Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Whole-Genome Phylogenetic Tree Building for xenoGI Input
2.3. xenoGI Parameters
2.4. AT Content Analysis
2.5. Hil Phylogenies
3. Results
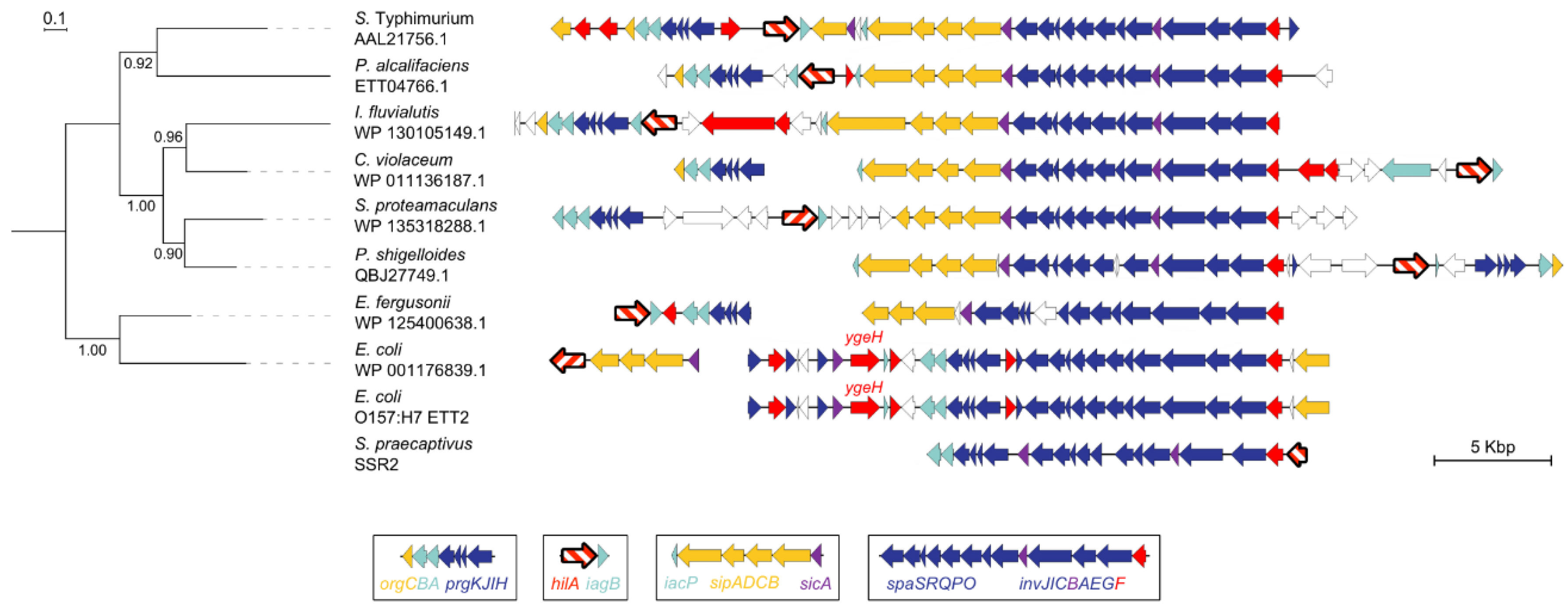
3.1. SPI-1 is a Mosaic of Gene Islands
3.2. A Cohesive SPI-1 Gene Set is Highly Conserved in Salmonella
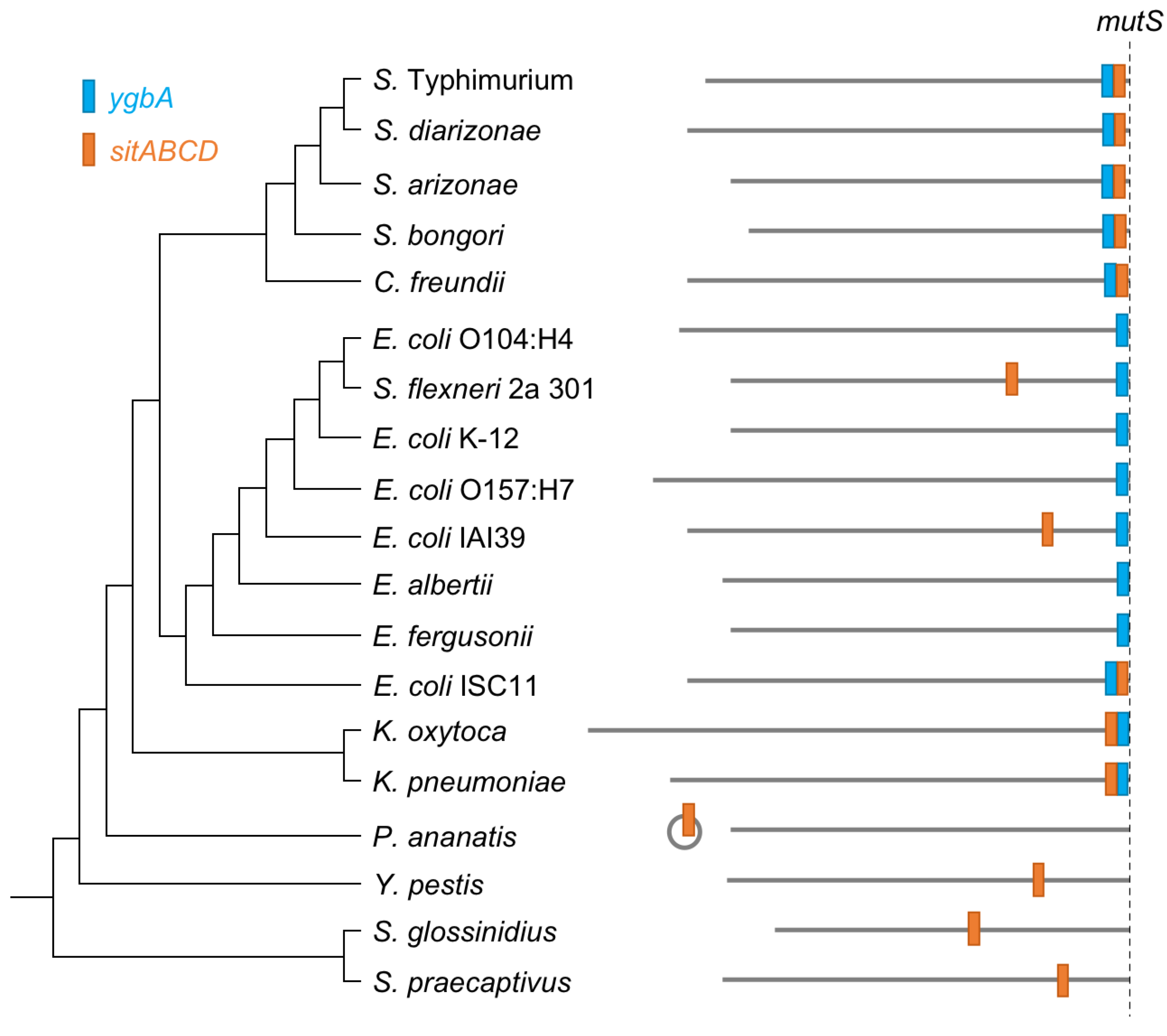
3.3. The ygbA and sitABCD Islands Predate Core SPI-1
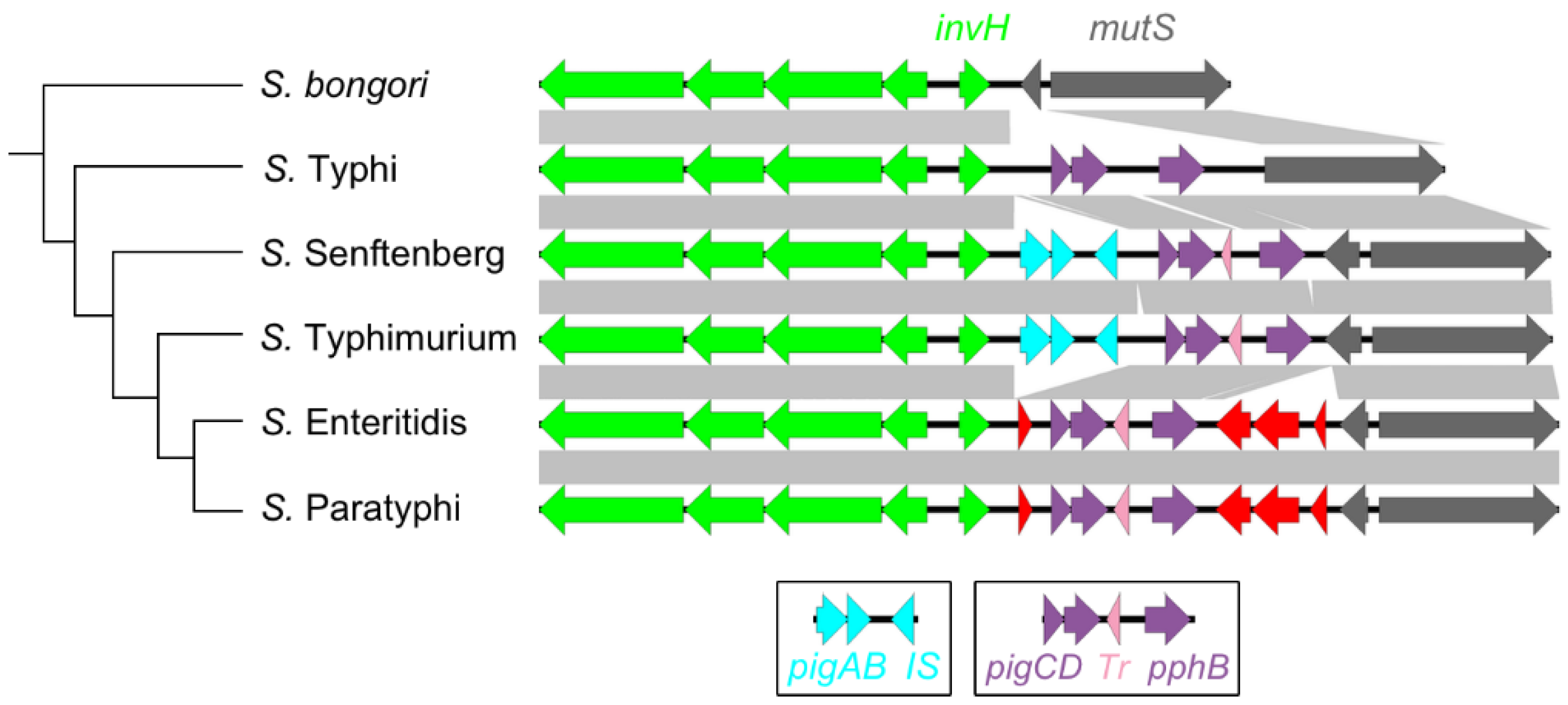
3.4. The Highly Variable mutS-Proximal Region
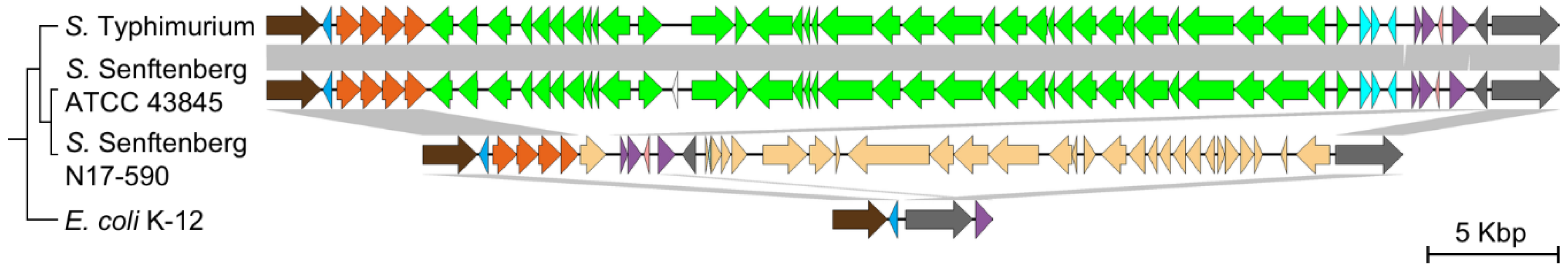
3.5. Decay and Loss of SPI-1
3.6. The fhlA/-/mutS Locus is a Hotspot for Island Acquisition
3.7. AT Nucleotide Content and the Evolution of Transcriptional Control
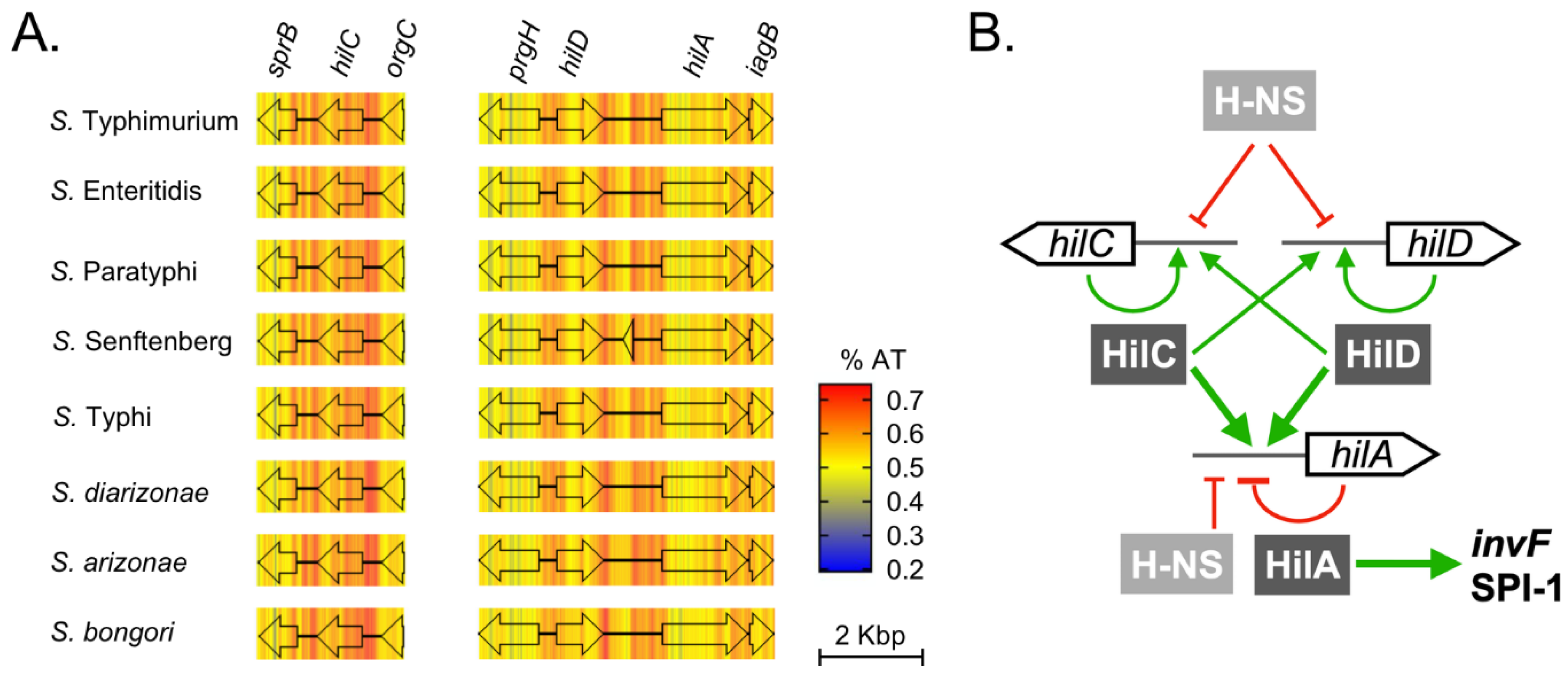
3.8. Evolution of Transcriptional Control: Acquisition of hilA
3.9. Evolution of Transcriptional Control: Addition of the HilC/D Paralogs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gonzalez-Alba, J.; Baquero, F.; Cantón, R.; Galán, J.E. Stratified reconstruction of ancestral Escherichia coli diversification. BMC Genom. 2019, 20, 936. [Google Scholar] [CrossRef]
- Maddamsetti, R.; Lenski, R.E. Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection. PLoS Genet. 2018, 14, e1007199. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Summers, A.O. Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 2006, 17, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Carniel, E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001, 21, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Blum-Oehler, G.; Tschäpe, H.; Mühldorfer, I. Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol. 1996, 23, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef]
- Schmidt, H.; Hensel, M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004, 17, 14–56. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell. Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef]
- Hacker, J.; Kaper, J. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef]
- Ou, H.-Y.; Chen, L.-L.; Lonnen, J.; Chaudhuri, R.R.; Thani, A.; Smith, R.; Garton, N.J.; Hinton, J.; Pallen, M.; Barer, M.R.; et al. A novel strategy for the identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Nucleic Acids Res. 2006, 34, 1–11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Groisman, E.A.; Ochman, H. Pathogenicity islands: Bacterial evolution in quantum leaps. Cell 1996, 87, 791–794. [Google Scholar] [CrossRef]
- Hochhut, B.; Jahreis, K.; Lengeler, J.W.; Schmid, K. CTnsct94, a conjugative transposon found in Enterobacteria. J. Bacteriol. 1997, 179, 2097–2102. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Trzebiatowski, J.R.; Cruickshank, R.W.; Gouzy, J.; Brown, S.D.; Elliot, R.M.; Fleetwood, D.J.; McCallum, N.G.; Rossbach, U.; Stuart, G.S.; et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 2002, 184, 3086–3095. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Schlievert, P.; Ruzin, A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001, 3, 585–594. [Google Scholar] [CrossRef]
- Fookes, M.; Schroeder, G.N.; Langridge, G.C.; Blondel, C.J.; Mammina, C.; Connor, T.R.; Seth-Smith, H.; Vernikos, G.S.; Robinson, K.S.; Sanders, M.; et al. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 2011, 7, e1002191. [Google Scholar] [CrossRef]
- Hayward, M.R.; AbuOun, M.; La Ragione, R.M.; Tchórzewska, M.A.; Cooley, W.A.; Everest, D.J.; Petrovska, L.; Jansen, V.; Woodward, M.J. SPI-23 of S. Derby: Role in adherence and invasion of porcine tissues. PLoS ONE 2014, 9, e107857. [Google Scholar] [CrossRef]
- Urrutia, I.M.; Fuentes, J.A.; Valenzuela, L.M.; Ortega, A.P.; Hidalgo, A.A.; Mora, G.C. Salmonella Typhi shdA: Pseudogene or allelic variant? Infect. Genet. Evol. 2014, 26, 146–152. [Google Scholar] [CrossRef]
- Wisner, A.; Desin, T.; White, A.; Köster, W. The Salmonella pathogenicity island-1 and -2 encoded type III secretion systems. In Salmonella—A Diversified Superbug; InTech: Rijeka, Croatia, 2012; pp. 469–494. [Google Scholar]
- Fàbrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Galán, J.E. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 1996, 20, 263–271. [Google Scholar] [CrossRef]
- Hensel, M.; Shea, J.E.; Gleeson, C.; Jones, M.D.; Dalton, E.; Holden, D.W. Simultaneous identification of bacterial virulence genes by negative selection. Science 1995, 269, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.M.; Bajaj, V.; Lee, C.A. A 40 kB chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region on the Escherichia coli K-12 chromosome. Mol. Microbiol. 1995, 15, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997, 5, 318–322. [Google Scholar] [CrossRef]
- Doolittle, R.; Feng, D.; Tsang, S.; Cho, G.; Little, E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 1996, 26, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Wilson, A. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 1987, 26, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.-P.; Chaudhuri, R.R.; Fivian, A.; Bailey, C.M.; Antonio, M.; Barnes, W.M.; Pallen, M.J. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 2004, 186, 3547–3560. [Google Scholar] [CrossRef]
- Betts, H.J.; Chaudhuri, R.R.; Pallen, M.J. An analysis of type-III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 2004, 12, 476–482. [Google Scholar] [CrossRef]
- Dale, C.; Jones, T.; Pontes, M. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 2005, 22, 758–765. [Google Scholar] [CrossRef]
- Haller, J.C.; Carlson, S.; Pederson, K.J.; Pierson, D.E. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 2000, 36, 1436–1446. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, H.; Cheng, X.; Shu, X.; White, A.; Stavrinides, J.; Köster, W.; Zhu, G.; Zhao, Z.; Wang, Y. A global survey of bacterial type III secretion systems and their effectors. Environ. Microbiol. 2017, 19, 3879–3895. [Google Scholar] [CrossRef]
- Kirzinger, M.W.B.; Butz, C.J.; Stavrinides, J. Inheritance of Pantoea type III secretion systems through both vertical and horizontal transfer. Mol. Genet. Genom. 2015, 290, 2075–2088. [Google Scholar] [CrossRef] [PubMed]
- Ménard, R.; Prévost, M.C.; Gounon, P.; Sansonetti, P.; Dehio, C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc. Natl. Acad. Sci. USA 1996, 93, 1254–1258. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.; da Silva Neto, J. Chromobacterium violaceum pathogenicity: Updates and insights from genome sequencing of novel Chromobacterium species. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Wren, B.W.; Thomson, N.R.; Titball, R.; Holden, M.; Prentice, M.B.; Sebaihia, M.; James, K.; Churcher, C.; Mungall, K.; et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 2001, 413, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Lumb, B.; Ryan, D.; Reeves, P.R. Molecular evolution of large virulence plasmid in Shigella and enteroinvasive Escherichia coli. Infect. Immun. 2001, 69, 6303–6309. [Google Scholar] [CrossRef] [PubMed]
- The, H.C.; Thanh, D.P.; Holt, K.E.; Thomson, N.R.; Baker, S. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 2016, 14, 235–250. [Google Scholar] [CrossRef]
- Yang, J.; Nie, H.; Chen, L.; Zhang, X.; Yang, F.; Xu, X.; Zhu, Y.; Yu, J.; Jin, Q. Revisiting the molecular evolutionary history of Shigella spp. J. Mol. Evol. 2007, 64, 71–79. [Google Scholar] [CrossRef]
- Burkinshaw, B.J.; Strynadka, N.C.J. Assembly and structure of the T3SS. Biochim. Biophys. Acta 2014, 1843, 1649–1663. [Google Scholar] [CrossRef]
- Correa, V.R.; Majerczak, D.R.; Ammar, E.-D.; Merighi, M.; Pratt, R.C.; Hogenhout, S.A.; Coplin, D.L.; Redinbaugh, M.G. The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl. Environ. Microbiol. 2012, 78, 6327–6336. [Google Scholar] [CrossRef]
- Foultier, B.; Troisfontaines, P.; Müller, S.; Opperdoes, F.R.; Cornelis, G.R. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 2002, 55, 37–51. [Google Scholar] [CrossRef]
- Hueck, C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998, 62, 379–433. [Google Scholar] [CrossRef] [PubMed]
- Pancetti, A.; Galán, J.E. Characterization of the mutS-proximal region of the Salmonella typhimurium SPI-1 identifies a group of pathogenicity island-associated genes. FEMS Microbiol. Lett. 2001, 197, 203–208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, D.; Hardt, W.D.; Galán, J.E. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 1999, 67, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Bush, E.C.; Clark, A.E.; DeRanek, C.A.; Eng, A.; Forman, J.; Heath, K.; Lee, A.B.; Stoebel, D.M.; Wang, Z.; Wilber, M.; et al. xenoGI: Reconstructing the history of genomic island insertions in clades of closely related bacteria. BMC Bioinform. 2018, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Kröger, C.; Colgan, A.; Srikumar, S.; Händler, K.; Sivasankaran, S.K.; Hammarlöf, D.L.; Canals, R.; Grissom, J.E.; Conway, T.; Hokamp, K.; et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 2013, 14, 683–695. [Google Scholar]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Srikumar, S.; Kröger, C.; Hébrard, M.; Colgan, A.; Owen, S.; Sivasankaran, S.K.; Cameron, A.; Hokamp, K.; Hinton, J. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015, 11, e1005262. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Freese, N.H.; Norris, D.C.; Loraine, A.E. Integrated genome browser: Visual analytics platform for genomics. Bioinformatics 2016, 32, 2089–2095. [Google Scholar] [CrossRef]
- Sullivan, M.; Petty, N.K.; Beatson, S. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Galán, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Nucleic Acids Res. 2014, 68, 415–438. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 15, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, J.L. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetic Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Venkatesan, M.M.; Goldberg, M.B.; Rose, D.J.; Grotbeck, E.J.; Burland, V.; Blattner, F.R. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 2001, 69, 3271–3285. [Google Scholar] [CrossRef]
- Matsumoto, H.; Young, G. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol. Microbiol. 2005, 59, 689–706. [Google Scholar] [CrossRef]
- Walker, K.; Miller, V.L. Regulation of the Ysa Type III secretion system of Yersinia enterocolitica by YsaE/SycB and YrsS/YsrR. J. Bacteriol. 2004, 186, 4056–4066. [Google Scholar] [CrossRef]
- Bertelli, C.; Tilley, K.; Brinkman, F. Microbial genomic island discovery, visualization and analysis. Brief. Bioinform. 2019, 20, 1685–1698. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Leong, H.W. Computational methods for predicting genomic islands in microbial genomes. Comput. Struct. Biotechnol. J. 2016, 14, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Groisman, E.A.; Ochman, H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993, 12, 3779–3787. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.; Porwollik, S.; Long, F.; Cheng, P.; Wollam, A.; Clifton, S.W.; Weinstock, G.M.; McClelland, M. Evolutionary genomics of Salmonella enterica subspecies. mBio 2013, 4, e00579-12. [Google Scholar] [CrossRef]
- Worley, J.; Meng, J.; Allard, M.; Brown, E.W.; Timme, R. Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. mBio 2018, 9, e02303-18. [Google Scholar] [CrossRef]
- El Ghany, M.; Shi, X.; Li, Y.; Ansari, H.; Hill-Cawthorne, G.; Ho, Y.; Naeem, R.; Pickard, D.; Klena, J.; Xu, X.; et al. Genomic and phenotypic analyses reveal the emergence of an atypical Salmonella enterica serovar Senftenberg variant in China. J. Clin. Microbiol. 2016, 54, 2014–2022. [Google Scholar] [CrossRef]
- Ginocchio, C.; Rahn, K.; Clarke, R.; Galán, J.E. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect. Immun. 1997, 65, 1267–1272. [Google Scholar] [CrossRef]
- Hu, Q.; Coburn, B.; Deng, W.; Li, Y.; Shi, X.; Lan, Q.; Wang, B.; Coombes, B.K.; Finlay, B.B. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J. Clin. Microbiol. 2008, 46, 1330–1336. [Google Scholar] [CrossRef]
- Morita, M.; Shimada, K.; Baba, H.; Morofuji, K.; Oda, S.; Izumiya, H.; Ohnishi, M. Genome sequence of a Salmonella enterica serotype Senftenberg strain lacking Salmonella Pathogenicity Island-1 and isolated in Japan. Microbiol. Resour. Announc. 2019, 9, e00653-19. [Google Scholar] [CrossRef]
- Rahn, K.; De Grandis, S.; Clarke, R.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss III, R.; Gyles, C. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detecting Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Stevens, M.; Zurfluh, K.; Althaus, D.; Corti, S.; Lehner, A.; Stephan, R. Complete and assembled genome sequence of Salmonella enterica subsp. enterica serotype Senftenberg N17-509, a strain lacking Salmonella Pathogen Island 1. Genome Announc. 2018, 6, e00156-18. [Google Scholar]
- Singh, K.; Milstein, J.; Navarre, W. Xenogeneic silencing and its impact on bacterial genomes. Nucleic Acids Res. 2016, 70, 199–213. [Google Scholar] [CrossRef]
- Dillon, S.C.; Cameron, A.; Hokamp, K.; Lucchini, S.; Hinton, J.; Dorman, C.J. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 2010, 76, 1250–1265. [Google Scholar] [CrossRef]
- Lucchini, S.; Rowley, G.; Goldberg, M.B.; Hurd, D.; Harrison, M.; Hinton, J. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef]
- Cameron, A.; Dorman, C.J. A fundamental regulatory mechanism operating through OmpR and DNA topology that controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 2012, 8, e1002615. [Google Scholar] [CrossRef] [PubMed]
- Schechter, L.M.; Lee, C.A. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 2001, 40, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Will, W.; Navarre, W.; Fang, F.C. Integrated circuits: How transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr. Opin. Microbiol. 2015, 23, 8–13. [Google Scholar] [CrossRef]
- Olekhnovich, I.; Kadner, R. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 2006, 357, 373–386. [Google Scholar] [CrossRef]
- Bajaj, V.; Hwang, C.; Lee, C.A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 1995, 18, 715–727. [Google Scholar] [CrossRef]
- Daly, R.; Lostroh, C. Genetic analysis of the Salmonella transcription factor HilA. Can. J. Microbiol. 2008, 54, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Lostroh, C.; Bajaj, V.; Lee, C.A. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 2000, 37, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Hüttener, M.; Dietrich, M.; Paytubi, S.; Juárez, A. HilA-like regulators in Escherichia coli pathotypes: The YgeH protein from the enteroaggregative strain 042. BMC Microbiol. 2014, 14, 268. [Google Scholar] [CrossRef] [PubMed]
- Pallen, M.J.; Francis, M.; Fütterer, K. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 2003, 223, 53–60. [Google Scholar] [CrossRef][Green Version]
- Carter, M.; Pham, A. Complete genome sequence of a natural Escherichia coli O145:H11 isolate that belongs to phylogroup A. Genome Announc. 2018, 6, e00349-18. [Google Scholar]
- Taniguchi, T.; Akeda, Y.; Haba, A.; Yasuda, Y.; Yamamoto, K.; Honda, T.; Tochikubo, K. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect. Immun. 2001, 69, 5864–5873. [Google Scholar] [CrossRef]
- Blokesch, M.; Schoolnik, G. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007, 3, e81. [Google Scholar] [CrossRef]
- Reiter, W.; Palm, P.; Yeats, S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989, 17, 1907–1914. [Google Scholar] [CrossRef]
- Brown, E.W.; LeClerc, E.; Li, B.; Payne, W.L.; Cebula, T.A. Phylogenetic evidence for horizontal transfer of mutS alleles among naturally occurring Escherichia coli strains. J. Bacteriol. 2001, 183, 1631–1644. [Google Scholar] [CrossRef]
- Brown, E.W.; Mammel, M.K.; LeClerc, E.; Cebula, T.A. Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens. Proc. Natl. Acad. Sci. USA 2003, 100, 15676–15681. [Google Scholar] [CrossRef]
- Cao, G.; Meng, J.; Strain, E.; Stones, R.; Pettengill, J.; Zhao, S.; McDermott, P.; Brown, E.; Allard, M. Phylogenetics and differentiation of Salmonella Newport lineages by whole genome sequencing. PLoS ONE 2013, 8, e55687. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Lecointre, G.; Darlu, P.; Tenaillon, O.; Acquaviva, C.; Sayada, C.; Sunjevaric, I.; Rothstein, R.; Elion, J.; Taddei, F.; et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 2000, 103, 711–721. [Google Scholar] [CrossRef]
- Funchain, P.; Yeung, A.; Stewart, J.L.; Lin, R.; Slupska, M.M.; Miller, J.H. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 2000, 154, 959–970. [Google Scholar] [PubMed]
- Kotewicz, M.L.; Li, B.; Levy, D.D.; LeClerc, E.; Shifflet, A.W.; Cebula, T.A. Evolution of multi-gene segments in the mutS-rpoS intergenic region of Salmonella enterica serovar Typhimurium LT2. Microbiology 2002, 148, 2531–2540. [Google Scholar] [CrossRef]
- Herbelin, C.; Chirillo, S.; Melnick, K.; Whittam, T.S. Gene conservation and loss in the mutS-rpoS genomic region of the pathogenic Escherichia coli. J. Bacteriol. 2000, 182, 5381–5390. [Google Scholar] [CrossRef]
- Brown, E.W.; Kotewicz, M.L.; Cebula, T.A. Detection of recombination among Salmonella enterica strains using the incongruence length difference test. Mol. Phylogenetics Evol. 2002, 24, 102–120. [Google Scholar] [CrossRef]
- LeClerc, J.; Li, B.; Payne, W.L.; Cebula, T.A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 1996, 274, 1208–1211. [Google Scholar] [CrossRef]
- Duong, D.A.; Stevens, A.M.; Jensen, R.V. Complete genome assembly of Pantoea stewartii subsp. stewartii DC283, a corn pathogen. Genome Announc. 2017, 5, e00435-17. [Google Scholar]
- Vernikos, G.S.; Thomson, N.R.; Parkhill, J. Genetic flux over time in the Salmonella lineage. Genome Biol. 2007, 8, R100. [Google Scholar] [CrossRef]
- Dorman, C.J.; Dorman, M. Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium. Environ. Microbiol. 2017, 19, 3834–3845. [Google Scholar] [CrossRef]
- Bustamante, V.; Martínez, L.; Santana, F.; Knodler, L.; Steele-Mortimer, O.; Puente, J.L. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. USA 2008, 105, 14591–14596. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Blum-Oehler, G.; Mühldorfer, I.; Tschäpe, H. Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol. 1997, 23, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Blum, G.; Ott, M.; Lischewski, A.; Ritter, A.; Imrich, H.; Tschäpe, H.; Hacker, J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 1994, 62, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Bach, S.; Buchrieser, C.; Prentice, M.; Guiyoule, A.; Msadek, T.; Carniel, E. The High-Pathogenicity Island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect. Immun. 1999, 67, 5091–5099. [Google Scholar] [CrossRef]
- Fisher, C.R.; Davies, N.; Wyckoff, E.E.; Feng, Z.; Oaks, E.V.; Payne, S.M. Genetics and virulence association of the Shigella flexneri Sit iron transport system. Infect. Immun. 2009, 77, 1992–1999. [Google Scholar] [CrossRef]
- Janakiraman, A.; Slauch, J.M. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 2000, 35, 1146–1155. [Google Scholar] [CrossRef]
- Sun, W.-S.W.; Syu, W.-J.; Ho, W.-L.; Lin, C.-N.; Tsai, S.-F.; Wang, S.-H. SitA contributes to the virulence of Klebsiella pneumoniae in a mouse infection model. Microbes Infect. 2014, 16, 161–170. [Google Scholar] [CrossRef]
- Pfeiffer, V.; Sittka, A.; Tomer, R.; Tedin, K.; Brinkmann, V.; Vogel, J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007, 66, 1174–1191. [Google Scholar] [CrossRef]
- Shapiro, B.; Polz, M.F. Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol. 2014, 22, 235–247. [Google Scholar] [CrossRef]
- Redfield, R.J.; Findlay, W.; Bossé, J.; Kroll, J.; Cameron, A.; Nash, J. Evolution of competence and DNA uptake specificity in the Pastuerellaceae. BMC Evol. Biol. 2006, 6, 82. [Google Scholar] [CrossRef] [PubMed]


| Salmonella SPI-1 | Pantoea PSI-2 | Shigella Mxi-Spa | Escherichia ETT2 | Escherichia LEE | Yersinia YSA | Yersinia YSC | Sodalis SSR1 | Sodalis SSR2 | Pseudomonas Psc/Pcr/Pop/Exs | Chromobacterium CPI-1 | Universal 2 | Function 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| avrA | yopP | Effector | ||||||||||
| sprB | Regulator | |||||||||||
| hilC | S0103 | Regulator | ||||||||||
| orgC | corC | Effector | ||||||||||
| orgB | mxiL/mxiN | orgB | escL | YE3555 | yscL | orgAb | pscL | corB | sctL | Needle assembly | ||
| orgA | mxiK | orgA | YE3554 | yscK | orgA | pscK | corA | sctK | Needle assembly | |||
| prgK | psaJ | mxiJ/yscJ | eprK | escJ | ysaJ | yscJ | ysaJ | prgK | pscJ | cprK | sctJ | Inner mb ring |
| prgJ | psaI | mxiI | eprJ | escI | ysaI | yscI | ysaI | prgJ | pscI | cprJ | sctI | Needle subunit |
| prgI | psaG | mxiH | eprI | escF | YE3551 | yscF | ysaG | prgI | pscF | cprI | sctF | Needle subunit |
| prgH | psaF | mxiG | eprH | escD | YE3550 | yscD | ysaF | prgH | pscD | cprH | sctD | Inner mb ring |
| hilD | Regulator | |||||||||||
| hilA | ygeH | hilA | cilA | Regulator | ||||||||
| iagB | psaH | ipgF | ipgF | etgA | ysaH | ysaH | iagB | Transglosylase | ||||
| sptP | yspP | yopH | CV0974 | Effector | ||||||||
| sicP | psa7 | Chaperone | ||||||||||
| iacP | psaC/acpM | ipgG | acpY | ysaC | iacP | Acyl carrier | ||||||
| STM2880 | Unknown | |||||||||||
| sipA | ipaA | yspA | yspA | cipA | Effector | |||||||
| sipD | pspD | ipaD | espA | yspD | lcrV | yspD | pcrV | cipD | Tip complex | |||
| sipC | pspC | ipaC | espB | yspC | yopD | yspC | popD | cipC | Effector | |||
| sipB | pspB | ipaB | espD | yspB | yopB | yspB | popB | cipB | Effector | |||
| sicA | pchA | ipgC | ygeG | cesD | sycB | sycD | sycB | sicA | pcrH | cicA | Chaperone | |
| spaS | psaU | spa40 | epaS | escU | ysaU | yscU | ysaU | spaS | pscU | cpaS | sctU | Export apparatus |
| spaR | psaT | spa29 | epaR | escT | ysaT | yscT | ysaT | spaR | pscT | cpaR | sctT | Export apparatus |
| spaQ | psaS | spa9 | epaQ | escS | ysaS | yscS | ysaS | spaQ | pscS | cpaQ | sctS | Export apparatus |
| spaP | psaR | spa24 | epaP | escR | ysaR | yscR | ysaR | spaP | pscR | cpaP | sctR | Export apparatus |
| spaO | psaQ | spa33 | epaO | escQ/sepQ | ysaQ | yscQ | ysaQ | spaO | pscQ/hrcQ | cpaO | sctQ | Cytoplasmic ring |
| invJ | psaP | spa32/spaN | eivJ | escP/orf16 | yspN | yscP | ysaP | spaN | pscP | cpaN | sctP | Needle assembly |
| invI | psaO | spa13/spaM | eivI | escO/escA/orf15 | YE3543A | yscO | ysaO | spaM | pscO | cpaM | sctO | Needle assembly |
| invC/spaL | psaN | spa47/spaL | eivC | escN | ysaN | yscN | ysaN | invC | pscN | civC | sctN | ATPase |
| invB | psaK | spa15/spaK | ysaK | ysaK | invB | civB | Chaperone | |||||
| invA | psaV | mxiA | eivA | escV | ysaV | yscV/lcrD | ysaV | invA | pcrD | civA | sctV | Export apparatus |
| invE | psaW | mxiC | eivE | sepL/sepD | ysaW | yopN/tyeA | ysaW | invE | popN | civE | sctW | Export regulator |
| invG | psaC | mxiD | eivG | escC | ysaC | yscC | ysaC | invG | pscC | civG | sctC | Outer mb ring |
| invF | mxiE/ysaE | mxiE | eivF | ysaE | virF | ysaE | invF | exsA | civF | Regulator | ||
| invH | mxiM | yscW | exsB | Export apparatus | ||||||||
| pigA | Unknown | |||||||||||
| pigB | Unknown | |||||||||||
| STM2903 | Insertion element | |||||||||||
| pigC | Unknown | |||||||||||
| pigD | Unknown | |||||||||||
| STM2906 | Transposase | |||||||||||
| pphB | Phosphatase | |||||||||||
| STM2908 | Unknown |
| Genus | Species | Strain | % GC | Mbp | Genbank Accession | Genbank Assembly Version |
|---|---|---|---|---|---|---|
| Citrobacter | freundii | CFNIH1 | 52.2 | 5.09 | NZ_CP007557.1 | GCA_000648515.1_ASM64851v1 |
| Citrobacter | koseri | ATCC BAA-895 | 53.8 | 4.72 | NC_009792.1 | GCA_000018045.1_ASM1804v1 |
| Enterobacter | cloacae subsp. cloacae | ATCC 13047 | 52.47 | 5.32 | NC_014121.1 | GCA_000025565.1_ASM2556v1 |
| Enterobacter | lignolyticus | SCF1 | 57.2 | 4.81 | NC_014618.1 | GCA_000164865.1_ASM16486v1 |
| Escherichia | albertii | KF1 | 49.7 | 4.7 | NZ_CP007025.1 | GCA_000512125.1_ASM51212v1 |
| Escherichia | coli | O157:H7 Sakai | 50.45 | 5.59 | NC_002695.1 | GCA_000008865.1_ASM886v1 |
| Escherichia | coli | K12 MG1655 | 50.8 | 4.64 | NC_000913.3 | GCA_000005845.2_ASM584v2 |
| Escherichia | coli | IAI39 | 50.6 | 5.13 | NC_011750.1 | GCA_000026345.1_ASM2634v1 |
| Escherichia | coli | O104:H4 str 2011C-3493 | 50.63 | 5.44 | NC_018658.1 | GCA_000299455.1_ASM29945v1 |
| Escherichia | fergusonii | ATCC 35469 | 49.88 | 4.64 | NC_011740.1 | GCA_000026225.1_ASM2622v1 |
| Klebsiella | pneumoniae subsp. pneumoniae | HS11286 | 57.14 | 5.68 | NC_016845.1 | GCA_000240185.2_ASM24018v2 |
| Klebsiella | oxytoca | CAV1374 | 55.26 | 7.23 | NZ_CP011636.1 | GCA_001022195.1_ASM102219v1 |
| Pantoea | agglomerans | IG1 | 55.02 | 5.12 | NZ_CP016889.1 | GCA_001709315.1_ASM170931v1 |
| Pantoea | ananatis | LMG 5342 | 53.4 | 4.14 | NC_016816.1 | GCA_000283875.1_ASM28387v1 |
| Pantoea | stewartii | DC283 | 54.2 | 4.53 | NZ_CP017581.1 | GCA_002082215.1_ASM208221v1 |
| Pseudomonas | aeruginosa | PAO1 | 66.6 | 6.26 | NC_002516.2 | GCA_000006765.1_ASM676v1 |
| Salmonella | bongori | NCTC 12419 | 51.3 | 4.46 | NC_015761.1 | GCA_000252995.1_ASM25299v1 |
| Salmonella | enterica subsp. enterica Enteritidis | P125109 | 52.2 | 4.69 | NC_011294.1 | GCA_000009505.1_ASM950v1 |
| Salmonella | enterica subsp. enterica Typhi | CT18 | 51.88 | 5.13 | NC_003198.1 | GCA_000195995.1_ASM19599v1 |
| Salmonella | enterica subsp. enterica Typhimurium | LT2 | 52.22 | 4.95 | NC_003197.2 | GCA_000006945.2_ASM694v2 |
| Salmonella | enterica subsp. enterica Paratyphi C | RKS4594 | 52.21 | 4.89 | NC_012125.1 | GCA_000018385.1_ASM1838v1 |
| Salmonella | enterica subsp. enterica Senftenberg | ATCC 43845 | 51.93 | 5.26 | CP019194.1 | GCA_000486525.2_ASM48652v2 |
| Salmonella | enterica subsp. arizonae | 62z23 RSK2980 | 51.4 | 4.6 | NZ_CP006693.1 | GCA_000018625.1_ASM1862v1 |
| Salmonella | enterica subsp. diarizonae | HZS154 | 51.4 | 5.09 | CP023345.1 | GCA_002794415.1_ASM279441v1 |
| Shigella | flexneri | 2a 301 | 50.67 | 4.83 | NC_004337.2 | GCA_000006925.2_ASM692v2 |
| Sodalis | glossinidius | mortisans | 54.51 | 4.29 | NC_007712.1 | GCA_000010085.1_ASM1008v1 |
| Sodalis | praecaptivus | HS1 | 57.13 | 4.29 | NZ_CP006569.1 | GCA_000517425.1_ASM51742v1 |
| Yersinia | enterocolitica subsp. enterocolitica | 8081 | 47.25 | 4.68 | NC_008800.1 | GCA_000009345.1_ASM934v1 |
| Yersinia | pestis | CO92 | 47.61 | 4.83 | NC_003143 | GCA_000009065.1_ASM906v1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lerminiaux, N.A.; MacKenzie, K.D.; Cameron, A.D.S. Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System. Microorganisms 2020, 8, 576. https://doi.org/10.3390/microorganisms8040576
Lerminiaux NA, MacKenzie KD, Cameron ADS. Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System. Microorganisms. 2020; 8(4):576. https://doi.org/10.3390/microorganisms8040576
Chicago/Turabian StyleLerminiaux, Nicole A., Keith D. MacKenzie, and Andrew D. S. Cameron. 2020. "Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System" Microorganisms 8, no. 4: 576. https://doi.org/10.3390/microorganisms8040576
APA StyleLerminiaux, N. A., MacKenzie, K. D., & Cameron, A. D. S. (2020). Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System. Microorganisms, 8(4), 576. https://doi.org/10.3390/microorganisms8040576





