Abstract
Dinoflagellates have some of the largest genomes, and their liquid-crystalline chromosomes (LCCs) have high degrees of non-nucleosomal superhelicity with cation-mediated DNA condensation. It is currently unknown if condensins, pentameric protein complexes containing structural maintenance of chromosomes 2/4, commonly involved in eukaryotic chromosomes condensation in preparation for M phase, may be involved in the LCC structure. We find that CcSMC4p (dinoflagellate SMC4 homolog) level peaked at S/G2 phase, even though LCCs do not undergo global-decondensation for replication. Despite the differences in the chromosomal packaging system, heterologous CcSMC4p expression suppressed conditional lethality of the corresponding fission yeast mutant, suggesting conservation of some canonical condensin functions. CcSMC4p-knockdown led to sustained expression of the S-phase marker PCNAp, S-phase impediment, and distorted nuclei in the early stage of CcSMC4p depletion. Prolonged CcSMC4p-knockdown resulted in aneuploidal cells and nuclear swelling with increasing LCC decompaction–decondensation. Cumulatively, our data suggested CcSMC4p function was required for dinoflagellate S-phase progression, and we propose that condensin-mediated higher-order compaction provisioning is involved in the provision of local rigidity for the replisome.
1. Introduction
Accumulating evidence suggests chromatin remodeling between interphase and mitosis goes through condensin-dependent stages to their higher-order organization of nucleosomal chromosomes []. Condensin dysfunction leads to phenotypes depending on the chromosomal interactive point of the cell cycle. Condensin structural maintenance of chromosomes 2 and 4 subunits (SMC2 and SMC4) are essential genes in nucleosomal eukaryotes.
Dinoflagellate liquid crystalline chromosomes (LCCs) are the only non-nucleosomal eukaryotic chromosome condensation-organization; despite having some of the largest genome sizes, micrococcal nuclease-digested LCC preparations do not exhibit a nucleosomal ladder pattern [,,]. Ultrastructural chromosome spread studies have revealed a high level of DNA superhelicity []. Strong birefringence and re-interpreted biophysical data support LCCs having liquid crystalline anisotropic organization in vivo [,]. Divalent cation-mediated condensation and DNA supercoils are synergistic in nurturing DNA liquid-crystalline phase transitions [], and LCCs have 2–3 fold higher chromosomal divalent cations when compared to nc-chromosomes []. Cation chelation led to the orchestrated remodeling of higher-order structures, contrasted with little changes in nucleosomal chromatin following similar treatment [,,]. LCCs recompacted upon re-introduction of divalent cations after mild chelation [], implicating cation-mediated superhelical subunit modularity [,].
Condensins are pentameric protein complexes composed of SMC2p (cut 14 is fission yeast homologue) and SMC4p (cut 3p) core subunits, and three non-SMC proteins (CapHp/cnd2p, CAP-D2p/cnd1p, and CAP-Gp/cnd3p) []. Each SMC protein has two terminal Walker domains, a central Hinge segment, and the long helix-loop-helix coiled-coils in between; SMC2 and SMC4 subunits fold back at central hinge domains and hetero-dimerize between their n- and c-terminal Walker domains, forming two head ATPase domains. In vivo degron experiments have suggested chromatins are restraint between long SMC2/4 heterodimeric helix–loop–helix domains, which form structures referred to as the condensin “rings” []. The Head domains bind to the three non-SMC proteins, chromosome associated protein H (CapHcnd2) and the two heat repeat-motif-containing proteins CAP-D2cnd1 and CAP-Gcnd3, forming a compressible belt around chromatin domains [,,]. There is much controversy regarding condensin molecular mechanisms of action, with an ongoing model involving DNA loop extrusion with condensin sliding [,], in addition to the linkage of condensin-entrapped domains []. Bacterial SMC homologs (MukBEF, MukB) organized nucleoid DNA into a repetitive stable structure [,].
Predicted polypeptides of CcSMC4p and CcSMC2p (from Crypthecodinium cohnii transcriptomes) contained all conserved domains of eukaryote SMC homologs (Figure S1), including the N- and C-terminal ATPase walker motifs and the central hinge region. Higher levels of chromosomal cations and cation-aided liquid-crystalline phase transition might have made condensins redundant. However, the high level of superhelicity and the absence of nucleosomal architecture inspired us to pioneer this functional investigation of a dinoflagellate chromosomal protein. We adopted multiple approaches to investigate possible functions of condensin subunit CcSMC4p, including the first gene-knockdown of a dinoflagellate chromosomal protein. Our data implicated dinoflagellate condensins being required for proper S-phase progression, likely related to their provision of replisome organizational rigidity.
2. Materials and Methods
2.1. Dinoflagellate Cell Cultures and Flow Cytometry
The heterotrophic dinoflagellate Crypthecodinium cohnii (University of Texas Culture Collection strain 1649) was maintained in the MLH minimum medium without light at 28 °C (Tuttle and Loeblich III, 1975). Cells for flow cytometry were prepared as previously described []. Knockdown experiments and cell-cycle studies were conducted with C. cohnii cells. There are established transfection and coccoid stage-swarmer release synchronization protocol [,].
2.2. In Silico Analysis
ORFs encoding CcSMC2 and CcSMC4 (Crypthecodiunium cohnii) were originally cloned by sequence homology and subsequently confirmed with contigs constructed with expressed sequence tags from transcriptomes. The predicted amino acid sequences of dinoflagellate CcSMC2 and CcSMC4 ORFs were compared with other eukaryotic homologs from selected eukaryotic homologs (Figure S1) using multiple alignment tools (https://www.ebi.ac.uk/Tools/msa/clustalo/). GenBank accession number: KC160503 for CcSMC2 and KC160504 for CcSMC4.
2.3. Immunological Techniques, Preparation of Cell Lysates and Molecular Biological Techniques
Purified bacterial expressed recombinant polypeptides (His-tagged, vector PQE30, Qiagen) containing N-terminal coiled-coil region (predicted 369th–769th amino acids of CcSMC4p) and C-terminal coiled-coil region (predicted 895th–1323rd amino acids of CcSMC4p) were the immunogens for the anti-CcSMC4p antibody. Baculoviral-expressed (Thermal Fisher Scientific) recombinant polypeptides encoding full-length CcSMC2p were the immunogens for the anti-CcSMC2p antibody. All immunological manipulations were carried out according to the published protocols [,]. The affinity-purified antibodies against CcSMC4p or CcSMC2p recognized immunoreactive bands with approximate expected molecular weights in C. cohnii cell lysates (~170 kDa and ~137 kDa, respectively; Figure S2B). Antigen affinity-purified antibodies were used in all immunological techniques; all cell lysate blots were pre-cleared with dried acetone bacteria extract before immunoblot analysis [,,].
All fluorescent photomicrographs were taken with a Leica fluorescent microscope (DMLS), and confocal images were taken with Leica SP8 (Leica microsystems, Witzlar, Germany). All molecular biology and protein preparation techniques adopted our previously published protocols, including the preparation of cell lysates [], which was based on a pressure-release cell disruption method (Cell Disruptor, Constant System, UK). Dinoflagellate proliferative cell nuclear antigen (PCNA), which had expression peak at S-phase [,], was the S-phase marker. The anti-PCNAp (PC10) and anti-α-tubulin monoclonal antibodies were from ZyMed Corporation (San Francisco, CA, USA). All chemicals were from Sigma Aldrich unless otherwise stated.
2.4. Functional Suppression of cut3-477 Conditional Lethality through CcSMC4p Expression
Potential CcSMC4p-mediated suppression of conditional lethality of cut3-477, a temperature-sensitive fission yeast (Schizosaccharomyces pombe) mutant in SMC4 homolog [,], was conducted for functional studies. Fission yeast strains and expression vectors were gifts from Prof. Paul Nurse (Oxford University) and Prof. Mitsuhiro Yanagida (Okinawa Institute of Science and Technology, Okinawa, Japan). The vector expressing CcSMC4 open reading frame, driven by the thiamine-repressible nmt1 promoter, was constructed in the Rep3x vector [] and transformed into fission yeast mutants under unrepressed (0 µM thiamine), fully-repressed (15 µM thiamine), or semi-repressed (2.5 µM thiamine) conditions. Fission yeast cell lysates were prepared from transformed cells incubated in supplemented Edinburgh minimal media (EMM )with or without thiamine (15 µM) for 24 h at 28 °C. All yeast manipulation and protein preparations followed established protocols [].
2.5. Antisense Oligonucleotide-Mediated Gene-Knockdown Experiment
We adopted an antisense-oligonucleotide based gene-knockdown protocol [] with lipofection-spheroplasts mediated transfection in Crypthecodinium cohnii [,], according to manufacturer protocol (Lipofectamine, Invitrogen). The design of antisense oligonucleotides was aided with unpaired bases predicted on CcSMC4 mRNA (http://rtools.cbrc.jp/centroidhomfold/) []. Transfecting a combination of two anti-sense ODNs (SMC4-79, 5′-AAGGTGCGGTCAGGTGGAAACC and SMC4-129, 5′-TAGATCGGGTTGTGACGGGCATGAC) gave the best knockdown reduction of CcSMC4p (Figure S2). Control-ODNs, encoding the sense (complementary) sequence of the SMC4-79 oligo, were used as a control in the mock-transfection. Many metazoans have two condensin complexes, differing in their heat-repeat subunits and chromosome functions [], our antisense-oligonucleotide design would have targeted both complexes as the SMC proteins are shared. All experiments were carried out in triplicates, and the representative results are presented.
3. Results
3.1. Cell-Cycle-Phased Expression of CcSMC4p
Condensin functions are regulated in the cell cycle. We followed CcSMC4p and CcSMC2p levels in C. cohnii cell cycle with immunoblot analysis (Figure 1). Despite no apparent LCC decompaction–recompaction cycles in dinoflagellates, low apparent early G1 level (T = 0–4 hr) was elevated at S phase (T = 5–6 hr), before leveling at the G2/M (T = 8–12 hr) (Figure 1). This S-G2 expression pattern implicated possible cell-cycle operations for both SMC homologs; in nucleosomal eukaryotes, the SMC2/SMC4 levels did not change appreciably during the cell cycle []. Further studies should investigate possible post-translational modifications and subcellular locations of condensins.
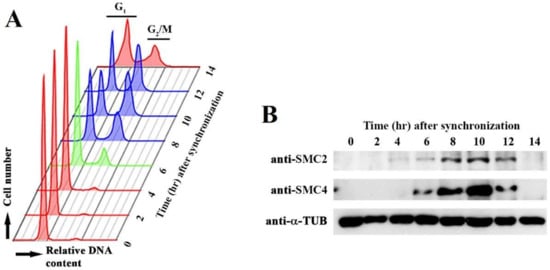
Figure 1.
CcSMC4p (dinoflagellate SMC4 homolog) expression in the cell cycle. (A) Flow cytograms of Crypthecodinium cohnii cells collected at different time points after synchronization at early G1. Red, G1; Green, S phase; purple G2/M cells. (B) Immunoblots of CcSMC2 and CcSMC4 collected at the corresponding time points.
3.2. Heterologous CcSMC4p Expression Suppressed Conditional Lethality of Fission Yeast cut3-477
Suppression of conditional lethality in their corresponding yeast mutants would be a good proof of functional conservation. In addition to substantial evolutionary distances between yeast and dinoflagellates, the heterologous CcSMC4p subunits need to form functional complexes with the yeast SMC2cut3 and non-SMC subunits. This may be too severe a test, but we were hopeful CcSMC4p might at least partially suppress the cut phenotype in the corresponding fission yeast temperature-sensitive mutant cut3-477 [].
Unexpectedly, we observed suppression of cut3-477 conditional lethality with CcSMC4p expression, at semi-repressive condition (2.5 µM thiamine and lesser so at 7.5 µM; Figure 2A,B). Fluorescent microscopy of DAPI-stained rescued cut3-477 cells showed well-compacted nuclei with no cut phenotype at semi-repressed (2.5 µM thiamine) expression, but not under fully repressed condition (15 µM thiamine []; Figure 2B). This phenotypic suppression suggested CcSMC4p had the required canonical SMC4 functions and implicated interactions with the corresponding components in the yeast complexes. Immunoblot confirmed CcSMC4p was expressed in the transformed cut3-477 cells (Figure 2C).
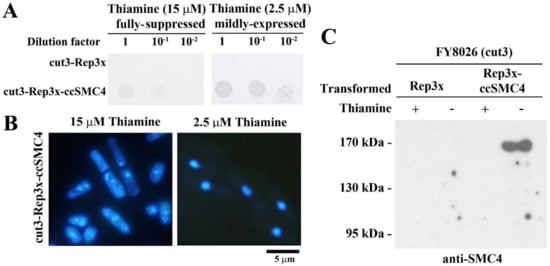
Figure 2.
CcSMC4p expression suppressed conditional lethality of fission yeast cut3-477. (A) Spot-dilution assay demonstrating functional suppression of conditional lethality in cut3-477 with CcSMC4 expression driven by thiamine-repressible nmt1 promotor []. Serial dilutions of overnight transformant cultures were spotted on EMM agar, supplemented with thiamine at fully repressed (15 µM) or partially repressed (2.5 µM) concentrations and incubated at a restrictive temperature of 36 °C for 3 days. (B) Fluorescent photomicrographs of DAPI-stained CcSMC4-transformed cut3-477 cells at partially repressed (2.5 µM) and fully repressed conditions (15 µM). Arrows pointed to septation without completion of genome replication. (C) Immunoblot of cell lysates prepared from CcSMC4-transformed cut3-477 cells with the anti-CcSMC4p antibody.
3.3. Prolong CcSMC4p-Knockdown Led to S-Phase Impediment with Nuclear Swelling
Bearing in mind SMC4-knockouts commonly cause lethality, we adopted a gene-knockdown approach to study possible CcSMC4 functions. Cell proliferation in antisense-ODN-treatment (ak-cells) dropped between 24–33 h when compared to the mock-transfected cells, which continued to proliferate during the experimental period (Figure 3A). This indicated the effect of CcSMC4p-knockdown being manifested early in the experiment and that most ak-cells had been transfected with the antisense-ODNs. Gene-knockdown would not have affected the pre-existing CcSMC4p, which in other systems have both cytoplasmic and nuclear populations []. Immunoblot suggested this CcSMC4p pool dropped to undetectable levels by T = 46–70 h(Figure 3B). However, there was a sustained level of the S-phase marker PCNAp, contrasted with the periodic expression in the mock-transfected cells (Figure 3B). A flow cytogram of ak-cells exhibited sustained sub-2N populations in all later samples, contrasted with the clear gaps between G1–G2 peaks peaks (yellow line, Figure 3C). There is no global decondensation during dinoflagellate DNA replication, a consequence of no normal S-phase bridge between G1–G2, and, hence, there are clear gaps between G1 and G2/M flow peaks [,].
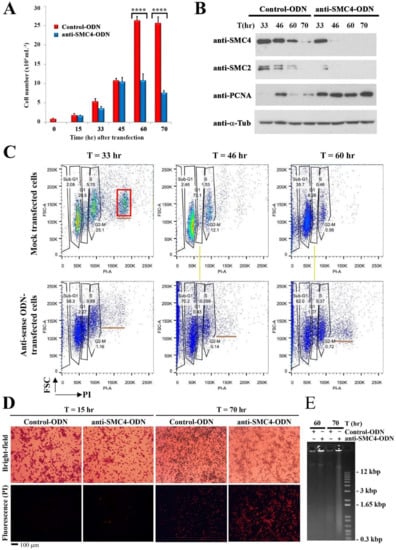
Figure 3.
CcSMC4p-knockdown led to S-phase impediment. (A) Cell number of mock-transfected and ak-cells observed during the experiment from T = 0 to 70 h. Asynchronous cells were used for large-scale experiments. Data represent means ± standard error of three replicate experiments. Asterisk indicates a significant (p < 0.0001) difference compared with the control. (B) Immunoblot analysis for CcSMC4p and PCNAp in cell lysates prepared from later time points (T = 33, 45, 60, 70 h) in mock-transfected and ak-cells harvested at indicated time points post-transfection. Data for earlier time points are presented in Figure 5D. (C) DNA flow cytograms of cells collected from T = 33 to T = 60 h. x-axis: propidium iodide staining; y-axis: forward scatter. (D) Propidium iodide staining of control-ODN/anti-SMC4-ODN-transfected C. cohnii cells at T = 15 and 70 h post-transfection. (E) Ethidium bromide-stained agarose gel showing large molecular weight DNAs extracted from control-ODN and ak-cells harvested at T = 60 and 70 h post-transfection. Red box indicated multiple fission cells39. Yellow lines align anuploidal ak-cells with the S-phase cells in mock transfection.
DNA-flow cytograms exhibited small cell populations with aneuploidy greater than the G2/M peak (Figure 4C, underlined; not 4N, red-lined for comparison). These cells were distinct from the normal clearly separated G1 and G2/M flow peaks, or the multiple fission cells (red square, T = 33 h), which is a special growth rate mediated divisional mode [].
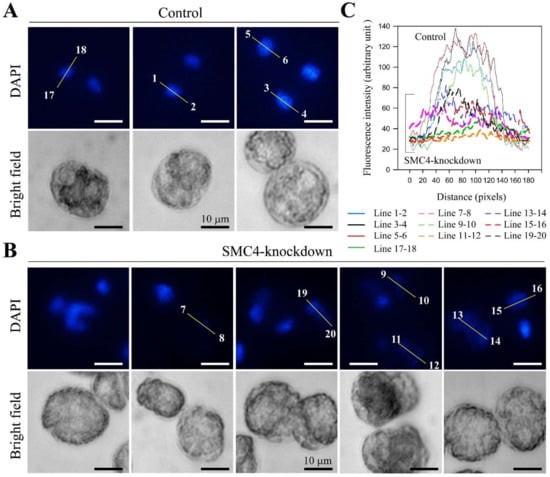
Figure 4.
CcSMC4p-knockdown led to enlarged nuclei with reduction in DAPI staining. Fluorescent photomicrographs of (A) control cells (mock-transfected with sense-ODN); (B) ak-cells (T = 33 h) stained with DAPI; (C) quantification of fluorescent level along transects in (A) and (B). Numbers on the fluorescent photomicrographs indicate the directions and line numbers for each line transect. In CcSMC4p-knockdown cells, much lower levels and spread-out of DAPI-staining suggested enlarged nuclei when compared to control nuclei of mock-transfected cells; some ak-nuclei (e.g., 11–12, 13–14) were more enlarged than others (e.g., 15–16) when contrasted with condensed nuclei in ak-cells and totally decompacted LCCs after AMSA treatment (Figure S3B).
Transect analysis of the DAPI-stained image confirmed that many ak-nuclei (e.g., transect 11–14) increased in volume when compared to regular sizes in control nuclei (1–6, 17–18); all AK-nuclei had level DAPI lower than half that of control nuclei, with some cells losing DAPI stainability (transect 7–8; Figure 4B,C). There were nuclei with different degrees of DAPI-staining, some with distorted nuclei and some with hardly detectable staining. This was consistent with LCC decompaction in some ak-cells, as observed in chelation-mediated LCC decompaction in EDTA-treated nuclei []. As an independent control, we conducted a set of experiments with topoisomerase II inhibitor (AMSA) that induced C. cohnii LCC decondensation []. As in the case of some ak-cells (T = 33, transects 11–12,13–14, 19–20, and T = 70; Figure S3A), the diffused DAPI-staining (after AMSA treatment) of the whole swelled nucleus filled the whole cells (Figure S3B), suggesting decondensed LCCs resulting from CcSMC4p knockdown. These progression from decompacted to decondensed nuclei (T = 33–70 hr), and the aneuploidal profiles (Figure 3C) were consistent with the extended period of CcSMC4p-knockdown leading to increasing mis-accessibility to the pre-replicative complex [], consistent with some ak-cells progressing to partial re-replication. Consistently, PI-staining suggested there were associated increases in cell deaths in latter timepoints associated with DNA breakage; the DNA streak (Figure 3E) LCC decondensation (Figure 4, Figure S3A) and the aneuploidal flow cytograms (Figure 3C) suggested they were not the results of apoptosis. Condensins are also involved in DNA damage repair responses [,] and increasing decompaction with condensin non-replacement will further aggravate replicative stress and amplify no-return effects on LCC decompaction-re-replication.
Continuous overlapping between 1N–2N DNA peaks in later timepoints, instead of clearly distinct G1 versus G2/M peaks in the ak-cells (yellow line), indicated DNA replication was impeded at intermediate stage. Multiple fission, having a 4N stage in addition to 1N and 2N, is a strictly growth-rate dependent mode of cell division in C. cohnii [], represented as a distinct third peak (4N) in DNA flow cytograms of mock-transfected cells (red square box, Figure 3C, but not in ak-cells. Instead of this growth-dependent division mode, the aneuploidal sub-4N (b, underlined) population, towards the latter part of the experiment, was consistent with origin misfiring and semi re-replication in ak-cells.
3.4. CcSMC4p-Knockdown Led to S-Phase Impediment from the First Cell Cycle
These observations suggest that CcSMC4p functions are required for proper replisome progression. We thus sought independent evidence to verify if CcSMC4p-knockdown may affect the first cell cycle, as residue CcSMC4p might not be depleted to a critical level. In experiments using synchronized G1 cells for transfection, retardation of large molecular weight DNAs that is indicative of replication intermediates [] was observed at T = 18 h, corresponding to the relative reduction in CcSMC4p in the mock-transfected cells, suggesting entry to S phase synchronously at between T = 15–18 h (Figure 5B). This did not occur in the ak-cells. CcSMC4p immuno-labeling in the mock-transfected cells (T = 15 h) labeled most cells strongly, with both cytoplasmic and overlapping distribution with DAPI-staining (Figure 3A). CcSMC4p levels in some ak-cells were undetectable, as in the immunoblot and with the pre-immune serum (Figure 5A,B). Most cells in both mock-transfected and ak-cells had condensed DAPI-staining, contrast with cells in later time points of ak-cells (Figure 4). As most cells did not enter the first S phase (cell proliferation halted between T = 24–33 h), this also registered the experimental timeframe, with apparent depletion of CcSMC4p by T = 33 h, to within one extended cell-cycle.
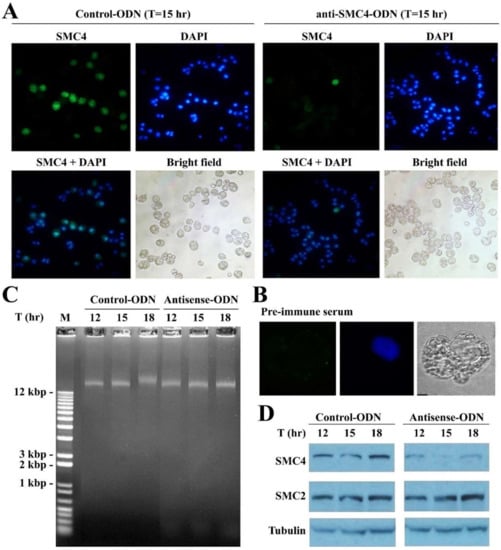
Figure 5.
CcSMC4p-knockdown in synchronized cells led to S-phase impediment. (A) Fluorescent photomicrographs of CcSMC4p immunofluorescent signals and DAPI-staining in double-stained mock-transfected and ak-cells collected at T = 15 h post-transfection. Green immunofluorescent signals correspond to CcSMC4p. Blue fluorescent signals corresponded to DAPI-staining. CcSMC4p + DAPI: Merged CcSMC4p signals with DAPI signals. Not all cells were labelled strongly, which might be related to the highly compacted state of LCCs, and the use of affinity-purified antibody. (B) Immunofluorescent labeling with pre-immune serum. (C) Total DNA preparations from samples in (A) were analyzed with 1% agarose gel post-stained with ethidium bromide. Large molecular weight DNA with retarded relative mobility was indicative of replication intermediates []. (D) Immunoblot analysis of the mock-transfected and ak-cells collected at T = 12th, 15th, 18th hr after transfections for CcSMC4p, CcSMC2p, and α--tubulin.
4. Discussion
No global decondensation occurs during dinoflagellate DNA replication, and LCCs only undergo local decompaction during S-phase. Nucleosomal chromatin rigidity is contributed by nucleosome–nucleosome interactions [], reinforced by this condensin-mediated chromosome domain association linker [,,,,]. Replisome maintenance involved strictly orchestrated nucleosome disassembly–reassembly; during chromosome replication, replisome components interacted with nucleosomal histones ahead of the replicon [,,]. Condensin association was important for replication forks converging at rRNA loci and was proposed to have a role in other loci [,]. Loss of chromosome rigidity resulted in conditional mutants of fission yeast cut3ts and cut14 ts [] and the ring structures of the condensin complexes were important in the maintenance of chromosomal rigidity []. The increasing nuclear size and reduction in chromosome DAPI-staining (Figure 3A, Figure 5A, and Figure S3A) suggest that extended CcSMC4p-knockdown leads to increasing non-replacement of chromosomal condensins, consequenced with decompacted LCC domains with reduced rigidity. CcSMC4p-knockdown-mediated S-phase impediment implicated some condensin functions were required for proper S-phase progression. This implicates a role of condensin-mediated LCC compaction in providing replisome rigidity, failure of which would likely lead to replisome stalling and replicative stresses.
The degree of eventual decondensation was comparable to those observed in cation-chelated [] or with topo II inhibitor-treated LCCs [] (Figure S3B) in which swelled nuclei filled the size of a normal cell. This suggested that condensins were involved in LCC compaction, likely in the restraint of proposed superhelical modules []. Our working model rests with CcSMC4p-knockdown leading to replisome impediment in the early part of the experiment, and further replicative malfunctions with extended CcSMC4p depletion (Figure 5), contributed from origin mis-firings as a result of unwarranted accessibility of pre-replicative complexes [] to cell cycle activators [,]. Premature replication termination led to nucleokinesis-cytokinesis as a result of licensing site exposures at un-recompacted replicons [,]. Condensins also participate in DNA damage responses, including single-strand DNA break repair [,], CcSMC4p-knockdown likely compromised the ability of DNA damage repair in ak-cells, contributing to sustained LCC lesions, further decompacted chromatins, and S-phase arrest. These were also manifested with increasing sub-G1 peaks, aneuploidal DNA peaks, and DNA breakages (T = 60, 70 h; Figure 3), as well as nuclear swelling with LCC decompaction (Figure 4B). Continued CcSMC4p-knockdown led to LCC decondensation in later time points (Figure S3A), in agreement with condensin-mediated compaction contributing to overall LCC compaction. As there was only local decompaction during S phase [], LCC decondensation and S-phase impediment with sustained CcSMC4p-knockdown also implicated condensin loading per replication throughout the LCCs.
Although there is no information relating to replication termination mechanisms [] in LCCs, continued origin mis-firing likely caused depletion of replication complex (e.g., cdc6, cdt1) from the nucleus and mis-signaling of replication termination to G2-coupled chromosome segregation. This is effectively the cut phenotype in which septation occurs without completing nucleokinesis [,,]. Similarly, the titan seed mutant of Arabidopsis ttn3 (Titan mutant of SMC2 homolog) was associated with a leaky phenotype of anaphase with remnant phragmoplasts and polyploid-like replication products []. Bacterial condensins and histone-like HU protein promote origin location and are required for genome segregation [,]. It is interesting that condensins are also required for liquid crystalline genomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/4/565/s1, Figure S1: Predicted amino acid sequences of dinoflagellate CcSMC2 and CcSMC4, Figure S2: Immunoblot analysis of CcSMC4p antisense-knockdown and CcSMC2/4 antibodies, Figure S3: CcSMC4p-knockdown led to nuclear swelling in extended timepoints.
Author Contributions
Conceptualization, J.T.Y.W., T.H.K.Y. and Z.W.; methodology, T.H.K.Y., A.C.M.K. and Z.W.; formal analysis, T.H.K.Y. and A.C.M.K.; investigation, T.H.K.Y., A.C.M.K. and Z.W.; data curation, T.H.K.Y.; writing—original draft preparation, J.T.Y.W., T.H.K.Y.; writing—review and editing, J.T.Y.W.; visualization, T.H.K.Y. and A.C.M.K.; supervision, J.T.Y.W.; project administration, J.T.Y.W.; funding acquisition, J.T.Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from grant HIA05/06.SC01 from the University Grant Council, CERG grant (HKUST6421/06 M) from the Research Grant Council of Hong Kong.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A pathway for mitotic chromosome formation. Science 2018, 359, eaao6135. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.J.; Nooden, L.D. Isolation and partial characterization of dinoflagellate chromatin. Biochim. Biophys. Acta 1974, 349, 402–414. [Google Scholar] [CrossRef]
- Shupe, K.; Rizzo, P.J. Nuclease digestion of dhromatin from the eukaryotic algae Olisthodiscus luteus, Peridinium balticum, and Crypthecodinium cohnii. J. Protozool. 1983, 30, 599–606. [Google Scholar] [CrossRef]
- Herzog, M.; Soyer, M.O. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species Prorocentrum micans E. Eur. J. Cell Biol. 1981, 23, 295–302. [Google Scholar]
- Spector, D.L. Dinoflagellate nuclei. In Dinoflagellates; Academic Press: San Diego, CA, USA, 1984; Volume 1, pp. 107–147. [Google Scholar]
- Chow, M.H.; Yan, K.T.H.; Bennett, M.J.; Wong, J.T.Y. Liquid crystalline chromosomes: Birefringence and DNA condensation. Eukaryot. Cell 2010, 10, 1577–1587. [Google Scholar] [CrossRef]
- Wong, J.T.Y. Architectural organization of dinoflagellate liquid crystalline chromosomes. Microorganisms 2019, 7, 27. [Google Scholar] [CrossRef]
- Ziv, R.; Levin-Zaidman, S.; Gutman, S.B.; Arad, T.; Minsky, A. Supercoiling-regulated liquid-crystalline packaging of topologically-constrained, nucleosome-free DNA molecules. Biochemistry 1984, 33, 14177–14184. [Google Scholar] [CrossRef]
- Levi-Setti, R.; Gavrilov, K.L.; Rizzo, P.J. Divalent cation distribution in dinoflagellate chromosomes imaged by high-resolution ion probe mass spectrometry. Eur. J. Cell Biol. 2008, 87, 963–976. [Google Scholar] [CrossRef]
- Strick, R.; Strissel, P.L.; Gavrilov, K.; Levi-Setti, R. Cation–chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 2001, 155, 899–910. [Google Scholar] [CrossRef]
- Herzog, M.; Soyer, M.O. The native structure of dinoflagellate chromosomes and their stabilization by Ca2+ and Mg2+ cations. Eur. J. Cell Biol. 1983, 30, 33–41. [Google Scholar]
- Sun, S.; Wong, J.T.; Liu, M.; Dong, F. Counterion-mediated decompaction of liquid crystalline chromosomes. DNA Cell Biol. 2012, 31, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.Y. Architectural Organization of Dinoflagellate Liquid Crystalline Chromosomes–the non-nucleosomal eukaryotic chromosome packaging system. Front. Microbiol. 2018. [Google Scholar] [CrossRef]
- Hirano, T. Condensins: Universal organizers of chromosomes with diverse functions. Genes Dev. 2012, 26, 1659–1678. [Google Scholar] [CrossRef] [PubMed]
- Cuylen, S.; Metz, J.; Haering, C.H. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 2011, 18, 894–901. [Google Scholar] [CrossRef]
- Hirano, T. SMC proteins and chromosome mechanics: From bacteria to humans. Philos. Trans. R. Soc. Lond B Biol. Sci. 2005, 360, 507–514. [Google Scholar] [CrossRef]
- Onn, I.; Aono, N.; Hirano, M.; Hirano, T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007, 26, 1024–1034. [Google Scholar] [CrossRef]
- Kschonsak, M.; Merkel, F.; Bisht, S.; Metz, J.; Rybin, V.; Hassler, M.C.H. Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell 2017, 17, 588–600. [Google Scholar] [CrossRef]
- Rana, V.; Bosco, G. Condensin regulation of genome architecture. J. Cell. Physiol. 2017, 232, 1617–1625. [Google Scholar] [CrossRef]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging of DNA loop extrusion by condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Case, R.B.; Chang, Y.P.; Smith, S.B.; Gore, J.; Cozzarelli, N.R.; Bustamante, C. The bacterial condensin MukBEF compacts DNA into a repetitive, stable structure. Science 2004, 305, 222–227. [Google Scholar] [CrossRef][Green Version]
- Petrushenko, Z.M.; Lai, C.-H.; Rai, R.; Rybenkov, V.V. DNA reshaping by MukB right-handed knoting and left-handed supercoiling. J. Biol. Chem. 2006, 281, 4606–4615. [Google Scholar] [CrossRef] [PubMed]
- Kwok, A.C.; Wong, J.T.Y. The activity of a wall-bound cellulase is required for and is coupled to cell cycle progression in the dinoflagellate Crypthecodinium Cohnii. Plant. Cell 2010, 22, 1281–1298. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.Y.; Whiteley, A. An improved method of cell cycle synchronisation for the heterotrophic dinoflagellate Crypthecodinium Cohnii Biecheler analyzed by flow cytometry. J. Exp. Mar. Biol. Ecol. 1996, 197, 91–99. [Google Scholar] [CrossRef]
- Kwok, A.C.; Mak, C.C.; Wong, F.T.; Wong, J.T. Novel method for preparing spheroplasts from cells with an internal cellulosic cell wall. Eukaryot. Cell 2007, 6, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Harlow, E.; Lane, D.P. Antibodies: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1988. [Google Scholar]
- Chan, W.S.; Kwok, A.C.M.; Wong, J.T.Y. Knockdown of dinoflagellate cellulose synthase CesA1 resulted in malformed intracellular cellulosic thecal plates and severely impeded cyst-to-swarmer transition. Front. Microbiol. 2019, 10, 546. [Google Scholar] [CrossRef] [PubMed]
- Leveson, A.C.; Wong, J.T.Y. PCNA-like proteins in dinoflagellates. J. Phycol. 1999, 35, 798–805. [Google Scholar] [CrossRef]
- Liu, J.; Jiao, N.; Hong, H.; Luo, T.; Cai, H. Proliferating cell nuclear antigen (PCNA) as a marker of cell proliferation in the marine dinoflagellate Prorocentrum donghaiense Lu and the green alga Dunaliella salina Teodoresco. J. Appl. Phycol. 2005, 17, 323–330. [Google Scholar] [CrossRef]
- Hirano, T.; Funahashi, S.; Uemura, T.; Yanagida, M. Isolation and characterization of Schizosaccharomyces pombe cut m.utants that block nuclear division but not cytokinesis. EMBO J. 1986, 5, 2973–2979. [Google Scholar] [CrossRef]
- Saka, Y.; Sutani, T.; Yamashita, Y.; Saitoh, S.; Takeuchi, M.; Nakaseko, Y.; Yanagida, M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994, 13, 4938–4952. [Google Scholar] [CrossRef]
- Maundrell, K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990, 265, 10857–10864. [Google Scholar]
- Forsburg, S.L.; Rhind, N. Basic methods for fission yeast. Yeast 2006, 23, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, A.; Kimura, K.; Yokoyama, S.; Hanaoka, F. Cell Cycle-dependent Phosphorylation, Nuclear Localization, and Activation of Human Condensin. J. Biol. Chem. 2004, 279, 4551–4559. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef]
- Ono, T.; Yamashita, D.; Hirano, T. Condensin II initiates sister chromatid resolution during S phase. J. Cell Biol. 2013, 200, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Mak, C.K.M.; Hung, V.K.L.; Wong, J.T.Y. Type II topoisomerase activities in both the G1 and G2/M phases of the dinoflagellate cell cycle. Chromosoma 2005, 114, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Yeung, P.K.; New, D.C.; Leveson, A.; Yam, C.H.; Poon, R.Y.; Wong, J.T. The spindle checkpoint in the dinoflagellate Crypthecodinium cohnii. Exp. Cell Res. 2000, 254, 120–129. [Google Scholar] [CrossRef]
- Lam, C.M.C.; Yeung, P.K.K.; Lee, H.C.; Wong, J.T. Cyclic ADP-ribose links metabolism to multiple fission in the dinoflagellate Crypthecodinium cohnii. Cell Calcium 2009, 45, 346–357. [Google Scholar] [CrossRef]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef]
- Heale, J.T.; Ball, A.R.; Schmiesing, J.A.; Kim, J.-S.; Kong, X.; Zhou, S.; Hudson, D.F.; Earnshaw, W.C.; Yokomori, K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell 2006, 21, 837–848. [Google Scholar] [CrossRef]
- Wu, N.; Yu, H. The Smc complexes in DNA damage response. Cell Biosci. 2012, 2, 5. [Google Scholar] [CrossRef]
- Waseem, N.H.; Labib, K.; Nurse, P.; Lane, D.P. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992, 11, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, K.; Tamura, S.; Shimamoto, Y. Chromatin as a nuclear spring. Biophys. Phys. 2018, 15, 189–195. [Google Scholar] [CrossRef]
- Marko, J.F. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008, 16, 469–497. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.H.; Hirano, T. HEAT repeats-versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 2016, 129, 3963–3970. [Google Scholar] [CrossRef] [PubMed]
- Houlard, M.; Godwin, J.; Metson, J.; Lee, J.; Hirano, T.; Nasmyth, K. Condensin confers the longitudinal rigidity of chromosomes. Nat. Cell Biol. 2015, 17, 771. [Google Scholar] [CrossRef]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 2018, 72, 140–151. [Google Scholar] [CrossRef]
- Foltman, M.; Evrin, C.; De Piccoli, G.; Jones, R.C.; Edmondson, R.D.; Katou, Y.; Nakato, R.; Shirahige, K.; Labib, K. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013, 3, 892–904. [Google Scholar] [CrossRef]
- Johzuka, K.; Terasawa, M.; Ogawa, H.; Ogawa, T.; Horiuchi, T. Condensin loaded onto the Replication Fork Barrier Site in the rRNA Gene Repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 2226–2236. [Google Scholar] [CrossRef]
- Bhaud, Y.; Guillebault, D.; Lennon, J.; Defacque, H.; Soyer-Gobillard, M.O.; Moreau, H. Morphology and behaviour of dinoflagellate chromosomes during the cell cycle and mitosis. J. Cell Sci. 2000, 113, 1231–1239. [Google Scholar]
- Woodward, A.M.; Göhler, T.; Luciani, M.G.; Oehlmann, M.; Ge, X.; Gartner, A.; Jackson, D.A.; Blow, J.J. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006, 173, 673–683. [Google Scholar] [CrossRef]
- Vas, A.; Mok, W.; Leatherwood, J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol. 2001, 21, 5767–5777. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.W.; Jallepalli, P.V.; Huneycutt, B.J.; Kelly, T.J. Interaction of the S phase regulator cdc18 with cyclin-dependent kinase in fission yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 6142–6147. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507. [Google Scholar] [CrossRef] [PubMed]
- Galarreta, A.; Lecona, E.; Valledor, P.; Ubieto, P.; Lafarga, V.; Specks, J.; Fernandez-Capetillo, O. USP7 couples DNA replication termination to mitotic entry. bioRxiv 2018, 1, 305318. [Google Scholar] [CrossRef]
- Sutani, T.; Yuasa, T.; Tomonaga, T.; Dohmae, N.; Takio, K.; Yanagida, M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Develop. 1999, 13, 2271–2283. [Google Scholar] [CrossRef]
- Samejima, I.; Matsumoto, T.; Nakaseko, Y.; Beach, D.; Yanagida, M. Identification of seven new cut genes involved in Schizosaccharomyces pombe mitosis. J. Cell Sci. 1993, 105, 135–143. [Google Scholar]
- Liu, C.-m.; McElver, J.; Tzafrir, I.; Joosen, R.; Wittich, P.; Patton, D.; Van Lammeren, A.A.M.; Meinke, D. Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant. J. 2002, 29, 405–415. [Google Scholar] [CrossRef]
- Lioy, V.S.; Cournac, A.; Marbouty, M.; Duigou, S.; Mozziconacci, J.; Espéli, O.; Boccard, F.; Koszul, R. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 2018, 172, 771–783. [Google Scholar] [CrossRef]
- Trojanowski, D.; Hołówka, J.; Zakrzewska-Czerwińska, J. Where and When Bacterial Chromosome Replication Starts: A Single Cell Perspective. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).