Phage Display Detection of Mimotopes that Are Shared Epitopes of Clinically and Epidemiologically Relevant Enterobacteria
Abstract
1. Introduction
2. Materials and Methods
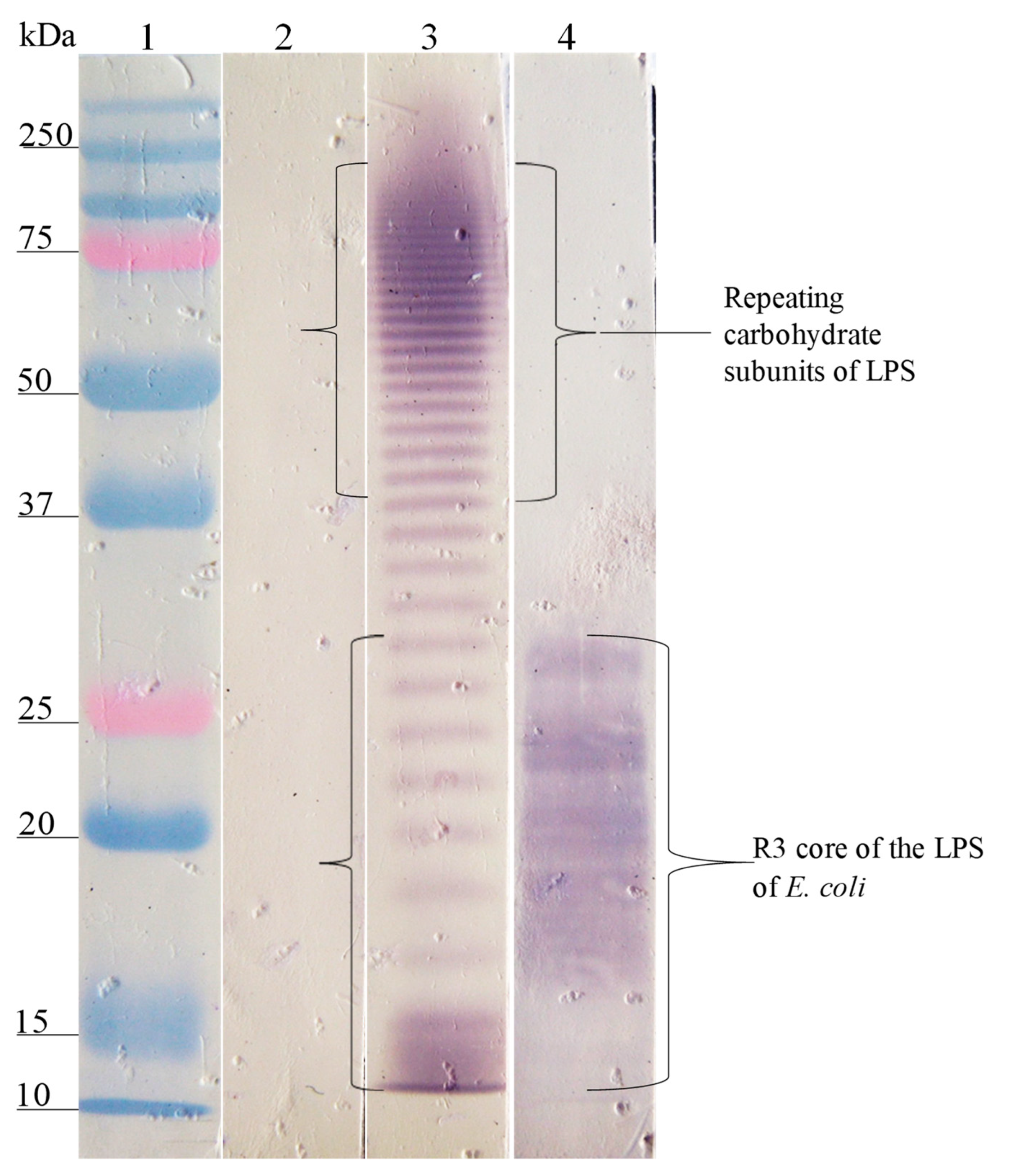
2.1. Lipopolysaccharides
2.2. Anti-Sera Production Against LPS
2.3. Purification of the IgG Antibodies
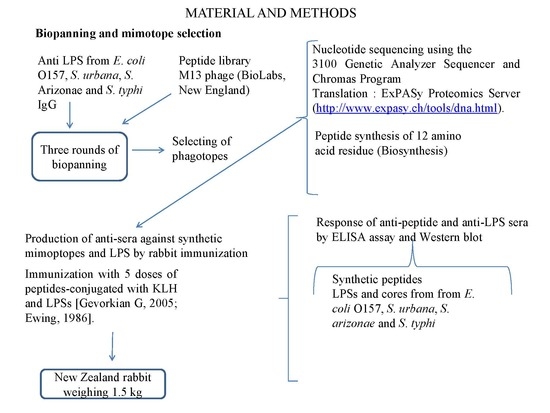
2.4. Biopanning and Mimotope Selection
2.5. Mimotope Synthesis
2.6. Production of Anti-Sera Against Synthetic Mimoptopes by Rabbit Immunization
2.7. Evaluation of Antibody Reactivity Against LPS and Synthetic Mimotope Using ELISA
2.8. Human Serum Analysis Against LPS and Synthetic Mimotopes Using ELISA
2.9. Interaction Site of Synthetic Antipeptide Sera
2.10. Statistical Analysis
3. Results
3.1. Mimotopes Selection
3.2. Anti-Peptide Sera Response
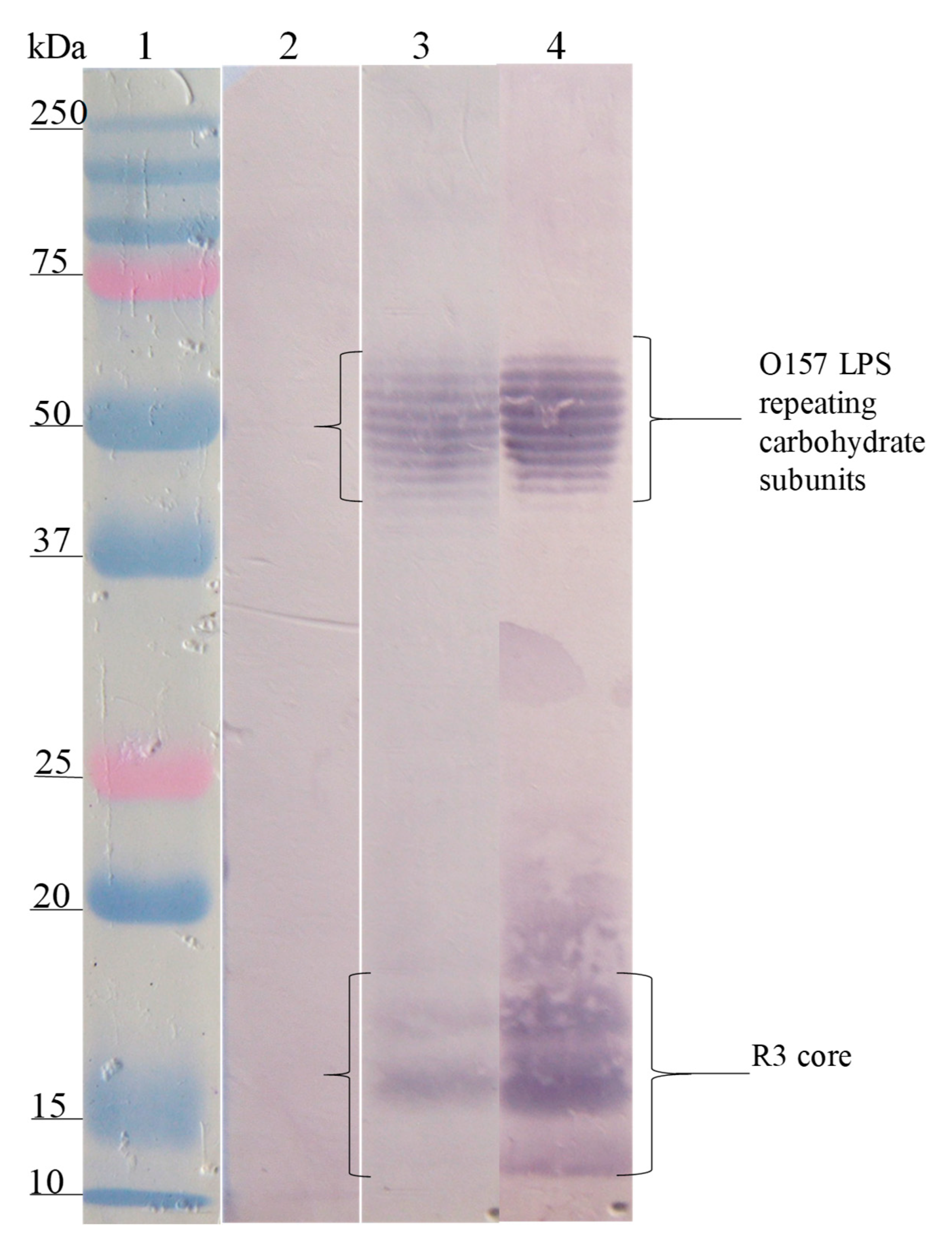
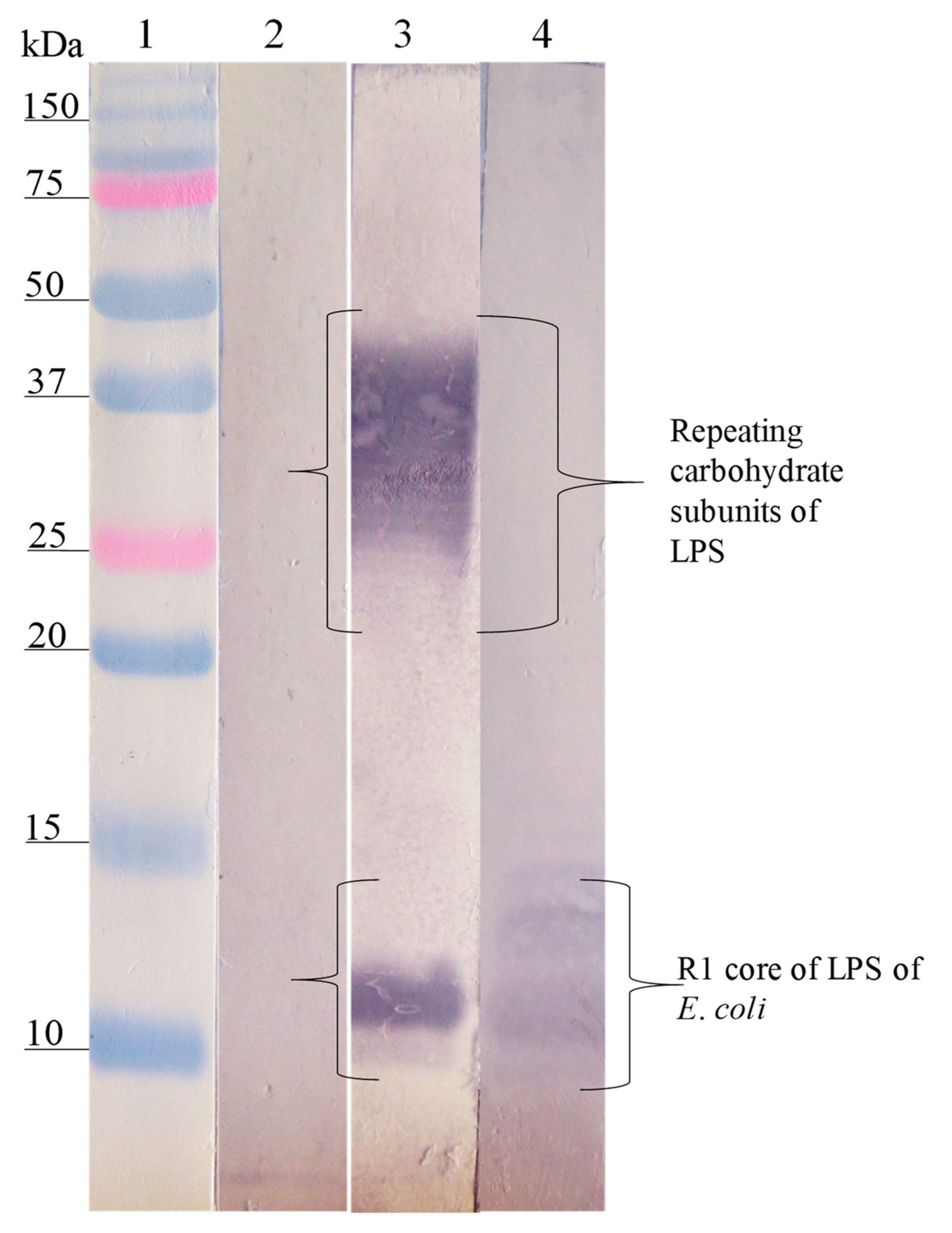
3.3. Interaction Site of Anti-Peptide Antibodies on the LPSs
3.4. Sera Response of General Population Against Synthetic Peptides and LPSs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bryce, J.; Boschi-Pinto, C.; Shibuya, K.; Black, R.E. WHO estimates of the causes of death in children. Lancet 2005, 365, 1147–1152. [Google Scholar] [CrossRef]
- GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.M.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z.; et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef]
- Anderson, J.D., IV; Bagamian, K.H.; Muhib, F.; Amaya, M.P.; Laytner, L.A.; Wierzba, T.; Rheingans, R. Burden of enterotoxigenic Escherichia coli and Shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: A modelling analysis. Lancet Glob. Health 2019, 7, e321–e330. [Google Scholar] [CrossRef]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Beutler, B.; Rietschel, E.T. Innate immune sensing and its roots: The story of endotoxin. Nat. Rev. Immunol. 2003, 3, 169–176. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef]
- Smith, G.P.; Scott, J.K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993, 217, 228–257. [Google Scholar] [CrossRef]
- Meyer, T.; Schirrmann, T.; Frenzel, A.; Miethe, S.; Stratmann-Selke, J.; Gerlach, G.F.; Strutzberg-Minder, K.; Dübel, S.; Hust, M. Identification of immunogenic proteins and generation of antibodies against Salmonella Typhimurium using phage display. BMC Biotechnol. 2012, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, S.; Hossany, R.B.; Pinto, B.M. Immunological Evidence for Functional Rather than Structural Mimicry by a Shigella flexneri Y Polysaccharide-Mimetic Peptide. Clin. Vaccine Immunol. 2008, 15, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Microbiol. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Crim, S.M.; Iwamoto, M.; Huang, J.Y.; Griffin, P.M.; Gilliss, D.; Cronquist, A.B.; Cartter, M.; Tobin-D’Angelo, M.; Blythe, D.; Smith, K. Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2006–2013. Morb. Mortal Wkly. Rep. 2014, 63, 328–332. [Google Scholar]
- DGE-SINAVE. Salud/Sistema de Notificación Semanal de casos nuevos de enfermedades. Boletín Epiedemiológico 2019, 36, 68. [Google Scholar]
- Gutiérrez-Cogco, L.; Montiel-Vázquez, E.; Aguilera-Pérez, P.; González-Andrade, M.D.C. Serotipos de Salmonella identificados en los servicios de salud de México. Salud Pública Méx. 2000, 42, 490–495. [Google Scholar] [CrossRef]
- Callaway, T.R.; Carr, M.A.; Edrington, T.S.; Anderson, R.C.; Nisbet, D.J. Diet, Escherichia coli O157:H7, and cattle: A review after 10 years. Curr. Issues Mol. Biol. 2009, 11, 67–79. [Google Scholar]
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086. [Google Scholar] [CrossRef]
- Noftall, K.; Taylor, M.; Hoang, L.; Galanis, E. Outbreaks: Shiga toxin–producing Escherichia coli in British Columbia, 2011–2017: Analysis to inform exclusion guidelines. Can. Commun. Dis. Rep. 2019, 45, 238–243. [Google Scholar] [CrossRef]
- Nielsen, E.M.; Scheutz, F. Characterisation of Escherichia coli O157 isolates from Danish cattle and human patients by genotyping and presence and variants of virulence genes. Vet. Microbiol. 2002, 88, 259–273. [Google Scholar] [CrossRef]
- Lopez, E.L.; Diaz, M.; Grinstein, S.; Devoto, S.; Mendilaharzu, F.; Murray, B.E.; Ashkenazi, S.; Rubeglio, E.; Woloj, M.; Vasquez, M.; et al. Hemolytic Uremic Syndrome and Diarrhea in Argentine Children: The Role of Shiga-like Toxins. J. Infect. Dis. 1989, 160, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Eslava, C.; de la Torre, G.G.; Leon, L.A.; Licona, D.; Leon, L.; Zarco, L.A.; Cravioto, A. Common epitopes in LPS of different Enterobacteriaceae are associated with an immune response against Escherichia coli O157 in bovine serum samples. J. Med. Microbiol. 2007, 56, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Hernández-Chiñas, U.; Licona-Moreno, D.; Zenteno, E.; Cravioto, A.; Eslava-Campos, C.A. Immunogenic peptide mimotopes from an epitope of Escherichia coli O157 LPS. Biochem. J. 2016, 473, 3791–3804. [Google Scholar] [CrossRef] [PubMed]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharide: Extraction with phenol-water and further applications of the procedure. Meth. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of Core Oligosaccharide Types in Lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- NOM NOM-062-ZOO-1999; ETP LA PRODUCCION. Norma Oficial Mexicana NOM-062-ZOO-1999: Especificaciones técnicas para la producción, cuidado y uso de los animales de laboratorio. D. Of. Fed. México 1999. [Google Scholar]
- Ewing, W.H. Edwards and Ewing’s Identification of Enterobacteriaceae; Elsevier Science Publishing Co., Inc.: New York, NY, USA, 1986; 536p. [Google Scholar]
- Ulises, H.-C.; Tatiana, G.; Karlen, G.; Guillermo, M.-H.; Juan, X.-C.; Carlos, E. Peptide sequences identified by phage display are immunodominant functional motifs of Pet and Pic serine proteases secreted by Escherichia coli and Shigella flexneri. Peptides 2009, 30, 2127–2135. [Google Scholar] [CrossRef]
- Wilson, R.K. High-throughput purification of M13 templates for DNA sequencing. Biotechniques 1993, 15, 414–416, 418–420, 422. [Google Scholar]
- Galfrè, G.; Monaci, P.; Nicosia, A.; Luzzago, A.; Felici, F.; Cortese, R. Immunization with phage-displayed mimotopes. In Methods in Enzymology; Elsevier; Academic Press: New York, NY, USA, 1996; Volume 267, pp. 109–115. [Google Scholar]
- Navarro, A.; Eslava, C.; Hernandez, U.; Navarro-Henze, J.L.; Aviles, M.; Garcia-de la Torre, G.; Cravioto, A. Antibody responses to Escherichia coli O157 and other lipopolysaccharides in healthy children and adults. Clin. Diagn. Lab. Immunol. 2003, 10, 797–801. [Google Scholar] [CrossRef][Green Version]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.N. Peptide-displaying phage technology in glycobiology. Glycobiology 2011, 22, 318–325. [Google Scholar] [CrossRef]
- Tsang, R.S.W.; Nielsen, K.; Diane Henning, M.; Schlecht, S.; Aleksić, S. A Murine Monoclonal Antibody that Recognizes a Genus-Specific Epitope in the Salmonella Lipopolysaccharid Outer Core. Zbl. Bakt. 1991, 274, 446–455. [Google Scholar] [CrossRef]
- Mansfield, L.P.; Forsythe, S.J. Demonstration of the Rb1 lipopolysaccharide core structure in Salmonella strains with the monoclonal antibody M105. J. Med. Microbiol. 2001, 50, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Kaniuk, N.; Frirdich, E. Molecular insights into the assembly and diversity of the outer core oligosaccharide in lipopolysaccharides from Escherichia coli and Salmonella. J. Endotoxin Res. 2003, 9, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, J.R.; da Silva, R.F.; Neale, W.A.; Smallcombe, C.C.; Clark, H.W.; Mackay, R.-M.A.; Watson, A.S.; Madsen, J.; Hood, D.W.; Burns, I.; et al. Structural definition of hSP-D recognition of Salmonella enterica LPS inner core oligosaccharides reveals alternative binding modes for the same LPS. PLoS ONE 2018, 13, e0199175. [Google Scholar] [CrossRef]
- MacArthur, M.W.; Thornton, J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991, 218, 397–412. [Google Scholar] [CrossRef]
- Harris, S.L.; Craig, L.; Mehroke, J.S.; Rashed, M.; Zwick, M.B.; Kenar, K.; Toone, E.J.; Greenspan, N.; Auzanneau, F.-I.; Marino-Albernas, J.-R.; et al. Exploring the basis of peptide–carbohydrate crossreactivity: Evidence for discrimination by peptides between closely related anti-carbohydrate antibodies. Proc. Natl. Acad. Sci. USA 1997, 94, 2454–2459. [Google Scholar] [CrossRef]
- Dharmasena, M.N.; Jewell, D.A.; Taylor, R.K. Development of peptide mimics of a protective epitope of Vibrio cholerae Ogawa O-antigen and investigation of the structural basis of peptide mimicry. J. Biol. Chem. 2007, 282, 33805–33816. [Google Scholar] [CrossRef]
- Borrelli, S.; Hossany, R.B.; Findlay, S.; Pinto, B.M. Group A Streptococcus Polysaccharide-Mimetic Peptide. Am. J. Immuna. 2006, 2, 77–87. [Google Scholar]
- James, J.A.; Gross, T.; Scofield, R.H.; Harley, J.B. Immunoglobulin epitope spreading and autoimmune disease after peptide immunization: Sm B/B′-derived PPPGMRPP and PPPGIRGP induce spliceosome autoimmunity. J. Exp. Med. 1995, 181, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-S.; Yu, J.; Lin, J.; Zhong, L.; Bren, K.L.; Nahm, M.H. Peptide mimotopes of pneumococcal capsular polysaccharide of 6B serotype: A peptide mimotope can bind to two unrelated antibodies. J. Immunol. 2002, 168, 6273–6278. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K.R.; Loganathan, D.; Goldstein, I.J.; Schultz, P.; Gallop, M.A. Peptide ligands for a sugar-binding protein isolated from a random peptide library. Proc. Natl. Acad. Sci. USA 1992, 89, 5393–5397. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Passo, C.L.; Scuderi, A.; Kolberg, J.; Baxendale, H.; Goldblatt, D.; Oggioni, M.R.; Felici, F.; Andrew, P.W. Peptide mimics of two pneumococcal capsular polysaccharide serotypes (6B and 9V) protect mice from a lethal challenge with Streptococcus pneumoniae. Eur. J. Immunol. 2009, 39, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.J.; Sharma, S.; Kumar, G.; Visweswariah, S.S.; Surolia, A. Biopanning of endotoxin-specific phage displayed peptides. Biochem. Biophys. Res. Commun. 2003, 307, 133–138. [Google Scholar] [CrossRef]
- Westerink, M.A.; Giardina, P.C.; Apicella, M.A.; Kieber-Emmons, T. Peptide mimicry of the meningococcal group C capsular polysaccharide. Proc. Natl. Acad. Sci. USA 1995, 92, 4021–4025. [Google Scholar] [CrossRef]
- Agostino, M.; Sandrin, M.S.; Thompson, P.E.; Farrugia, W.; Ramsland, P.A.; Yuriev, E. Carbohydrate-mimetic peptides: Structural aspects of mimicry and therapeutic implications. Expert Opin. Biol. Ther. 2011, 11, 211–224. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Q.; Sales, D.; Bianco, A.E.; Craig, A. Vaccination with peptide mimotopes produces antibodies recognizing bacterial capsular polysaccharides. Vaccine 2010, 28, 6425–6435. [Google Scholar] [CrossRef]
- Heinrichs, D.E.; Yethon, J.A.; Whitfield, C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998, 30, 221–232. [Google Scholar] [CrossRef]
- Jansson, P.-E.; Lindberg, B.; Lindberg, A.A.; Wollin, R. Structural Studies on the Hexose Region of the Core in Lipopolysaccharides from Enterobacteriaceae. Eur. J. Biochem. 1981, 115, 571–577. [Google Scholar] [CrossRef] [PubMed]
 ); E. coli O157, S. urbana, S. arizonae, and S. typhi LPSs (
); E. coli O157, S. urbana, S. arizonae, and S. typhi LPSs ( ); and the Ra core (
); and the Ra core ( ) was evaluated by the ELISA test, as was previously mentioned in the Methods section. The pre-immune sera (
) was evaluated by the ELISA test, as was previously mentioned in the Methods section. The pre-immune sera ( ) response is the average of the obtained results in the different assays.
) response is the average of the obtained results in the different assays.
 ); E. coli O157, S. urbana, S. arizonae, and S. typhi LPSs (
); E. coli O157, S. urbana, S. arizonae, and S. typhi LPSs ( ); and the Ra core (
); and the Ra core ( ) was evaluated by the ELISA test, as was previously mentioned in the Methods section. The pre-immune sera (
) was evaluated by the ELISA test, as was previously mentioned in the Methods section. The pre-immune sera ( ) response is the average of the obtained results in the different assays.
) response is the average of the obtained results in the different assays.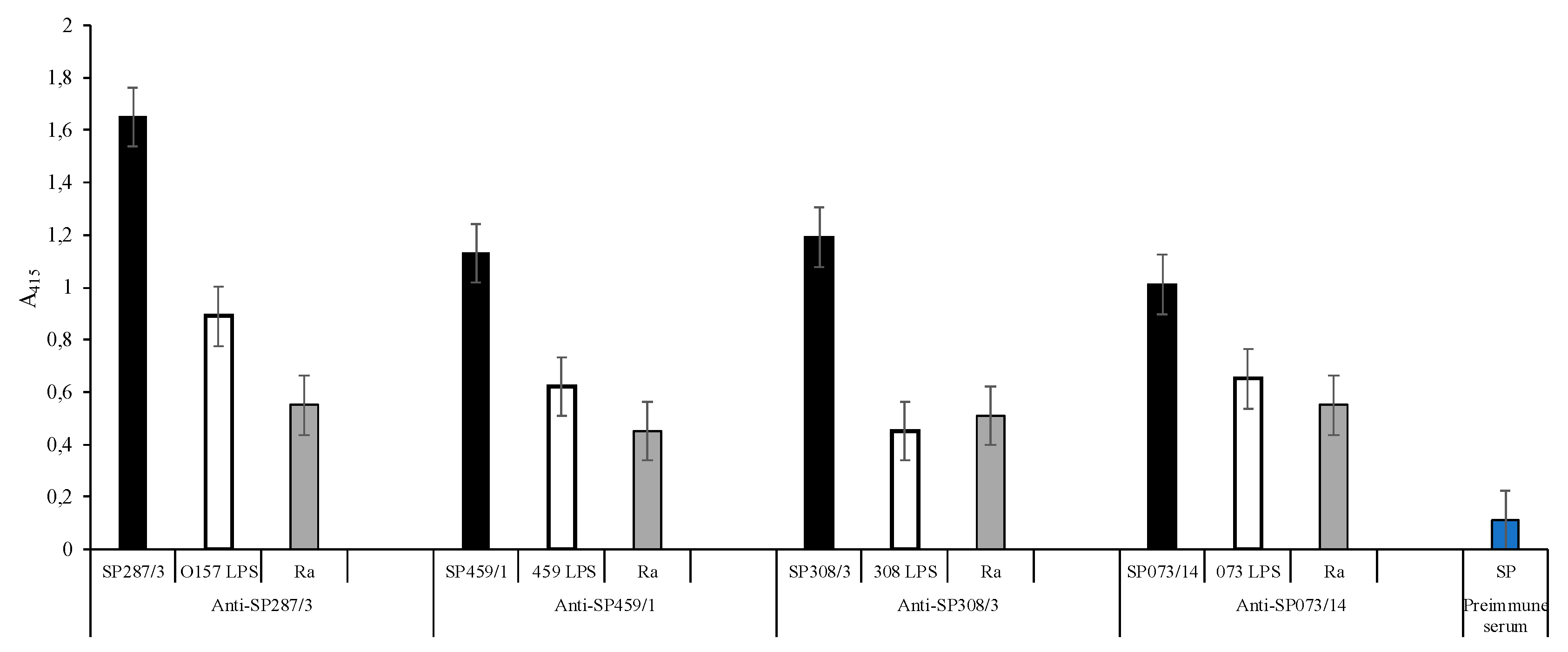

 ) of E. coli O157 (FMU287), S. urbana (FMU459), S. arizonae (FMU308), and S. typhi (FMU073), as well as the synthetic peptides (
) of E. coli O157 (FMU287), S. urbana (FMU459), S. arizonae (FMU308), and S. typhi (FMU073), as well as the synthetic peptides ( ) SP287/3, SP459/1, SP308/3, and SP073/14 were analyzed independently in an ELISA test assay against community serum samples, as described previously in the Methods section.
) SP287/3, SP459/1, SP308/3, and SP073/14 were analyzed independently in an ELISA test assay against community serum samples, as described previously in the Methods section.
 ) of E. coli O157 (FMU287), S. urbana (FMU459), S. arizonae (FMU308), and S. typhi (FMU073), as well as the synthetic peptides (
) of E. coli O157 (FMU287), S. urbana (FMU459), S. arizonae (FMU308), and S. typhi (FMU073), as well as the synthetic peptides ( ) SP287/3, SP459/1, SP308/3, and SP073/14 were analyzed independently in an ELISA test assay against community serum samples, as described previously in the Methods section.
) SP287/3, SP459/1, SP308/3, and SP073/14 were analyzed independently in an ELISA test assay against community serum samples, as described previously in the Methods section.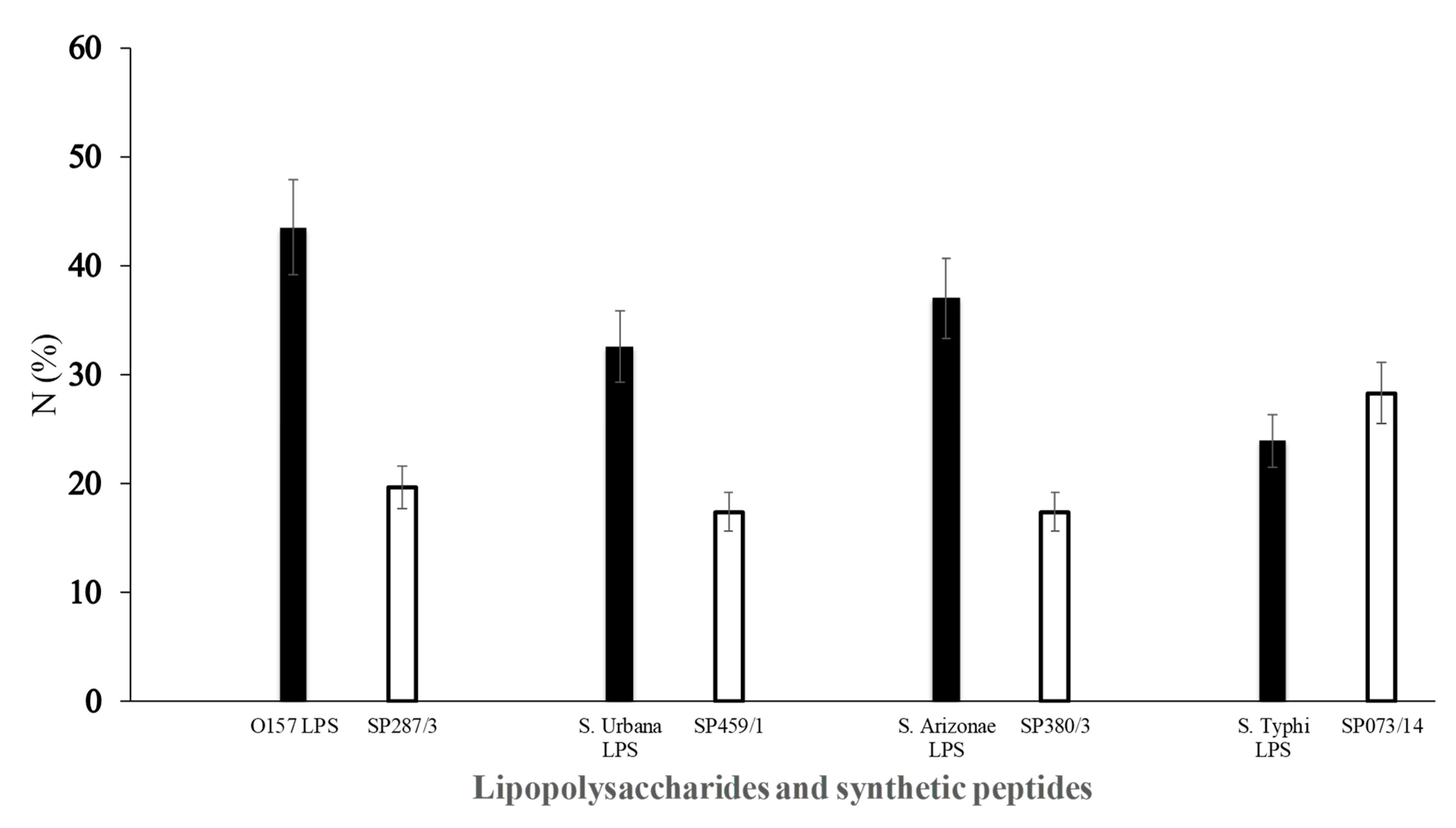
| Phage Clones | Amino Acid Analysis and Sequence * | Molecular Weight | Recognition by IgG anti-LPS 1 (A405 Mean) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SP287/3 | S | T | L | N | Y | M | Y | X | A | H | P | F | 1430.6 | 1.64 | |||||||||
| SP073/14 | I | S | L | S | N | I | V | D | S | Q | T | P | 1273.4 | 1.10 | |||||||||
| SP459/1 | G | F | S | V | I | T | G | A | A | M | F | E | 1229.4 | 1.4 | |||||||||
| SP308/3 | H | N | P | F | T | F | F | G | P | M | F | Y | 1504.7 | 0.63 | |||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro, A.; Licona-Moreno, D.; Monsalvo-Reyes, A.; Hernández-Chiñas, U.; Eslava-Campos, C.A. Phage Display Detection of Mimotopes that Are Shared Epitopes of Clinically and Epidemiologically Relevant Enterobacteria. Microorganisms 2020, 8, 780. https://doi.org/10.3390/microorganisms8050780
Navarro A, Licona-Moreno D, Monsalvo-Reyes A, Hernández-Chiñas U, Eslava-Campos CA. Phage Display Detection of Mimotopes that Are Shared Epitopes of Clinically and Epidemiologically Relevant Enterobacteria. Microorganisms. 2020; 8(5):780. https://doi.org/10.3390/microorganisms8050780
Chicago/Turabian StyleNavarro, Armando, Delia Licona-Moreno, Alejandro Monsalvo-Reyes, Ulises Hernández-Chiñas, and Carlos A. Eslava-Campos. 2020. "Phage Display Detection of Mimotopes that Are Shared Epitopes of Clinically and Epidemiologically Relevant Enterobacteria" Microorganisms 8, no. 5: 780. https://doi.org/10.3390/microorganisms8050780
APA StyleNavarro, A., Licona-Moreno, D., Monsalvo-Reyes, A., Hernández-Chiñas, U., & Eslava-Campos, C. A. (2020). Phage Display Detection of Mimotopes that Are Shared Epitopes of Clinically and Epidemiologically Relevant Enterobacteria. Microorganisms, 8(5), 780. https://doi.org/10.3390/microorganisms8050780