Investigation of Intestinal Microbiota and Fecal Calprotectin in Non-Toxigenic and Toxigenic Clostridioides difficile Colonization and Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Microbiome Analysis
2.3. Calprotectin Measurement
2.4. Statistical Analysis
3. Results
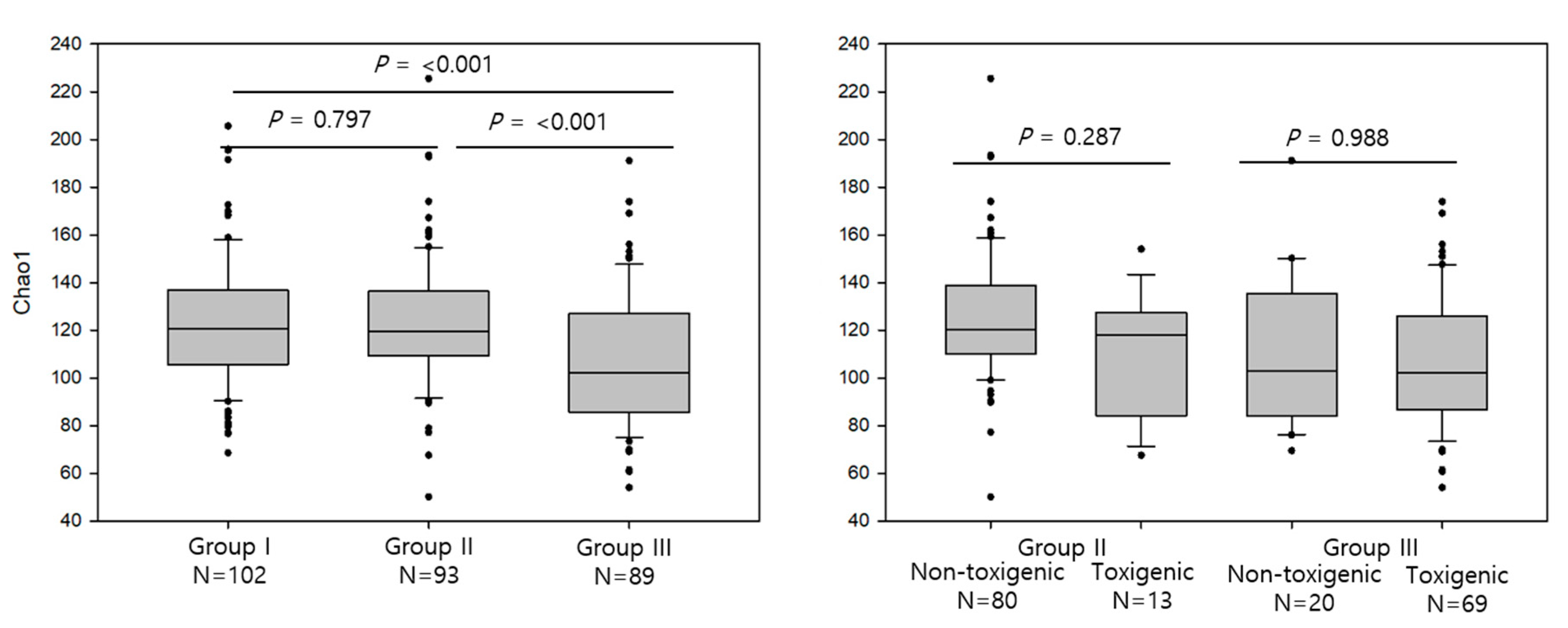
3.1. Comparison of Alpha Diversity of the Intestinal Microbiota Across Different Groups
3.2. Comparison of Beta Diversity of the Intestinal Microbiota Across Different Groups
3.3. Comparison of Mean Relative Abundance in Each Group at the Phylum Level
3.4. Comparison of Mean Relative Abundance in Each Group at the Genus Level
3.5. Comparison of Calprotectin Levels between Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lawson, P.A.; Citron, D.M.; Tyrrell, K.L.; Finegold, S.M. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 2016, 40, 95–99. [Google Scholar] [CrossRef]
- Balsells, E.; Shi, T.; Leese, C.; Lyell, I.; Burrows, J.; Wiuff, C.; Campbell, H.; Kyaw, M.H.; Nair, H. Global burden of Clostridium difficile infections: A systematic review and meta-analysis. J. Glob. Health 2019, 9, 1–20. [Google Scholar] [CrossRef]
- Bagdasarian, N.; Rao, K.; Malani, P.N. Diagnosis and treatment of Clostridium difficile in adults: A systematic review. JAMA 2015, 313, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.S.; Monaghan, T.M.; Wilcox, M.H. Clostridium difficile infection: Epidemiology, diagnosis and understanding transmission. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.B.; Buffie, C.G.; Carter, R.A.; Leiner, I.; Toussaint, N.C.; Miller, L.C.; Gobourne, A.; Ling, L.; Pamer, E.G. Loss of Microbiota-Mediated Colonization Resistance to Clostridium difficile Infection With Oral Vancomycin Compared With Metronidazole. J. Infect. Dis. 2015, 212, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Seekatz, A.M.; Young, V.B. Clostridium difficile and the microbiota. J. Clin. Investig. 2014, 124, 4182–4189. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Ticinesi, A.; Gerritsen, J.; Nouvenne, A.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Duranti, S.; Mangifesta, M.; Viappiani, A.; et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: A metagenomic study. Sci. Rep. 2016, 6, 25945. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Jiang, C.; Li, Z.; Wang, X.; Peng, Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 2015, 34, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef]
- Isaac, S.; Scher, J.U.; Djukovic, A.; Jimenez, N.; Littman, D.R.; Abramson, S.B.; Pamer, E.G.; Ubeda, C. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J. Antimicrob. Chemother. 2016. [Google Scholar] [CrossRef]
- Louie, T.J.; Byrne, B.; Emery, J.; Ward, L.; Krulicki, W.; Nguyen, D.; Wu, K.; Cannon, K. Differences of the Fecal Microflora with Clostridium difficile Therapies. Clin. Infect. Dis. 2015, 60 (Suppl. 2), S91–S97. [Google Scholar] [CrossRef]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef]
- Crobach, M.J.T.; Baktash, A.; Duszenko, N.; Kuijper, E.J. Diagnostic Guidance for C. difficile Infections. Adv. Exp. Med. Biol. 2018, 1050, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.T.; Vernon, J.J.; Loo, V.G.; Kong, L.Y.; Pechine, S.; Wilcox, M.H.; Kuijper, E.J. Understanding Clostridium difficile Colonization. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.; Oh, H.J.; Kim, H.S.; Hwang, Y.J.; Yong, D.; Jeong, S.H.; Lee, K. Fecal Calprotectin Level Reflects the Severity of Clostridium difficile Infection. Ann. Lab. Med. 2017, 37, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Barbut, F.; Gouot, C.; Lapidus, N.; Suzon, L.; Syed-Zaidi, R.; Lalande, V.; Eckert, C. Faecal lactoferrin and calprotectin in patients with Clostridium difficile infection: A case-control study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.; Planche, T.; Eckert, C.; Barbut, F.; Terveer, E.M.; Dekkers, O.M.; Wilcox, M.H.; Kuijper, E.J. European Society of Clinical Microbiology and Infectious Diseases: Update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2016, 22 (Suppl. 4), S63–S81. [Google Scholar] [CrossRef]
- Cohen, S.H.; Gerding, D.N.; Johnson, S.; Kelly, C.P.; Loo, V.G.; McDonald, L.C.; Pepin, J.; Wilcox, M.H. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect. Control. Hosp. Epidemiol. 2010, 31, 431–455. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Barbut, F.; Day, N.; Bouee, S.; Youssouf, A.; Grandvoinnet, L.; Lalande, V.; Couturier, J.; Eckert, C. Toxigenic Clostridium difficile carriage in general practice: Results of a laboratory-based cohort study. Clin. Microbiol. Infect. 2019, 25, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, E.; Smollan, G.; Huppert, A.; Fluss, R.; Tal, I.; Gilboa, M.; Zilberman-Daniels, T.; Keller, N.; Rahav, G.; Regev-Yochay, G.; et al. Universal screening for Clostridioides difficile in a tertiary hospital: Risk factors for carriage and clinical disease. Clin. Microbiol. Infect. 2019, 25, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Kim, H.; Moon, H.W.; Hur, M.; Yun, Y.M. Toxin positivity and tcdB gene load in broad-spectrum Clostridium difficile infection. Infection 2018, 46, 113–117. [Google Scholar] [CrossRef]
- Han, S.H.; Yi, J.; Kim, J.H.; Lee, S.; Moon, H.W. Composition of gut microbiota in patients with toxigenic Clostridioides (Clostridium) difficile: Comparison between subgroups according to clinical criteria and toxin gene load. PLoS ONE 2019, 14, e0212626. [Google Scholar] [CrossRef]
- Moon, H.W.; Kim, H.N.; Hur, M.; Shim, H.S.; Kim, H.; Yun, Y.M. Comparison of Diagnostic Algorithms for Detecting Toxigenic Clostridium difficile in Routine Practice at a Tertiary Referral Hospital in Korea. PLoS ONE 2016, 11, e0161139. [Google Scholar] [CrossRef]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef]
- Miquel, S.; Martin, R.; Rossi, O.; Bermudez-Humaran, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Theriot, C.M.; Young, V.B. Interactions between the Gastrointestinal Microbiome and Clostridium difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef]
- Stecher, B.; Maier, L.; Hardt, W.D. ‘Blooming’ in the gut: How dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013, 11, 277–284. [Google Scholar] [CrossRef]
- Vincent, C.; Miller, M.A.; Edens, T.J.; Mehrotra, S.; Dewar, K.; Manges, A.R. Bloom and bust: Intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.T.; Ducarmon, Q.R.; Terveer, E.M.; Harmanus, C.; Sanders, I.; Verduin, K.M.; Kuijper, E.J.; Zwittink, R.D. The Bacterial Gut Microbiota of Adult Patients Infected, Colonized or Noncolonized by Clostridioides difficile. Microorganisms 2020, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Steinbakk, M.; Naess-Andresen, C.F.; Lingaas, E.; Dale, I.; Brandtzaeg, P.; Fagerhol, M.K. Antimicrobial actions of calcium binding leucocyte L1 protein, calprotectin. Lancet 1990, 336, 763–765. [Google Scholar] [CrossRef]
- Caccaro, R.; D’Inca, R.; Sturniolo, G.C. Clinical utility of calprotectin and lactoferrin as markers of inflammation in patients with inflammatory bowel disease. Expert Rev. Clin. Immunol. 2010, 6, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Swale, A.; Miyajima, F.; Roberts, P.; Hall, A.; Little, M.; Beadsworth, M.B.; Beeching, N.J.; Kolamunnage-Dona, R.; Parry, C.M.; Pirmohamed, M. Calprotectin and lactoferrin faecal levels in patients with Clostridium difficile infection (CDI): A prospective cohort study. PLoS ONE 2014, 9, e106118. [Google Scholar] [CrossRef]
- Manceau, H.; Chicha-Cattoir, V.; Puy, H.; Peoc’h, K. Fecal calprotectin in inflammatory bowel diseases: Update and perspectives. Clin. Chem. Lab. Med. 2017, 55, 474–483. [Google Scholar] [CrossRef]
- Sipponen, T.; Kolho, K.L. Fecal calprotectin in diagnosis and clinical assessment of inflammatory bowel disease. Scand. J. Gastroenterol. 2014, 50, 74–80. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Di Rienzi, S.C.; Poole, A.C.; Koren, O.; Walters, W.A.; Caporaso, J.G.; Knight, R.; Ley, R.E. Conducting a microbiome study. Cell 2014, 158, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63. [Google Scholar] [CrossRef]
- Werner, J.J.; Koren, O.; Hugenholtz, P.; DeSantis, T.Z.; Walters, W.A.; Caporaso, J.G.; Angenent, L.T.; Knight, R.; Ley, R.E. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012, 6, 94–103. [Google Scholar] [CrossRef]


| Groups | Source | Number | Sex (M/F) | Age, Years (Median, IQR) |
|---|---|---|---|---|
| Group I | General examination | 102 | 52/50 | 57.0, 46.8–63.3 |
| C. difficile- non colonized | ||||
| Group II | General examination | 93 | 61/32 | 54.0, 47.0–63.0 |
| C. difficile-colonized | ||||
| Group III | Hospitalized patients | 89 | 49/40 | 67.0, 49–79 * |
| Diarrhea with C. difficile |
| Phylum | Group I | Group II | Group III | p | ||
|---|---|---|---|---|---|---|
| (n = 102) | (n = 93) | (n = 89) | I vs. II | I vs. III | II vs. III | |
| Bacteroidetes | 43.85 | 43.22 | 32.41 | 0.911 | <0.001 | 0.001 |
| Firmicutes | 33.55 | 29.25 | 32.45 | 0.065 | 0.924 | 0.542 |
| Proteobacteria | 20.91 | 25.75 | 32.67 | 0.096 | <0.001 | 0.077 |
| Actinobacteria | 0.98 | 0.81 | 1.1 | 0.564 | 0.952 | 0.75 |
| Fusobacteria | 0.64 | 0.89 | 0.97 | 0.753 | 0.724 | 0.987 |
| Phylum | Group II | p | Group III | p | ||
|---|---|---|---|---|---|---|
| Non-Toxigenic | Toxigenic | Non-Toxigenic | Toxigenic | |||
| (n = 80) | (n = 13) | (n = 20) | (n = 69) | |||
| Bacteroidetes | 42.85 | 45.46 | 0.491 | 32.21 | 32.47 | 0.968 |
| Firmicutes | 31.15 | 17.57 | <0.001 | 31.62 | 32.69 | 0.865 |
| Proteobacteria | 24.63 | 32.67 | 0.137 | 34.73 | 32.08 | 0.668 |
| Acinetobacter | 0.87 | 0.44 | 0.151 | 0.48 | 1.28 | 0.371 |
| Fusobacteria | 0.42 | 3.83 | 0.059 | 0.62 | 1.08 | 0.624 |
| Genus | Group I | Group II | Group III | p | ||
|---|---|---|---|---|---|---|
| (n = 102) | (n = 93) | (n = 89) | I vs. II | I vs. III | II vs. III | |
| Bacteroides | 26.68 | 26.44 | 23.48 | 0.993 | 0.448 | 0.543 |
| Enterobacteriaceae * | 11.42 | 16.59 | 22.1 | 0.036 | <0.001 | 0.067 |
| Prevotella | 10.09 | 8.94 | 1.36 | 0.756 | <0.001 | <0.001 |
| Ruminococcaceae * | 8.08 | 6.13 | 1.68 | 0.022 | <0.001 | <0.001 |
| Lachnospiraceae * | 7.16 | 6.06 | 3.49 | 0.118 | <0.001 | 0.001 |
| Sutterella | 3.13 | 1.97 | 1.38 | 0.012 | 0.002 | 0.441 |
| Rikenellaceae * | 1.95 | 1.58 | 1.06 | 0.35 | 0.013 | 0.215 |
| Lachnospira | 1.84 | 1.36 | 0.27 | 0.055 | <0.001 | <0.001 |
| Blautia | 1.62 | 1.15 | 0.5 | 0.003 | <0.001 | <0.001 |
| Ruminococcus | 1.61 | 0.79 | 0.14 | 0.002 | <0.001 | <0.001 |
| Phascolarctobacterium | 1.54 | 1.76 | 0.45 | 0.87 | <0.001 | 0.008 |
| Coprococcus | 1.25 | 1.05 | 0.2 | 0.514 | <0.001 | 0.514 |
| Parabacteroides | 1.42 | 1.92 | 4.23 | 0.215 | 0.001 | 0.011 |
| Serratia | 1.34 | 1.5 | 3.79 | 0.842 | <0.001 | <0.001 |
| Faecalibacterium | 1.25 | 0.88 | 0.38 | 0.001 | <0.001 | 0.002 |
| Oscillospira | 1.08 | 0.98 | 1.41 | 0.574 | 0.562 | 0.379 |
| Bilophila | 1.08 | 0.69 | 0.3 | 0.027 | <0.001 | 0.024 |
| Veillonella | 0.3 | 0.45 | 2.25 | 0.51 | 0.003 | 0.007 |
| Succinivibrio | 0.3 | 1.12 | 0 | 0.248 | 0.341 | 0.05 |
| Lactobacillus | 0.16 | 0.46 | 1.41 | 0.33 | 0.113 | 0.185 |
| Clostridium | 0.11 | 0.13 | 1.34 | 0.835 | 0.264 | 0.276 |
| Eubacterium | 0.08 | 0.21 | 1.41 | 0.822 | 0.291 | 0.239 |
| Enterococcus | 0.03 | 0.46 | 9.13 | 0.542 | <0.001 | <0.001 |
| Genus | Group II | p | Group III | p | ||
|---|---|---|---|---|---|---|
| Non-Toxigenic | Toxigenic | Non-Toxigenic | Toxigenic | |||
| (n = 80) | (n = 13) | (n = 20) | (n = 69) | |||
| Bacteroides | 25.03 | 35.11 | 0.035 | 19.23 | 24.71 | 0.312 |
| Enterobacteriaceae * | 15.26 | 24.75 | 0.044 | 25.77 | 21.03 | 0.368 |
| Prevotella | 9.83 | 3.47 | 0.014 | 0.7 | 1.55 | 0.445 |
| Ruminococcaceae * | 6.85 | 1.73 | <0.001 | 2.62 | 1.4 | 0.22 |
| Lachnospiraceae * | 6.36 | 4.24 | 0.073 | 2.65 | 3.73 | 0.423 |
| Sutterella | 2.1 | 1.19 | 0.05 | 0.74 | 1.56 | 0.407 |
| Phascolarctobacterium | 2.02 | 0.21 | <0.001 | 0.54 | 0.46 | 0.713 |
| Rikenellaceae * | 1.67 | 0.98 | 0.204 | 1.37 | 0.97 | 0.501 |
| Parabacteroides | 1.66 | 3.52 | 0.265 | 6.93 | 3.45 | 0.166 |
| Lachnospira | 1.43 | 0.94 | 0.25 | 0.49 | 0.2 | 0.395 |
| Serratia | 1.41 | 2.08 | 0.32 | 3.67 | 3.81 | 0.902 |
| Succinivibrio | 1.3 | 0 | 0.019 | 0 | 0 | 0.33 |
| Ruminococcus | 1.3 | 1.59 | 0.127 | 0.16 | 0.14 | 0.753 |
| Blautia | 1.26 | 0.45 | 0.006 | 0.3 | 0.56 | 0.297 |
| Coprococcus | 1.12 | 0.63 | 0.199 | 0.12 | 0.22 | 0.462 |
| Oscillospira | 0.91 | 1.42 | 0.158 | 1.53 | 1.38 | 0.84 |
| Pseudomonas | 0.43 | 1.57 | 0.161 | 0.71 | 0.6 | 0.865 |
| Fusobacteriim | 0.4 | 3.66 | 0.062 | 0.56 | 0.82 | 0.702 |
| Veillonella | 0.35 | 1.1 | 0.102 | 0.39 | 2.79 | 0.002 |
| Enterococcus | 0.5 | 0.19 | 0.793 | 15.23 | 7.36 | 0.278 |
| Acinetobacter | 0.3 | 0.04 | 0.662 | 1.17 | 0.21 | 0.306 |
| Lactobacillus | 0.52 | 0.05 | 0.051 | 0.13 | 3.54 | 0.043 |
| Clostridium | 0.12 | 0.16 | 0.636 | 0.05 | 1.71 | 0.378 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.-H.; Yi, J.; Kim, J.-H.; Moon, H.-W. Investigation of Intestinal Microbiota and Fecal Calprotectin in Non-Toxigenic and Toxigenic Clostridioides difficile Colonization and Infection. Microorganisms 2020, 8, 882. https://doi.org/10.3390/microorganisms8060882
Han S-H, Yi J, Kim J-H, Moon H-W. Investigation of Intestinal Microbiota and Fecal Calprotectin in Non-Toxigenic and Toxigenic Clostridioides difficile Colonization and Infection. Microorganisms. 2020; 8(6):882. https://doi.org/10.3390/microorganisms8060882
Chicago/Turabian StyleHan, Sung-Hee, Joowon Yi, Ji-Hoon Kim, and Hee-Won Moon. 2020. "Investigation of Intestinal Microbiota and Fecal Calprotectin in Non-Toxigenic and Toxigenic Clostridioides difficile Colonization and Infection" Microorganisms 8, no. 6: 882. https://doi.org/10.3390/microorganisms8060882
APA StyleHan, S.-H., Yi, J., Kim, J.-H., & Moon, H.-W. (2020). Investigation of Intestinal Microbiota and Fecal Calprotectin in Non-Toxigenic and Toxigenic Clostridioides difficile Colonization and Infection. Microorganisms, 8(6), 882. https://doi.org/10.3390/microorganisms8060882




