 (F = 3.798, p < 0.001) | T | 8.88 | 0.89 | 0.46 | 0.48 | 0.003 |
| ORP | 2.15 | −0.82 | 0.57 | 0.2 | 0.123 | |
| EC | 3.9 | 0.81 | 0.58 | 0.13 | 0.206 | |
| DO | 5.47 | −0.78 | −0.63 | 0.1 | 0.325 | |
| NO3−-N | 4.88 | −1.00 | −0.05 | 0.56 | 0.001 | |
| NH4+-N | 6.33 | −0.97 | −0.24 | 0.33 | 0.02 | |
| TP | 2.02 | −0.96 | 0.29 | 0.22 | 0.078 | |
| CODMn | 2.09 | 0.99 | −0.16 | 0.07 | 0.488 | |
| Fe | 1.68 | −0.36 | −0.93 | 0.39 | 0.008 | |
| Mn | 1.86 | −0.99 | −0.11 | 0.47 | 0.001 | |
| Functional composition based on Tax4Fun (F = 1.922, p < 0.05) | pH | 1.81 | −0.17 | −0.99 | 0.3 | 0.022 |
| DO | 1.76 | −0.58 | −0.81 | 0.37 | 0.007 | |
| CHl-α | 2.37 | −0.55 | −0.83 | 0.13 | 0.236 | |
| NO3−-N | 3.02 | −0.99 | −0.16 | 0.14 | 0.194 | |
| NH4+-N | 2.89 | −0.59 | 0.81 | 0.23 | 0.069 | |
| TP | 1.66 | −0.90 | 0.43 | 0.12 | 0.241 | |
| CODMn | 1.68 | 0.2 | 0.98 | 0.2 | 0.087 | |
| Fe | 1.35 | −0.98 | −0.19 | 0.11 | 0.298 | |
| Mn | 1.87 | −0.17 | 0.99 | 0.09 | 0.365 | |
| Functional composition based on FARPROTAX (F = 1.533, p < 0.05) | T | 10.94 | −0.31 | 0.95 | 0.22 | 0.068 |
| pH | 1.99 | 0.71 | −0.71 | 0.51 | 0.001 | |
| EC | 3.86 | −0.81 | 0.59 | 0.41 | 0.006 | |
| DO | 6.8 | 0.77 | −0.64 | 0.43 | 0.003 | |
| CHl-α | 3.73 | 0.51 | −0.86 | 0.1 | 0.314 | |
| NO3−-N | 4.69 | −0.28 | −0.96 | 0.48 | 0.003 | |
| NH4+-N | 5.12 | −0.88 | −0.48 | 0.15 | 0.159 | |
| TP | 1.95 | −0.84 | −0.54 | 0.25 | 0.043 | |
| CODMn | 2.37 | −0.19 | 0.98 | 0.15 | 0.146 | |
| Fe | 1.47 | 0.88 | −0.48 | 0.13 | 0.214 | |
| Mn | 1.9 | −0.38 | −0.93 | 0.34 | 0.018 |
| Components | Ex/Em | Description and Source Assignment | Reference |
|---|---|---|---|
| C1 | 275/325 | Protein-like substance | 275/330 (Li et al., 2020); 275/340 (Ziegmann et al., 2010) |
| C2 | 225/345 | Tryptophan-like DOM | 230/355 (Li et al., 2020); 230/330 (Stedmon et al., 2003) |
| C3 | 250/410 | Humic-like Substance (UVC) | 240/415 (Cory and Mcknight, 2005; Stedmon et al., 2003); 260(355)/434 (Murphy et al., 2008) |
| Empirical Network | Random Network | |||||||
|---|---|---|---|---|---|---|---|---|
| Type1 | Type2 | Type3 | Type4 | Type1 | Type2 | Type3 | Type4 | |
| Nodes | 471 | 373 | 281 | 50 | 471 | 373 | 281 | 50 |
| Edges | 3962 | 12470 | 7659 | 89 | 3962 | 12470 | 7659 | 89 |
| Modularity | 0.43 | 0.234 | 0.363 | 0.68 | 0.205 ± 0.004 | 0.205 ± 0.005 | 0.205 ± 0.005 | 0.205 ± 0.009 |
| Clustering coefficient | 0.38 | 0.69 | 0.64 | 0.61 | 0.036 ± 0.001 | 0.036 ± 0.005 | 0.036 ± 0.005 | 0.036 ± 0.001 |
| Network diameter | 5.72 | 5.63 | 3.91 | 6.01 | 4.000 ± 0.032 | 4.000 ± 0.063 | 4.000 ± 0.032 | 4.000 ± 0.063 |
| Average path length | 3.14 | 2.33 | 2.14 | 3.30 | 2.493 ± 0.001 | 2.493 ± 0.021 | 2.493 ± 0.021 | 2.494 ± 0.024 |
| Closeness centrality | 0.17 | 0.005 | 0.18 | 0.023 | 0.090 ± 0.011 | 0.090 ± 0.010 | 0.090 ± 0.010 | 0.090 ± 0.011 |
| Network density | 0.04 | 0.180 | 0.195 | 0.073 | 0.036 ± 0.000 | 0.036 ± 0.005 | 0.036 ± 0.005 | 0.036 ± 0.000 |
| Betweenness centrality | 0.03 | 0.04 | 0.019 | 0.16 | 0.007 ± 0.001 | 0.007 ± 0.001 | 0.007 ± 0.001 | 0.007 ± 0.005 |
| Degree centralization | 0.09 | 0.27 | 0.17 | 0.13 | 0.028 ± 0.004 | 0.029 ± 0.004 | 0.029 ± 0.004 | 0.028 ± 0.004 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, S.; Sun, Y.; Yu, M.; Shi, Z.; Zhang, H.; Peng, R.; Li, Z.; Cui, J.; Luo, X. Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake, Northern China. Microorganisms 2020, 8, 883. https://doi.org/10.3390/microorganisms8060883
Zhou S, Sun Y, Yu M, Shi Z, Zhang H, Peng R, Li Z, Cui J, Luo X. Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake, Northern China. Microorganisms. 2020; 8(6):883. https://doi.org/10.3390/microorganisms8060883
Chicago/Turabian StyleZhou, Shilei, Yue Sun, Minghui Yu, Zhenpeng Shi, Hang Zhang, Ruizhe Peng, Zaixing Li, Jiansheng Cui, and Xiao Luo. 2020. "Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake, Northern China" Microorganisms 8, no. 6: 883. https://doi.org/10.3390/microorganisms8060883
APA StyleZhou, S., Sun, Y., Yu, M., Shi, Z., Zhang, H., Peng, R., Li, Z., Cui, J., & Luo, X. (2020). Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake, Northern China. Microorganisms, 8(6), 883. https://doi.org/10.3390/microorganisms8060883






