Growth and Behavior of North American Microbes on Phragmites australis Leaves
Abstract
1. Introduction
2. Methods
2.1. Isolation and Identification of Live Microbe Cultures Taken from Phragmites Tissues
2.2. Mature Leaf Assay
2.3. Phragmites Seed Collection
2.4. Seedling Leaf Assay
2.5. Saprophyte Assay
2.6. Literature-Based Microbiome Comparison
3. Results
3.1. Microbe Identification
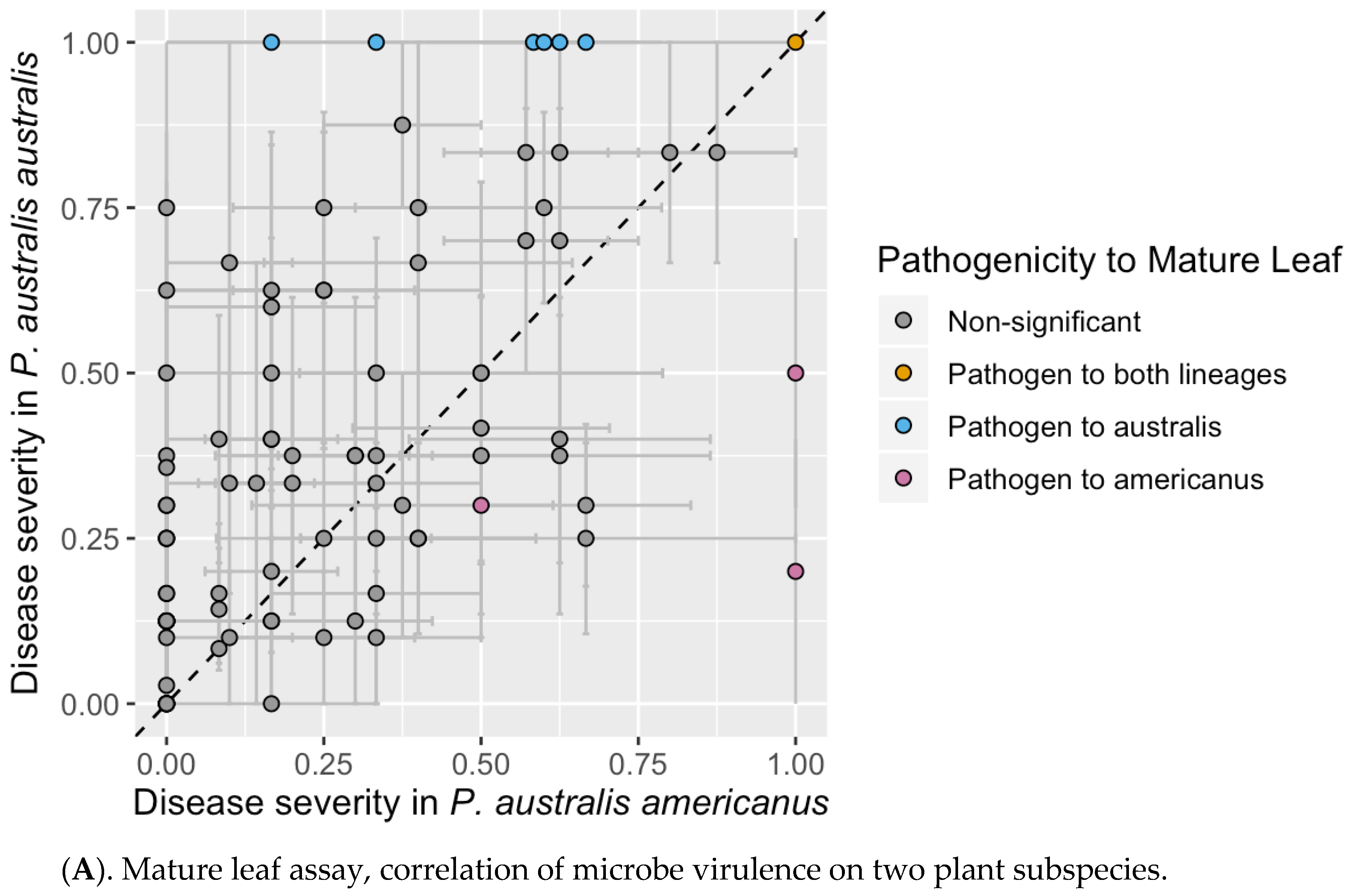
3.2. Mature Leaf Assay
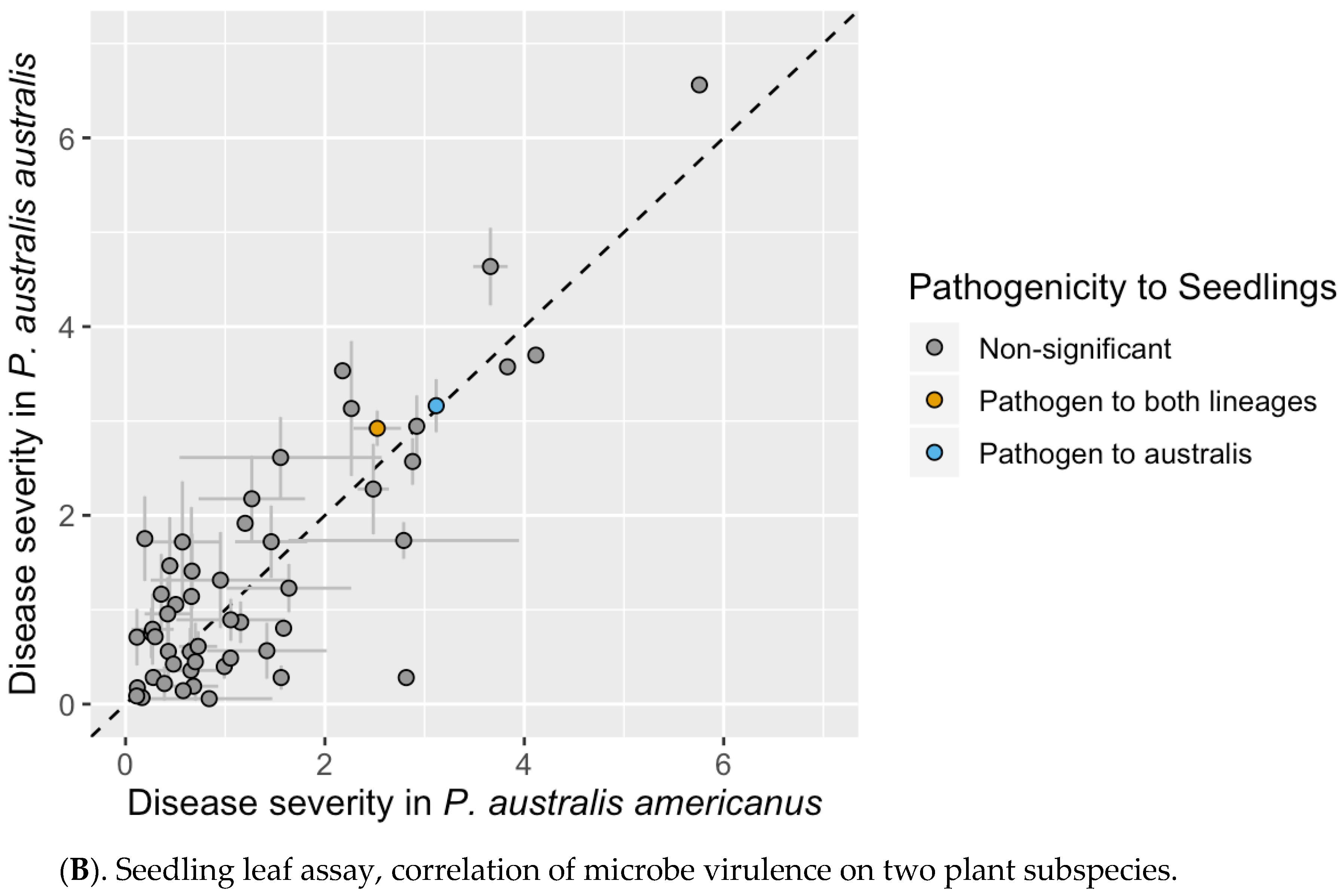
3.3. Seedling Leaf Assay
3.4. Saprophyte Assay
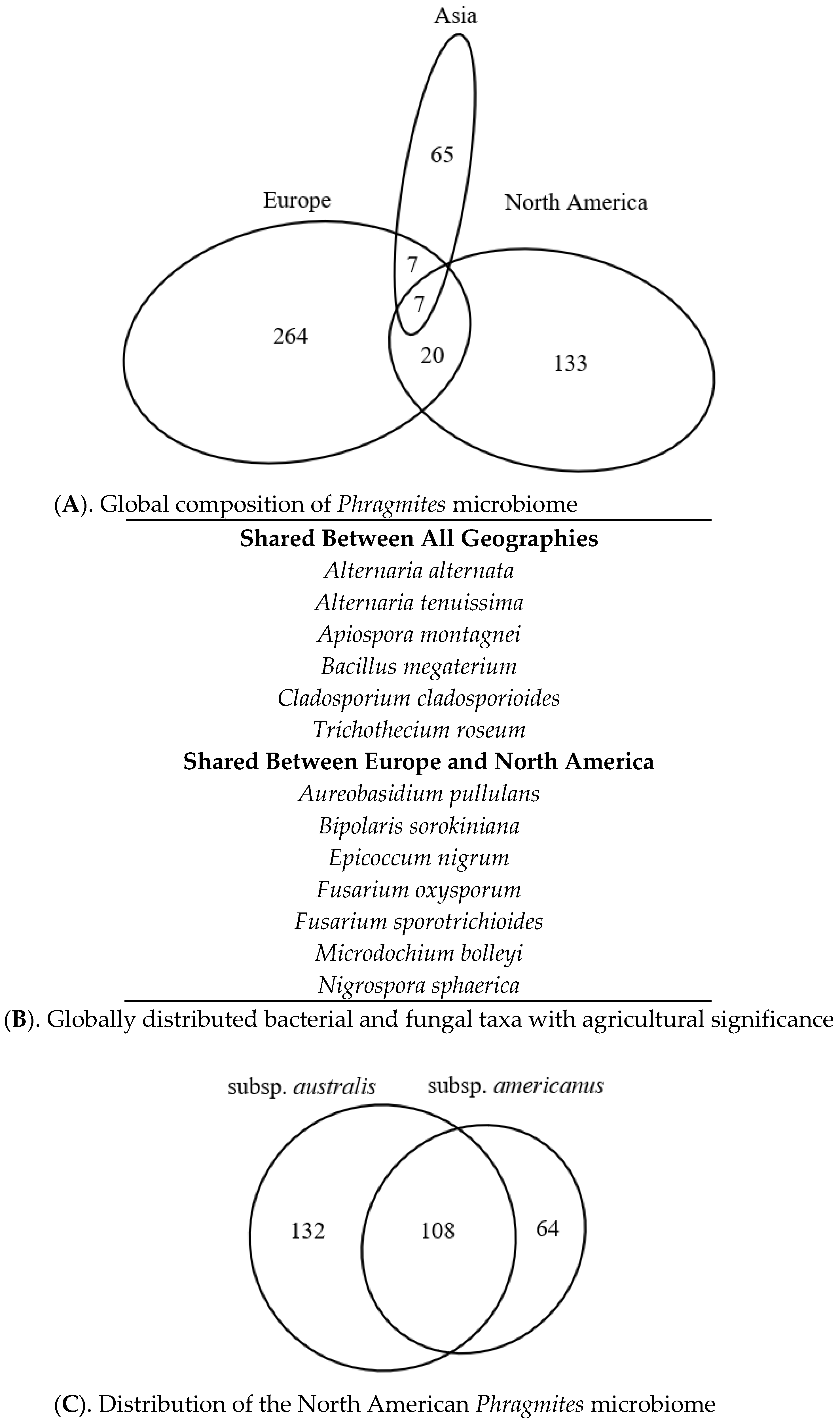
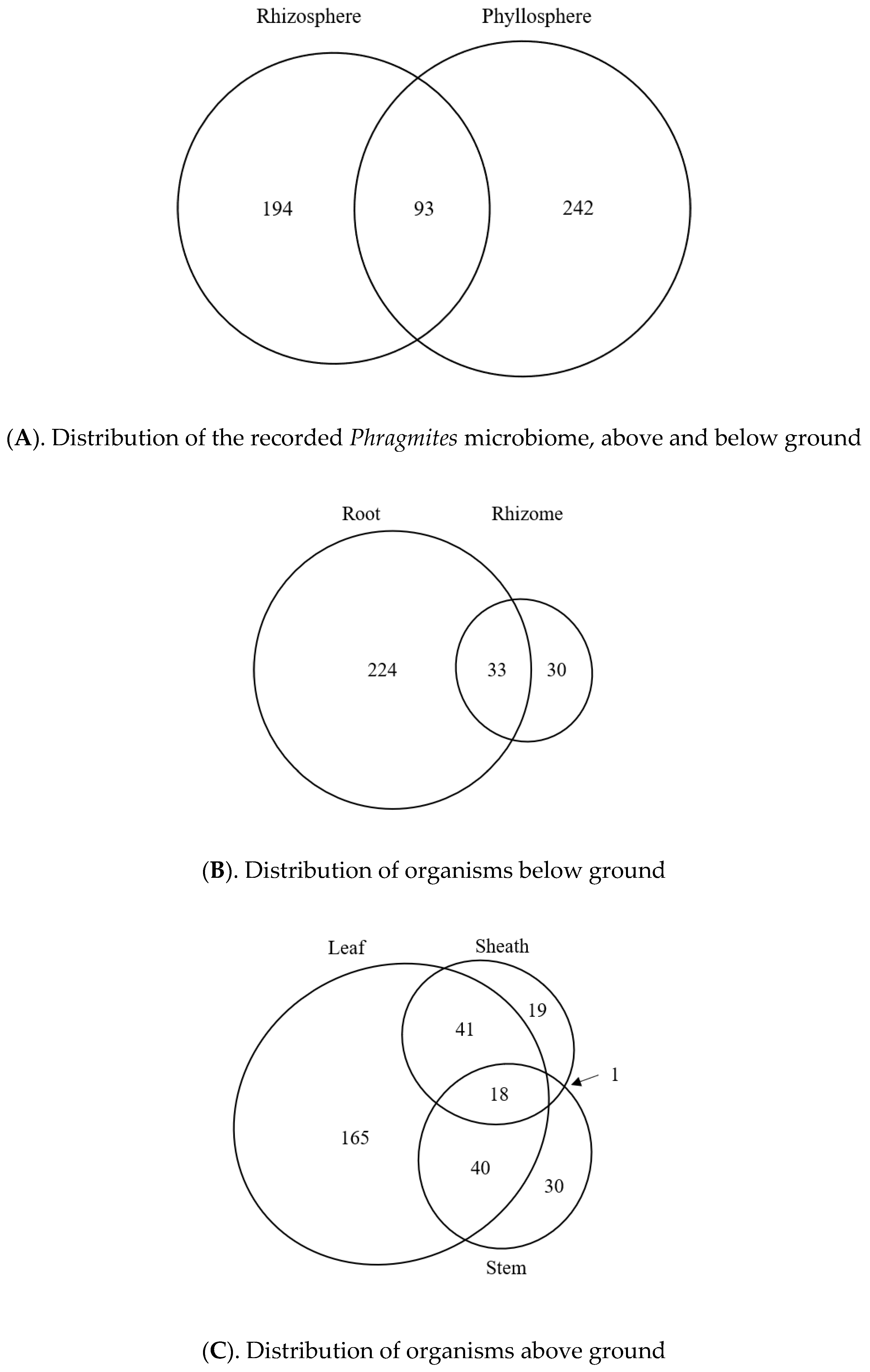
3.5. Literature-Based Microbiome Comparison
3.6. Taxonomic Distribution of the Phragmites Microbiome
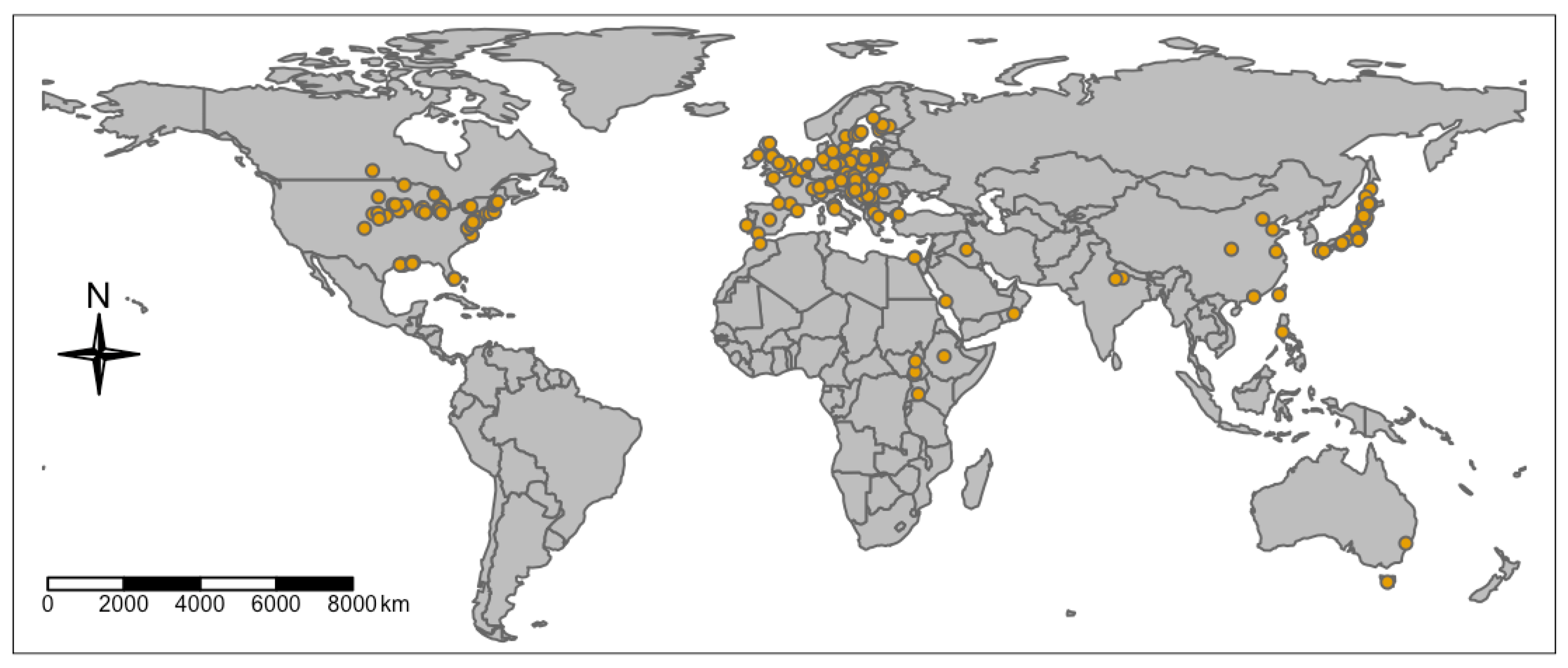
3.7. Geographic Distribution of the Published Phragmites Microbiome
3.8. Potential Tissue-Specific Patterns
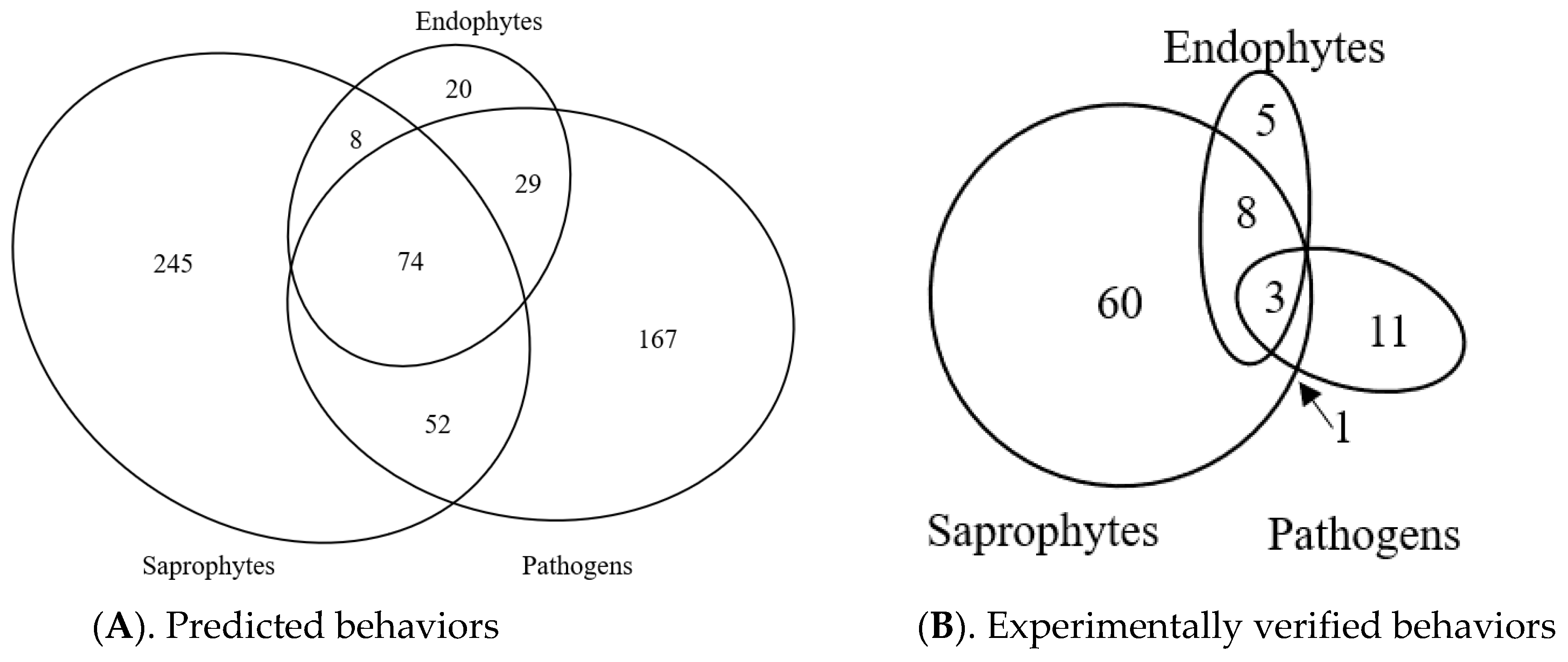
3.9. Predicted Trophic Feeding Behavior
4. Discussion
Implications for Reed Biocontrol
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Putten, W.H. Die-back of Phragmites australis in European wetlands: An overview of the European Research Programme on Reed Die-back and Progression (1993–1994). Aquat. Bot. 1997, 59, 263–275. [Google Scholar] [CrossRef]
- Saltonstall, K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc. Natl. Acad. Sci. USA 2002, 99, 2445–2449. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, C.; Mendelssohn, I.A.; Gustafsson, M.H.G.; Olesen, B.; Riis, T.; Sorrell, B.K.; Brix, H. Tracing the origin of Gulf Coast Phragmites (Poaceae): A story of long-distance dispersal and hybridization. Am. J. Bot. 2012, 99, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Saltonstall, K.; Burdick, D.; Miller, S.; Smith, B. Native and Non-native Phragmites: Challenges in Identification, Research, and Management of the Common Reed; National Estuarine Research Reserve Technical Report Series; September 2005. Available online: https://coast.noaa.gov/data/docs/nerrs/Research_TechSeries_Phrag_Final_2009.pdf (accessed on 7 May 2020).
- Kowalski, K.P.; Bacon, C.; Bickford, W.; Braun, H.; Clay, K.; Leduc-Lapierre, M.; Lillard, E.; McCormick, M.K.; Nelson, E.; Torres, M.; et al. Advancing the science of microbial symbiosis to support invasive species management: A case study on Phragmites in the Great Lakes. Front. Microbiol. 2015, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.B.; Karp, M.A. Soil pathogen communities associated with native and non-native Phragmites australis populations in freshwater wetlands. Ecol. Evol. 2013, 3, 5254–5267. [Google Scholar] [CrossRef] [PubMed]
- Clay, K.; Shearin, Z.R.; Bourke, K.A.; Bickford, W.A.; Kowalski, K.P. Diversity of fungal endophytes in non-native Phragmites australis in the Great Lakes. Biol. Invasions 2016, 18, 2703–2716. [Google Scholar] [CrossRef]
- Allen, W.J.; DeVries, A.E.; Bologna, N.A.; Bickford, W.A.; Kowalski, K.P.; Meyerson, L.A.; Cronin, J.T. Intraspecific and biogeographic variation in foliar fungal communities and pathogen damage of native and invasive Phragmites australis. Glob. Ecol. Biogeogr. 2020. [Google Scholar] [CrossRef]
- Shearin, Z.R.C.; Filipek, M.; Desai, R.; Desai, R.; Bickford, W.A.; Kowalski, K.P.; Clay, K. Fungal endophytes from seeds of invasive, non-native Phragmites australis and their potential role in germination and seedling growth. Plant Soil 2018, 422, 183–194. [Google Scholar] [CrossRef]
- Cerri, M.; Reale, L.; Moretti, C.; Buonaurio, R.; Coppi, A.; Ferri, V.; Foggi, B.; Gigante, D.; Lastrucci, L.; Quaglia, M.; et al. Claviceps arundinis identification and its role in the die-back syndrome of Phragmites australis populations in central Italy. Plant Biosyst. 2018, 152, 818–824. [Google Scholar] [CrossRef]
- Ernst, M.; Mendgen, K.W.; Wirsel, S.G.R. Endophytic Fungal Mutualists: Seed-Borne Stagonospora Spp. Enhance Reed Biomass Production in Axenic Microcosms. Mol. Plant-Microbe Interact. 2003, 16, 580–587. [Google Scholar] [CrossRef]
- Soares, M.A.; Li, H.-Y.; Kowalski, K.; Bergen, M.; Torres, M.S.; White, J.F. Evaluation of the functional roles of fungal endophytes of Phragmites australis from high saline and low saline habitats. Biol. Invasions 2016, 18, 2689–2702. [Google Scholar] [CrossRef]
- Cerri, M.; Sapkota, R.; Coppi, A.; Ferri, V.; Foggi, B.; Gigante, D.; Lastrucci, L.; Selvaggi, R.; Venanzoni, R.; Nicolaisen, M.; et al. Oomycete Communities Associated with Reed Die-Back Syndrome. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.; Armstrong, W. An overview of the effects of phytotoxins on Phragmites australis in relation to die-back. Aquat. Bot. 2001, 69, 251–268. [Google Scholar] [CrossRef]
- Yarwood, S.A.; Baldwin, A.H.; Gonzalez Mateu, M.; Buyer, J.S. Archaeal rhizosphere communities differ between the native and invasive lineages of the wetland plant Phragmites australis (common reed) in a Chesapeake Bay subestuary. Biol. Invasions 2016, 18, 2717–2728. [Google Scholar] [CrossRef]
- Bowen, J.L.; Kearns, P.J.; Byrnes, J.E.K.; Wigginton, S.; Allen, W.J.; Greenwood, M.; Tran, K.; Yu, J.; Cronin, J.T.; Meyerson, L.A. Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Bickford, W.A.; Goldberg, D.E.; Kowalski, K.P.; Zak, D.R. Root endophytes and invasiveness: No difference between native and non-native Phragmites in the Great Lakes Region. Ecosphere 2018, 9. [Google Scholar] [CrossRef]
- Bickford, W.A.; Zak, D.R.; Kowalski, K.P.; Goldberg, D.E. Soil Microbial Communities Do Not Explain the Invasiveness of Non-Native Phragmites australis. Ecol. Evol. in review.
- Rudrappa, T.; Bonsall, J.; Gallagher, J.L.; Seliskar, D.M.; Bais, H.P. Root-secreted allelochemical in the noxious weed Phragmites australis deploys a reactive oxygen species response and microtubule assembly disruption to execute rhizotoxicity. J. Chem. Ecol. 2007, 33, 1898–1918. [Google Scholar] [CrossRef]
- Weidenhamer, J.D.; Li, M.; Allman, J.; Bergosh, R.G.; Posner, M. Evidence does not support a role for gallic acid in Phragmites australis invasion success. J. Chem. Ecol. 2013, 39, 323–332. [Google Scholar] [CrossRef]
- Mozdzer, T.J.; Zieman, J.C. Ecophysiological differences between genetic lineages facilitate the invasion of non-native Phragmites australis in North American Atlantic coast wetlands. J. Ecol. 2010, 98, 451–458. [Google Scholar] [CrossRef]
- Spaepen, S. Plant Hormones Produced by Microbes. In Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture; Lugtenberg, B., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 247–256. [Google Scholar]
- Armstrong, W.; Cousins, D.; Armstrong, J.; Turner, D.W.; Beckett, P.M. Oxygen distribution in wetland plant roots and permeability barriers to gas-exchange with the rhizosphere: A microelectrode and modelling study with Phragmites australis. Ann. Bot. 2000, 86, 687–703. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Sninsky, J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Hopple, J.S.; Vilgalys, R. Phylogenetic Relationships in the Mushroom Genus Coprinus and Dark-Spored Allies Based on Sequence Data from the Nuclear Gene Coding for the Large Ribosomal Subunit RNA: Divergent Domains, Outgroups, and Monophyly. Mol. Phylogenet. Evol. 1999, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Links, M.G.; Dumonceaux, T.J.; Hemmingsen, S.M.; Hill, J.E. The Chaperonin-60 Universal Target Is a Barcode for Bacteria That Enables De Novo Assembly of Metagenomic Sequence Data. PLoS ONE 2012, 7, e49755. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program fro windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Robert, V.; Vu, D.; Amor, A.B.H.; van de Wiele, N.; Brouwer, C.; Jabas, B.; Szoke, S.; Dridi, A.; Triki, M.; Daoud, S.B.; et al. MycoBank gearing up for new horizons. IMA Fungus 2013, 4, 371–379. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Taylor, A.F.S.; Bates, S.T.; Thomas, D.; Bengtsson-palme, J.; Callaghan, T.M.; Douglas, B.; Griffith, G.W.; Ucking, R.L.; Suija, A.V.E.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Reimer, L.C.; Vetcininova, A.; Carbasse, J.S.; Söhngen, C.; Gleim, D.; Ebeling, C.; Overmann, J. BacDive in 2019: Bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019, 47, D631–D636. [Google Scholar] [CrossRef]
- Saltonstall, K. A rapid method for identifying the origin of North American Phragmites populations using RFLP analysis. Wetlands 2003, 23, 1043–1047. [Google Scholar] [CrossRef]
- Martinez, J.; Wang, K. A Sterilization Protocol for Field-Harvested Maize Seed Used for In Vitro Culture and Genetic Transformation; Maize Genetics Cooperation Newsletter: Ames, IA, USA, 2009; pp. 1–3. [Google Scholar]
- DeVries, A.E.; Bickford, W.A.; Kowalski, K.P. The effects of North American fungi and bacteria on Phragmites australis leaves 2017–2019, with comparisons to the global Phragmites microbiome: U.S. Geological Survey data release. Unpublished work. 2020. [Google Scholar] [CrossRef]
- Lambertini, C.; Sorrell, B.K.; Riis, T.; Olesen, B.; Brix, H. Exploring the borders of European Phragmites within a cosmopolitan genus. AoB Plants 2012, 2012, pls020. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, C.; Gustafsson, M.H.G.; Frydenberg, J.; Lissner, J.; Speranza, M.; Brix, H. A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Syst. Evol. 2006, 258, 161–182. [Google Scholar] [CrossRef]
- Hauber, D.P.; Saltonstall, K.; White, D.A.; Hood, C.S. Genetic Variation in the Common Reed, Phragmites australis, in the Mississippi River Delta Marshes: Evidence for Multiple Introductions. Estuaries Coasts 2011, 34, 851–862. [Google Scholar] [CrossRef]
- Micallef, L.; Rodgers, P. euler APE: Drawing area-proportional 3-Venn diagrams using ellipses. PLoS ONE 2014, 9, e101717. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2015, 20, 241–248. [Google Scholar] [CrossRef]
- Willson, K.G.; Perantoni, A.N.; Berry, Z.C.; Eicholtz, M.I.; Tamukong, Y.B.; Yarwood, S.A.; Baldwin, A.H. Influences of reduced iron and magnesium on growth and photosynthetic performance of Phragmites australis subsp. americanus (North American common reed). Aquat. Bot. 2017, 137, 30–38. [Google Scholar] [CrossRef]
- Llirós, M.; Trias, R.; Borrego, C.; Bañeras, L. Specific archaeal communities are selected on the root surfaces of ruppia spp. and Phragmites australis. Wetlands 2014, 34, 403–411. [Google Scholar] [CrossRef]
- Poon, M.O.K.; Hyde, K.D. Biodiversity of intertidal estuarine fungi on Phragmites at Mai Po Marshes, Hong Kong. Bot. Mar. 1998, 41, 141–155. [Google Scholar] [CrossRef]
- Komínková, D.; Kuehn, K.; Büsing, N.; Steiner, D.; Gessner, M. Microbial biomass, growth, and respiration associated with submerged litter of Phragmites australis decomposing in a littoral reed stand of a large lake. Aquat. Microb. Ecol. 2000, 22, 271–282. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Bickford, W.A. Plant Invasions and Microbes: The Interactive Effects of Plant-Associated Microbes on Invasiveness of Phragmites Australis; University of Michigan: Ann Arbor, MI, USA, 2020. [Google Scholar]
- Crocker, E.V.; Karp, M.A.; Nelson, E.B. Virulence of oomycete pathogens from Phragmites australis -invaded and noninvaded soils to seedlings of wetland plant species. Ecol. Evol. 2015, 5, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Keane, R.M.; Crawley, M.J. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 2002, 17, 164–170. [Google Scholar] [CrossRef]
| OTU | Subsp. of Original Host from Field Collection | Original Host Tissue | Putative Status of Original Host Tissue | Identification | Species Hypothesis (UNITE) | Pathogen Determination Mature Leaf | Disease Severity Seedlings | ||
|---|---|---|---|---|---|---|---|---|---|
| subsp. australis | subsp. americanus | subsp. australis (mean D.I.) | subsp. americanus (mean D.I.) | ||||||
| USGS23 | americanus | Seed | Diseased | Curvularia inaequalis | SH1890305.08FU | Strong Pathogen | Pathogen | 6.6 | 5.8 |
| USGS32 | australis | Leaf | Diseased | Bipolaris sorokiniana | SH1526399.08FU | Strong Pathogen | Weak Pathogen | 3.7 | 4.1 |
| USGS30 | australis | Leaf | Diseased | Pleosporaceae sp. | SH1547057.08FU | Strong Pathogen | Strong Pathogen | 3.5 | 2.2 |
| LSU0038 | americanus | Leaf | Diseased | Pleosporales sp. | SH1525096.08FU | Strong Pathogen | Pathogen | 3.2 | 3.1 |
| USGS15 | australis | Seed | Diseased | Septoriella hubertusii | SH2176229.08FU | Weak Pathogen | Strong Pathogen | 2.9 | 2.5 |
| USGS17 | americanus | Seed | Diseased | Alternaria sp. | SH1526398.08FU | Strong Pathogen | Nonpathogen | 2.6 | 2.9 |
| LSU0361 | Gulf lineage * | Leaf | Diseased | Colletotrichum sp. | SH1543705.08FU | Strong Pathogen | Pathogen | 2.6 | 1.6 |
| LSU0719 | australis | Leaf | Diseased | Epicoccum sorghinum | SH1547058.08FU | Pathogen | Strong Pathogen | 1.2 | 1.6 |
| LSU0107 | australis | Leaf | Diseased | Stagonospora neglecta | SH1525143.08FU | Strong Pathogen | Pathogen | 0.2 | n/d |
| LSU0172 | australis | Leaf | Diseased | Stagonospora neglecta | SH1525143.08FU | Strong Pathogen | Strong Pathogen | 2.9 | 2.9 |
| USGS07 | americanus | Stem | Diseased | Gibberella fujikuroi | SH1610159.08FU | Strong pathogen | Pathogen | 1.7 | 2.8 |
| LSU1247 | Gulf lineage * | Leaf | Healthy | Unknown fungus | n/a | Pathogen | Weak Pathogen | 3.6 | 3.8 |
| USGS34 | australis | Leaf | Diseased | Curvularia sp. | SH1526408.08FU | Pathogen | Weak Pathogen | 3.1 | 2.3 |
| USGS42 | australis | Leaf | Diseased | Fusarium sporotrichioides | SH2456045.08FU | Pathogen | Nonpathogen | 0.3 | 2.8 |
| LSU0677 | Gulf lineage * | Leaf | Diseased | Microdochium sp. | SH1555458.08FU | Weak Pathogen | Weak Pathogen | 4.6 | 3.7 |
| USGS02 | australis | Leaf | Diseased | Stagonospora neglecta | SH1525143.08FU | Weak Pathogen | Pathogen | 2.2 | 1.3 |
| LSU0240 | americanus | Leaf | Diseased | Gibberella fujikuroi | SH1610159.08FU | Nonpathogen | Not tested | 2.3 | 2.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeVries, A.E.; Kowalski, K.P.; Bickford, W.A. Growth and Behavior of North American Microbes on Phragmites australis Leaves. Microorganisms 2020, 8, 690. https://doi.org/10.3390/microorganisms8050690
DeVries AE, Kowalski KP, Bickford WA. Growth and Behavior of North American Microbes on Phragmites australis Leaves. Microorganisms. 2020; 8(5):690. https://doi.org/10.3390/microorganisms8050690
Chicago/Turabian StyleDeVries, Aaron E., Kurt P. Kowalski, and Wesley A. Bickford. 2020. "Growth and Behavior of North American Microbes on Phragmites australis Leaves" Microorganisms 8, no. 5: 690. https://doi.org/10.3390/microorganisms8050690
APA StyleDeVries, A. E., Kowalski, K. P., & Bickford, W. A. (2020). Growth and Behavior of North American Microbes on Phragmites australis Leaves. Microorganisms, 8(5), 690. https://doi.org/10.3390/microorganisms8050690





