Quantifying Plant-Borne Carbon Assimilation by Root-Associating Bacteria
Abstract
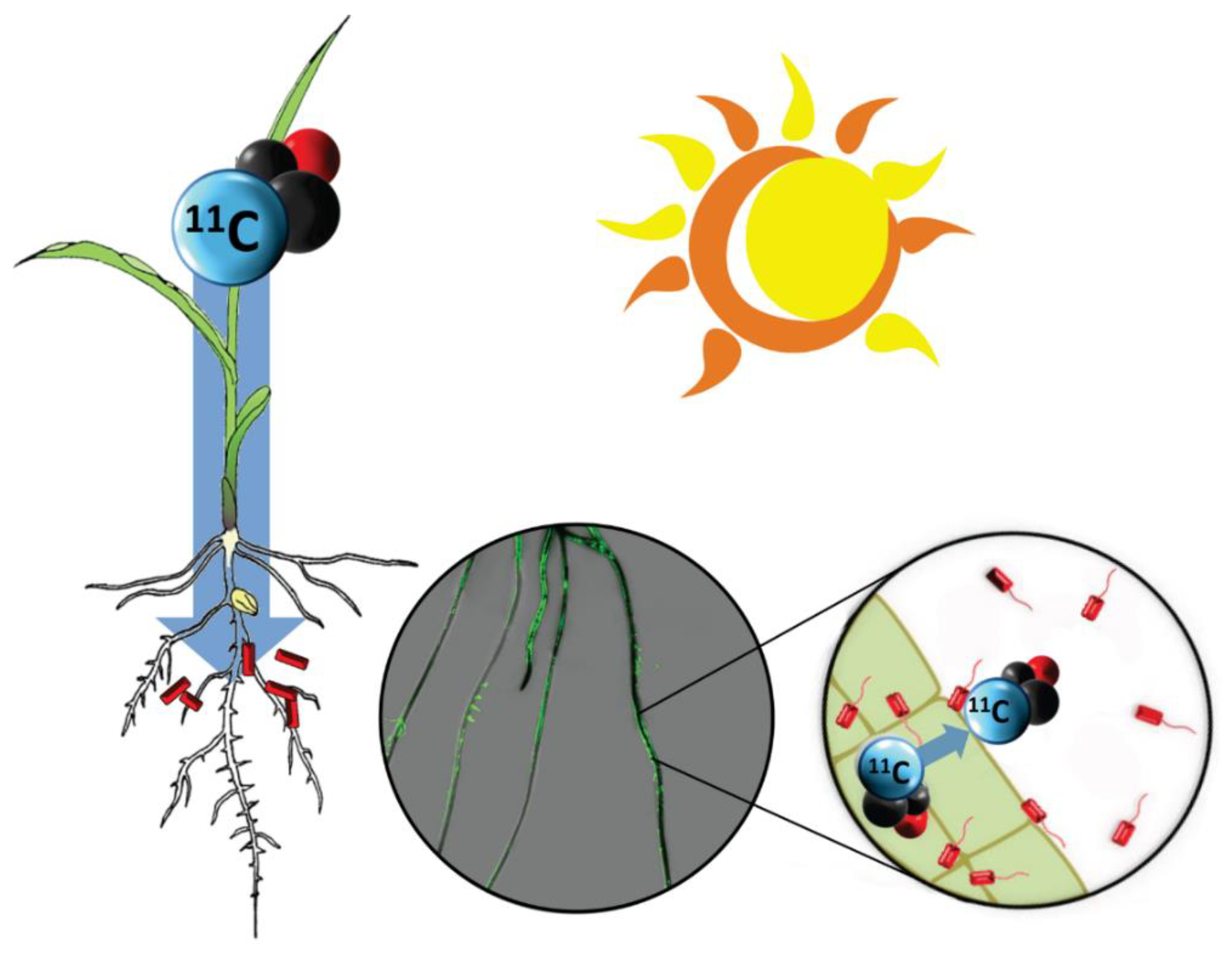
1. Introduction
2. Materials and Methods
2.1. Plant Growth
2.2. Bacteria Growth and Root Inoculation
2.3. Production and Administration of Radioactive 11CO2
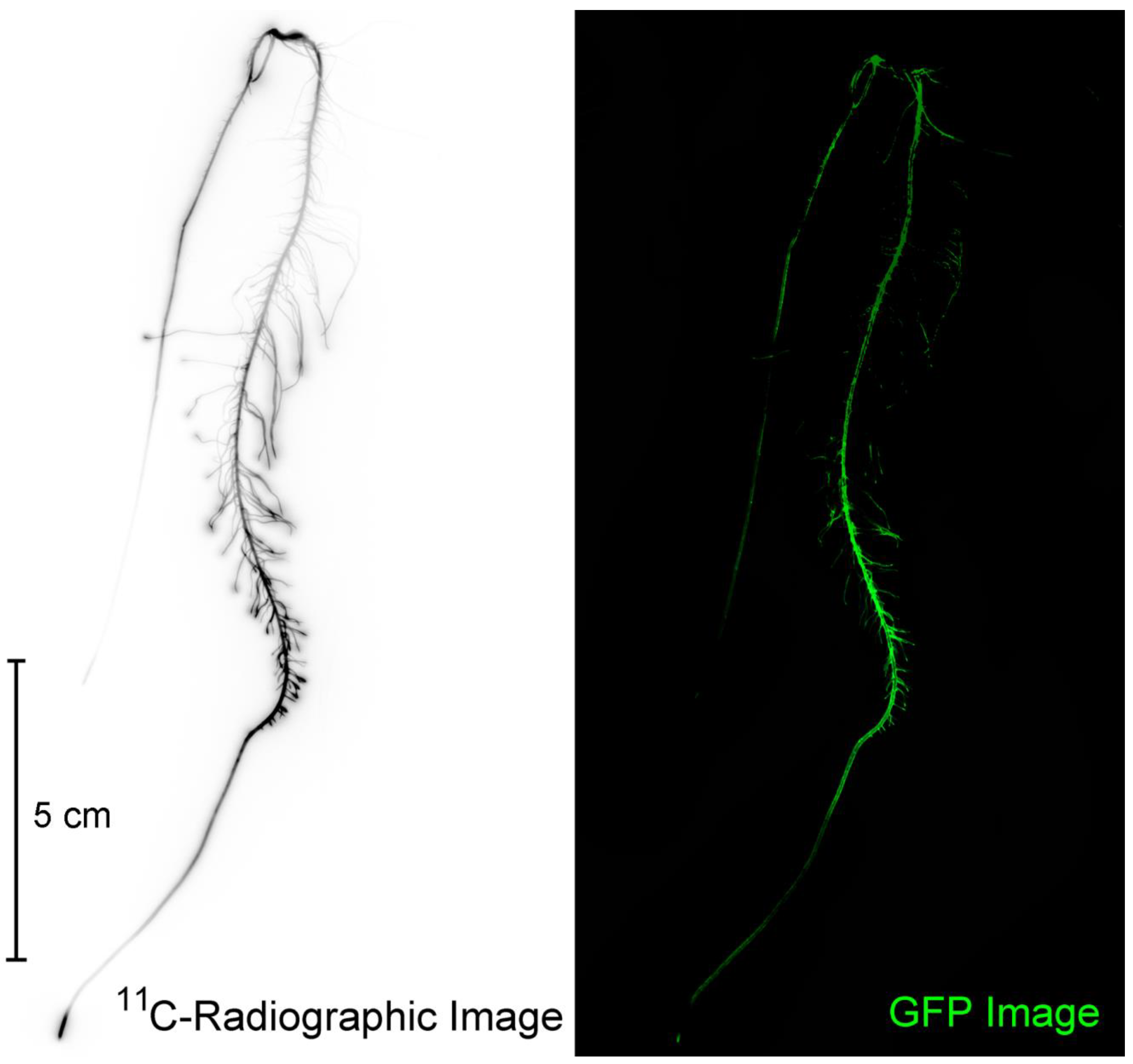
2.4. Autoradiography
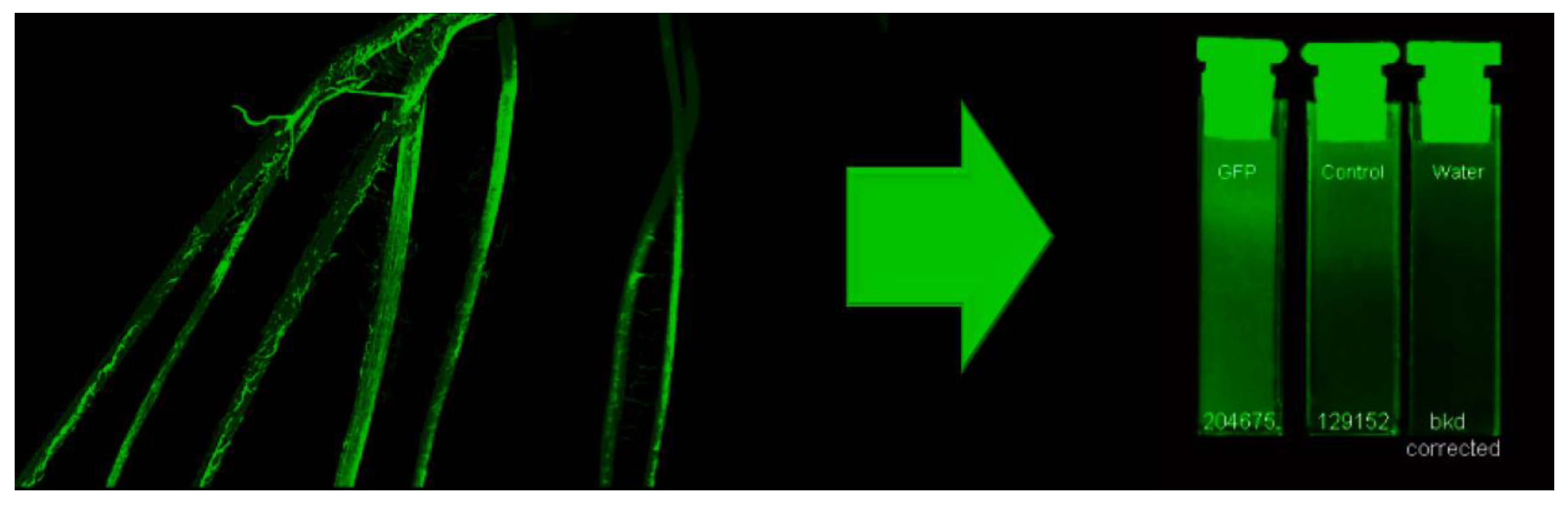
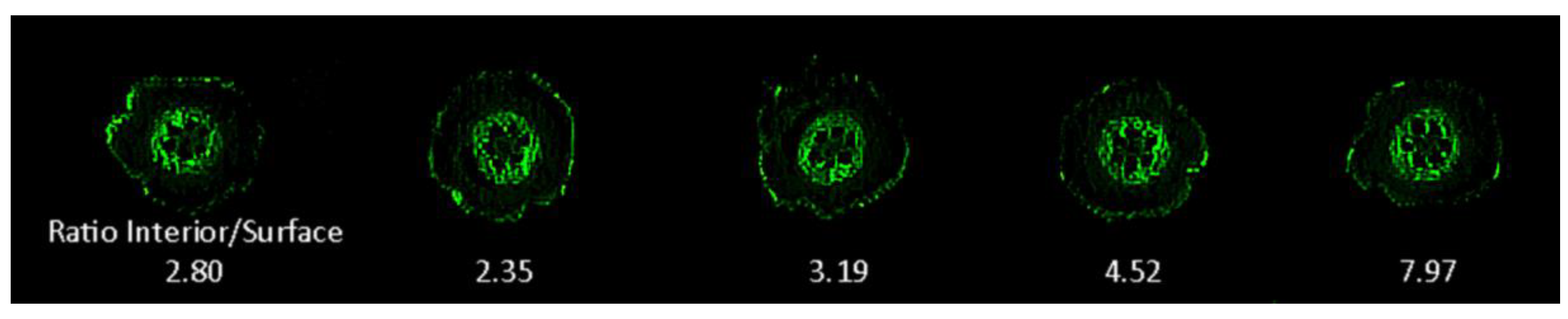
2.5. Fluorescence Imaging
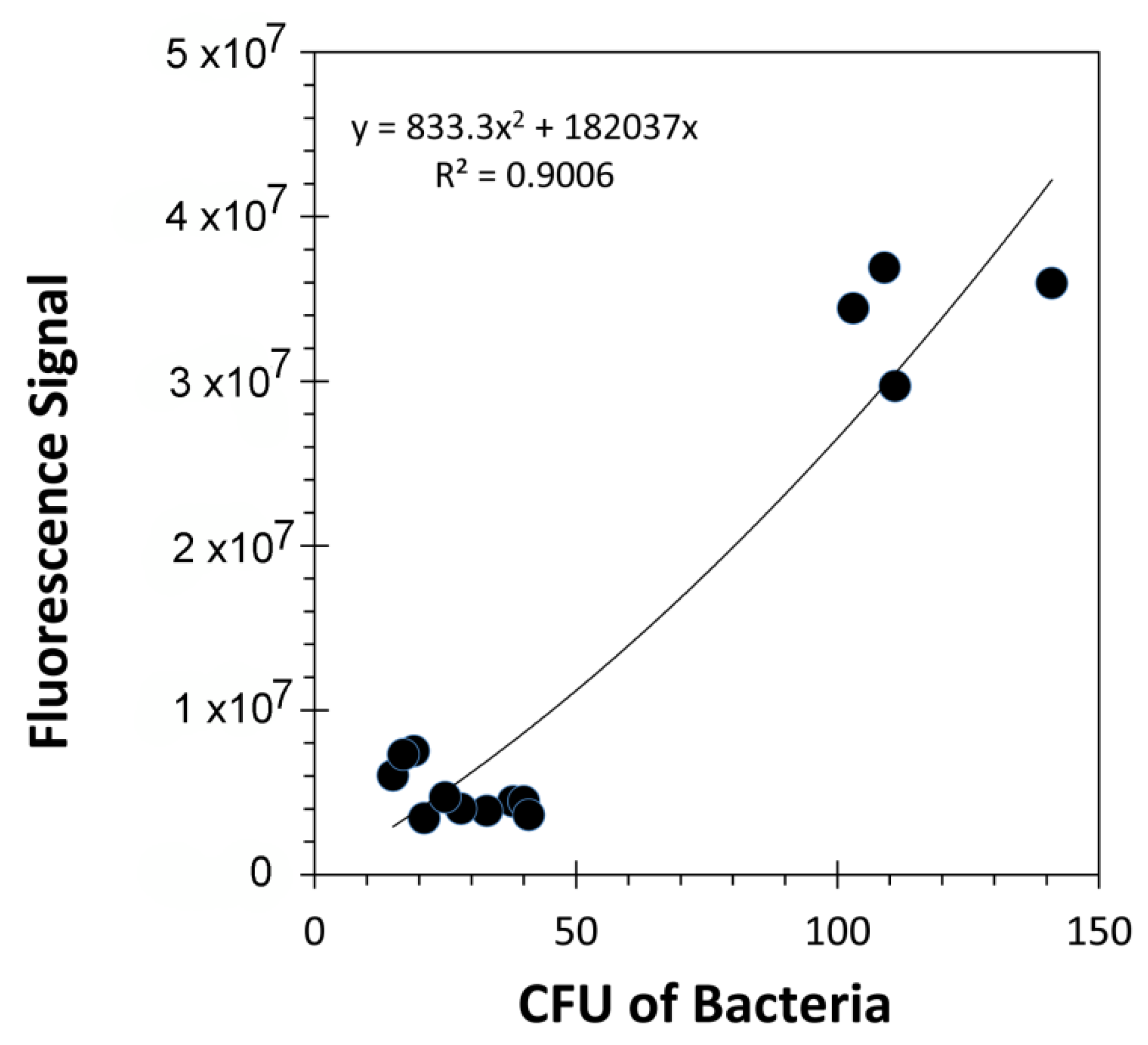
2.6. Bacteria Quantification—Drop Plate Assay
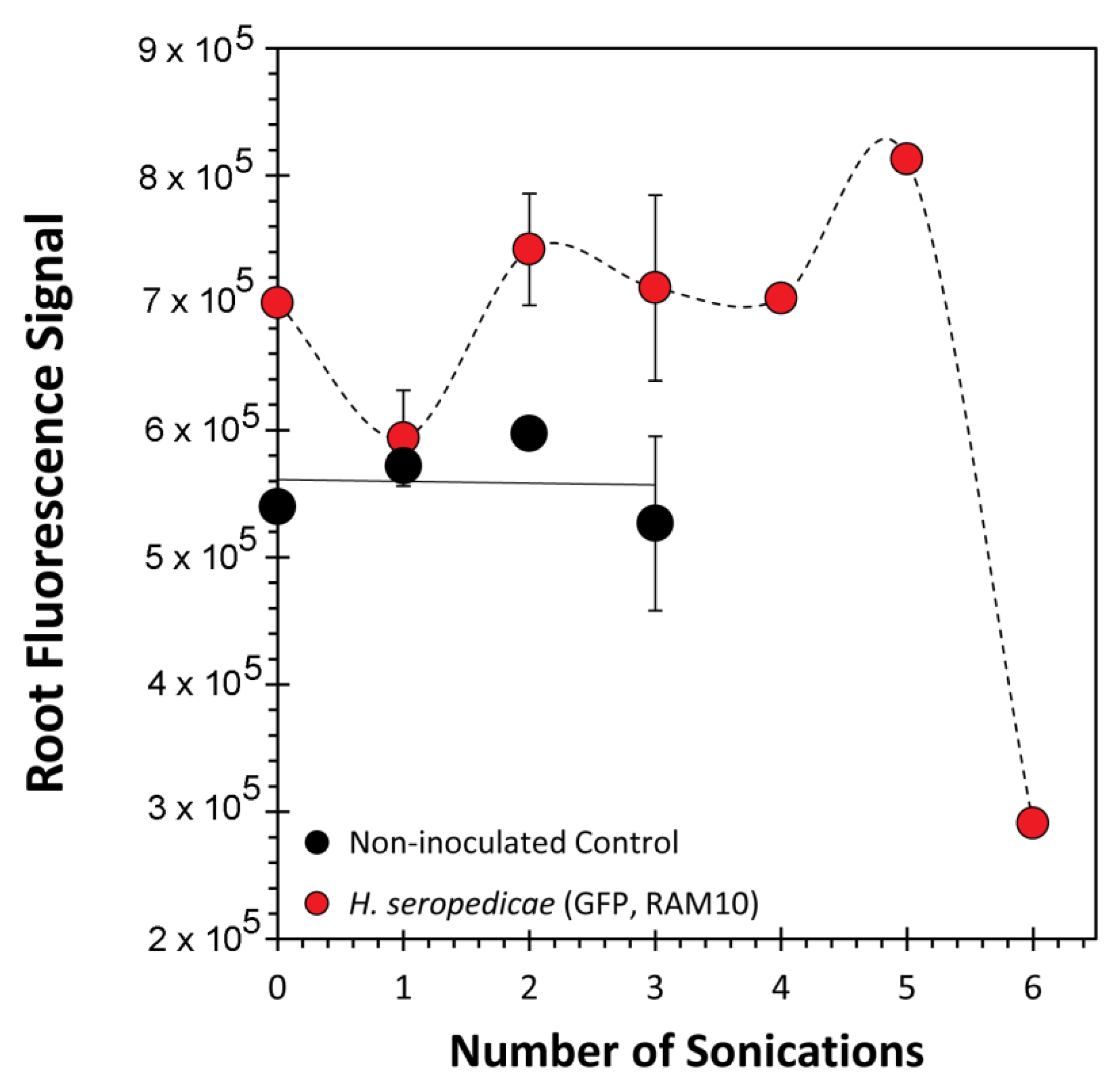
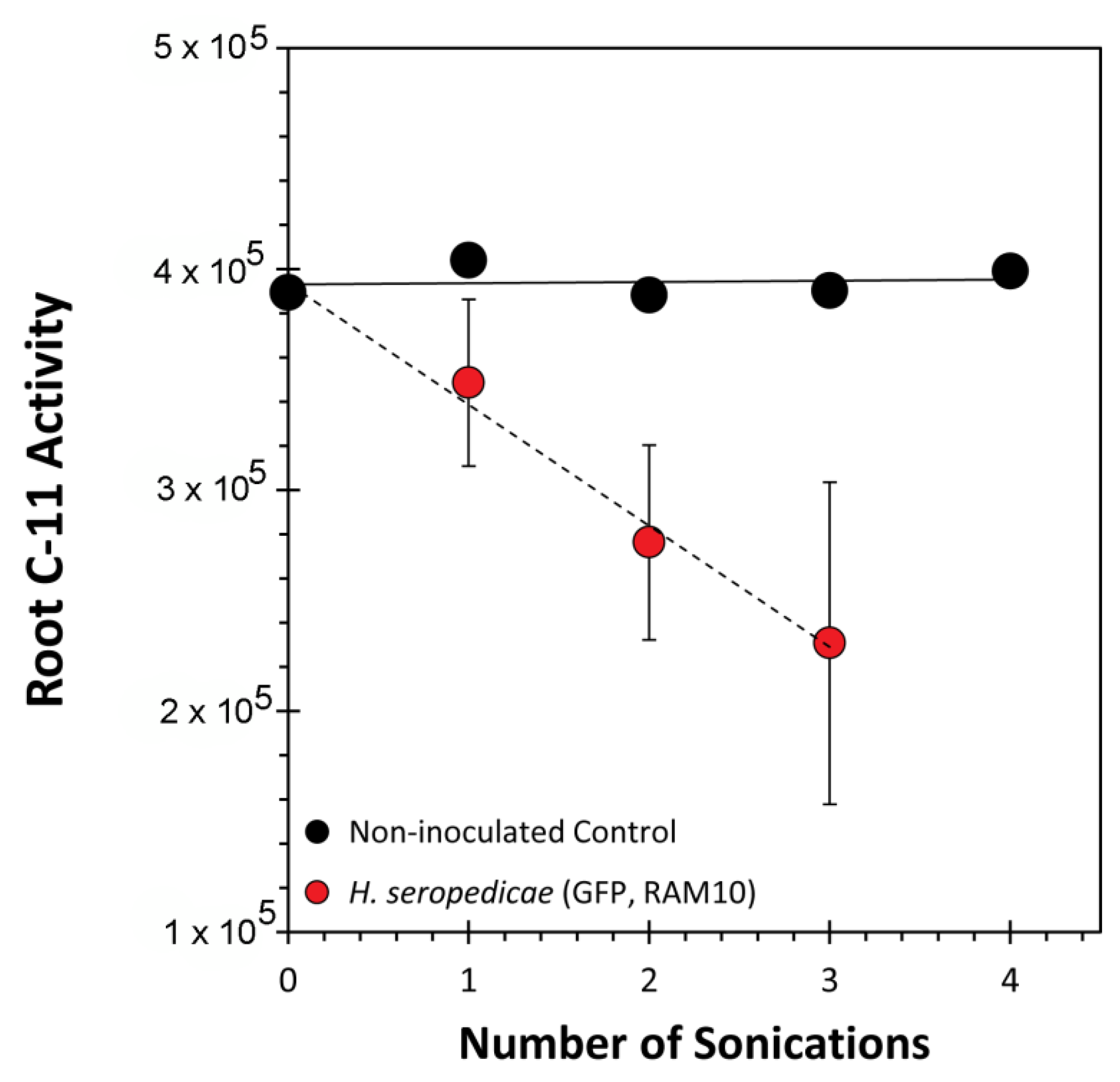
2.7. Root Sonication
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raun, W.; Johnson, G.V. Improving Nitrogen Use Efficiency for Cereal Production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef]
- Edmonds, D.E.; Tubana, B.S.; Kelly, J.P.; Crain, J.L.; Edmonds, M.D.; Solie, J.B.; Taylor, R.; Raun, W. Maize grain yield response to variable row nitrogen fertilization. J. Plant Nutr. 2013, 36, 1013–1024. [Google Scholar] [CrossRef]
- Ferrieri, R.; Herman, E.; Babst, B.; Schueller, M.J. Managing the Soil Nitrogen Cycle in Agroecosystems. In Soil Nitrogen Uses and Environmental Impacts; Taylor and Francis Group, CRC Press: Boca Raton, FL, USA, 2018; pp. 343–360. [Google Scholar]
- Westhoff, P.; Emerich, D.; Krishnan, H. The Economics of Biological Nitrogen Fixation in the Global Economy. Micrometeorol. Agric. Syst. 2015, 52, 309–328. [Google Scholar] [CrossRef]
- Dobermann, A. Nutrient use efficiency–measurement and management. In Fertilizer Best Management Practices; International Fertilizer Industry Association: Brussels, Belgium, 2007; pp. 1–22. [Google Scholar]
- Sutton, M.A.; Oenema, O.; Erisman, J.W.; Leip, A.; Van Grinsven, H.; Winiwarter, W. Too much of a good thing. Nature 2011, 472, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Panwar, J.; Akhtar, M.S.; Jha, P.N. Endophytic Nitrogen-Fixing Bacteria as Biofertilizer. Sustain. Agric. Rev. 2012, 11, 183–221. [Google Scholar] [CrossRef]
- Tkacz, A.; Poole, P.S. Role of root microbiota in plant productivity. J. Exp. Bot. 2015, 66, 2167–2175. [Google Scholar] [CrossRef]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Boil. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Amaral, F.P.D.; Agtuca, B.J.; Stacey, G. Setaria Root–Microbe Interactions. In Genetics and Genomics of Setaria; Springer International Publishing: Cham, Switzerland, 2016; Volume 19, pp. 239–250. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Life in grasses: Diazotrophic endophytes. Trends Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Boil. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Pankievicz, V.; Amaral, F.P.D.; Santos, K.F.D.N.; Agtuca, B.; Xu, Y.; Schueller, M.J.; Arisi, A.C.M.; Steffens, M.B.; De Souza, E.M.; Pedrosa, F.O.; et al. Robust biological nitrogen fixation in a model grass-bacterial association. Plant J. 2015, 81, 907–919. [Google Scholar] [CrossRef]
- Montaño, F.P.; Alias-Villegas, C.; Bellogín, R.; Del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; Baena, F.J.L.; Ollero, F.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Olsson, P.A.; Johnson, N.C. Tracking carbon from the atmosphere to the rhizosphere. Ecol. Lett. 2005, 8, 1264–1270. [Google Scholar] [CrossRef]
- Whiteley, A.S.; Thomson, B.; Lueders, T.; Manefield, M. RNA stable-isotope probing. Nat. Protoc. 2007, 2, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Mahé, S.; Ineson, P.; Staddon, P.; Ostle, N.; Cliquet, J.-B.; Francez, A.-J.; Fitter, A.H.; Young, J.P.W. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc. Natl. Acad. Sci. USA 2007, 104, 16970–16975. [Google Scholar] [CrossRef]
- Boschker, H.T.S.; Nold, S.C.; Wellsbury, P.; Bos, D.; De Graaf, W.; Pel, R.; Parkes, R.J.; Cappenberg, T.E. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 1998, 392, 801–805. [Google Scholar] [CrossRef]
- Drigo, B.; Pijl, A.S.; Duyts, H.; Kielak, A.M.; Gamper, H.A.; Houtekamer, M.J.; Boschker, H.T.S.; Bodelier, P.; Whiteley, A.S.; Van Veen, J.A.; et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc. Natl. Acad. Sci. USA 2010, 107, 10938–10942. [Google Scholar] [CrossRef]
- Behie, S.W.; Moreira, C.C.; Sementchoukova, I.; Barelli, L.; Zelisko, P.M.; Bidochka, M.J. Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat. Commun. 2017, 8, 14245. [Google Scholar] [CrossRef]
- Ferrieri, R.A.; Wolf, A.P. The Chemistry of Positron Emitting Nucleogenic (Hot) Atoms with Regard to Preparation of Labelled Compounds of Practical Utility. Radiochim. Acta 1983, 34, 69–83. [Google Scholar] [CrossRef]
- Ferrieri, R.A. Production and application of synthetic precursors labeled with carbon-11 and fluorine-18. In Handbook of Radiopharmaceuticals: Radiochemistry and Applications; John Wiley & Sons, Ltd.: Chichester, UK, 2003. [Google Scholar]
- Ferrieri, R.; Gray, D.W.; Babst, B.A.; Schueller, M.J.; Schlyer, D.J.; Thorpe, M.R.; Orians, C.M.; Lerdau, M. Use of carbon-11 in Populus shows that exogenous jasmonic acid increases biosynthesis of isoprene from recently fixed carbon. Plant Cell Environ. 2005, 28, 591–602. [Google Scholar] [CrossRef]
- Richter-Heitmann, T.; Eickhorst, T.; Knauth, S.; Friedrich, M.W.; Schmidt, H. Evaluation of Strategies to Separate Root-Associated Microbial Communities: A Crucial Choice in Rhizobiome Research. Front. Microbiol. 2016, 7, 929. [Google Scholar] [CrossRef] [PubMed]
- Roshchina, V.V. Vital Autofluorescence: Application to the Study of Plant Living Cells. Int. J. Spectrosc. 2012, 2012, 1–14. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waller, S.; Wilder, S.L.; Schueller, M.J.; Housh, A.B.; Ferrieri, R.A. Quantifying Plant-Borne Carbon Assimilation by Root-Associating Bacteria. Microorganisms 2020, 8, 700. https://doi.org/10.3390/microorganisms8050700
Waller S, Wilder SL, Schueller MJ, Housh AB, Ferrieri RA. Quantifying Plant-Borne Carbon Assimilation by Root-Associating Bacteria. Microorganisms. 2020; 8(5):700. https://doi.org/10.3390/microorganisms8050700
Chicago/Turabian StyleWaller, Spenser, Stacy L. Wilder, Michael J. Schueller, Alexandra B. Housh, and Richard A. Ferrieri. 2020. "Quantifying Plant-Borne Carbon Assimilation by Root-Associating Bacteria" Microorganisms 8, no. 5: 700. https://doi.org/10.3390/microorganisms8050700
APA StyleWaller, S., Wilder, S. L., Schueller, M. J., Housh, A. B., & Ferrieri, R. A. (2020). Quantifying Plant-Borne Carbon Assimilation by Root-Associating Bacteria. Microorganisms, 8(5), 700. https://doi.org/10.3390/microorganisms8050700




