Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
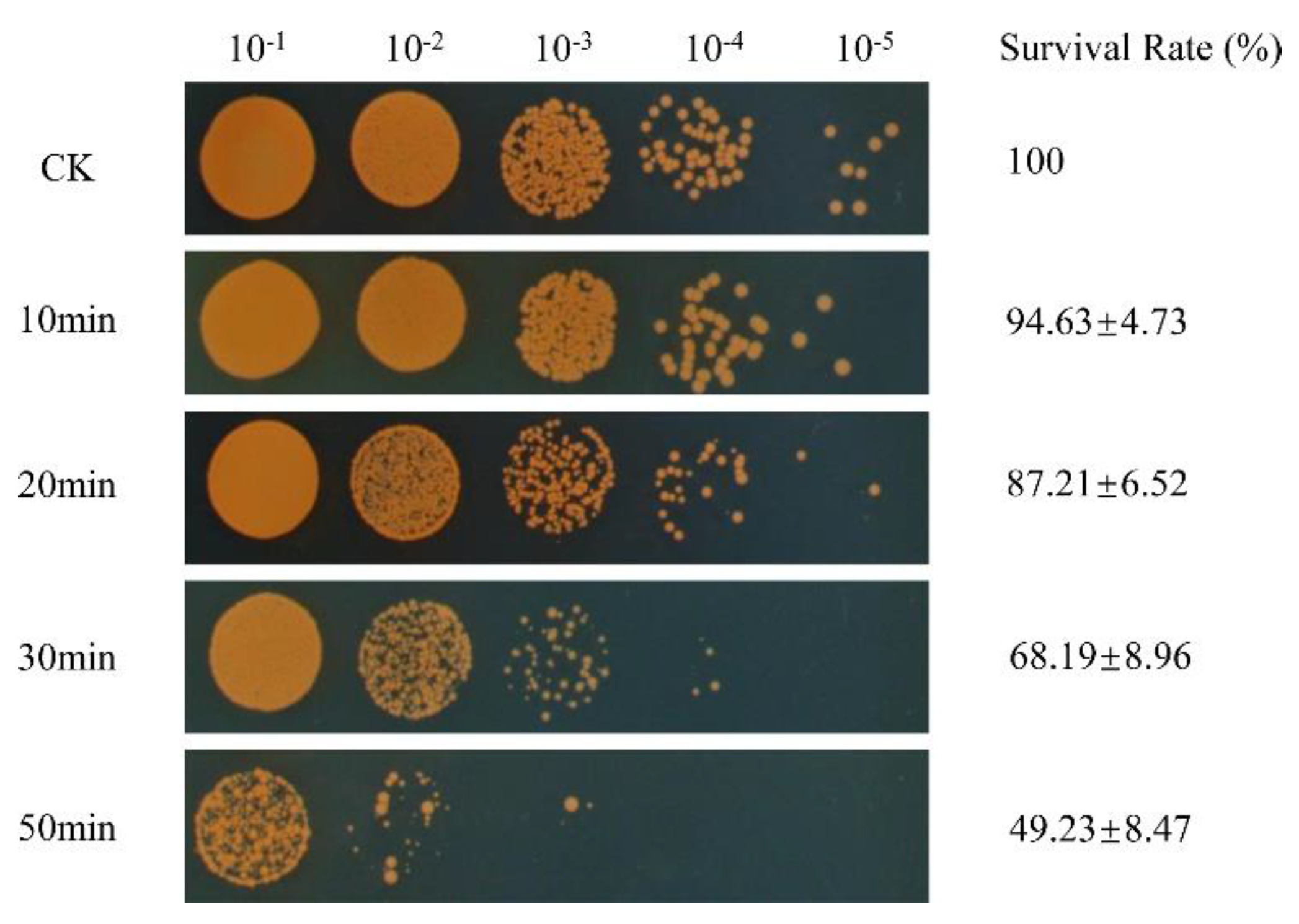
2.2. Cell Survival under Oxidative Stress
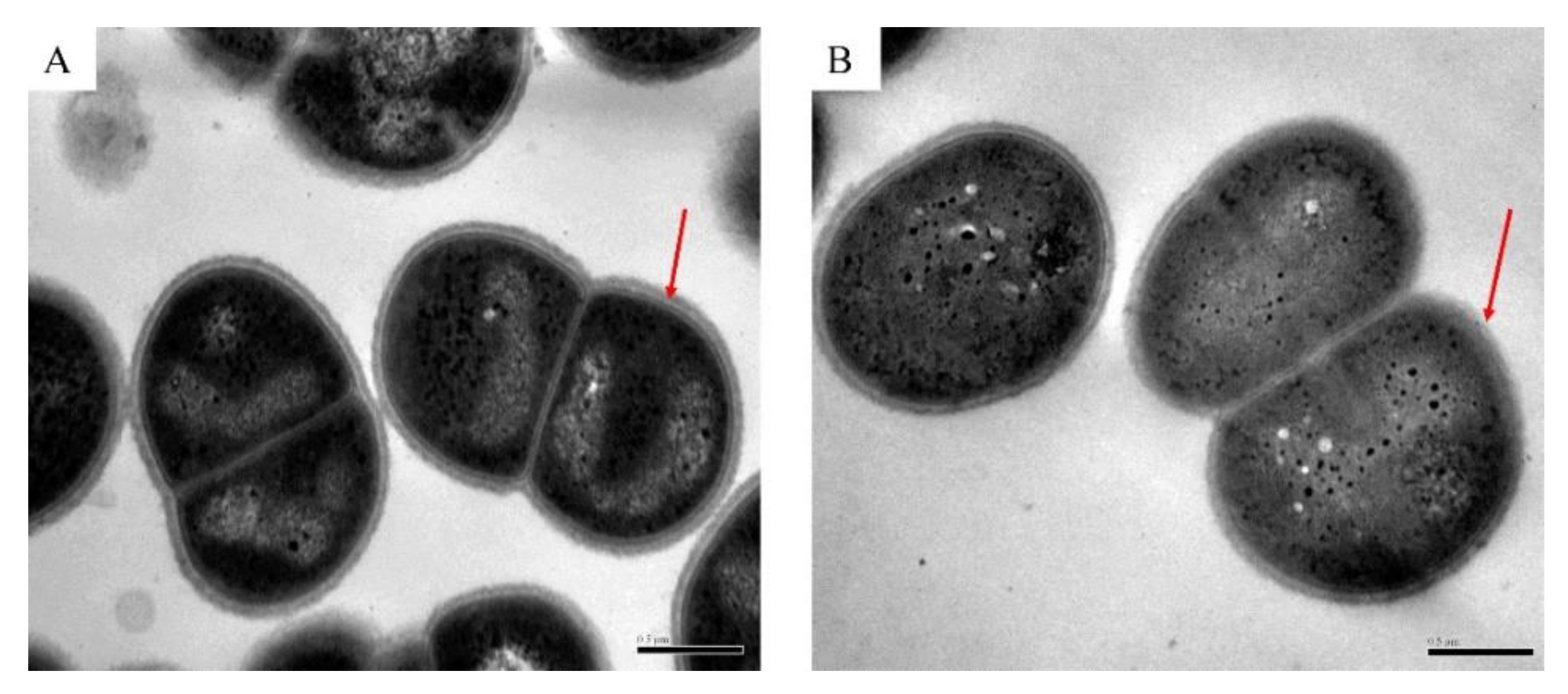
2.3. Transmission Electron Microscope (TEM)
2.4. Protein Extraction and Digestion
2.5. LC–MS/MS (Liquid Chromatography-Mass Spectrometry/Mass Spectrometry) Analysis by Q-Exactive and Sequence Database Searching
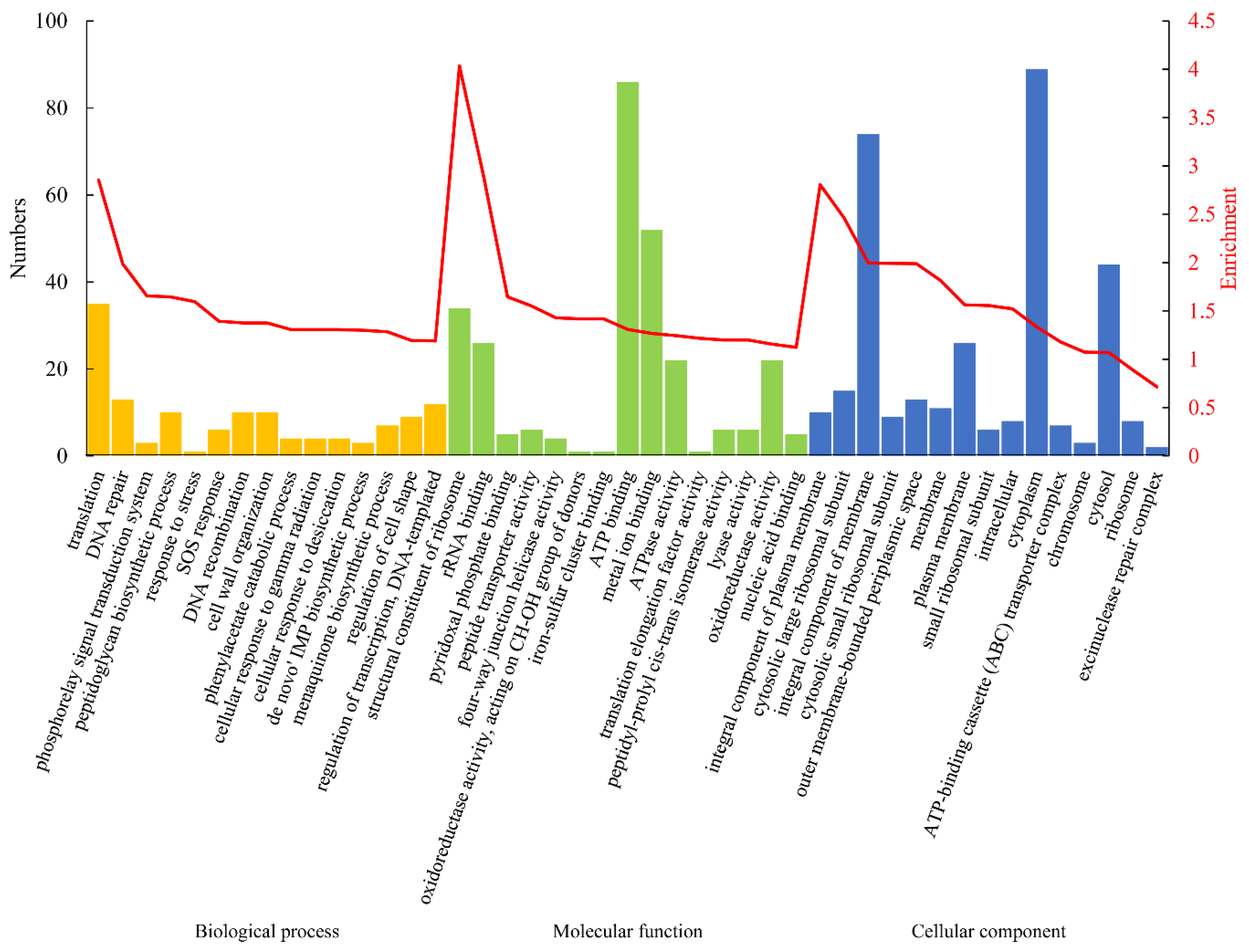
2.6. GO and KEGG Enrichment Analysis
2.7. Protein–Protein Interaction Prediction
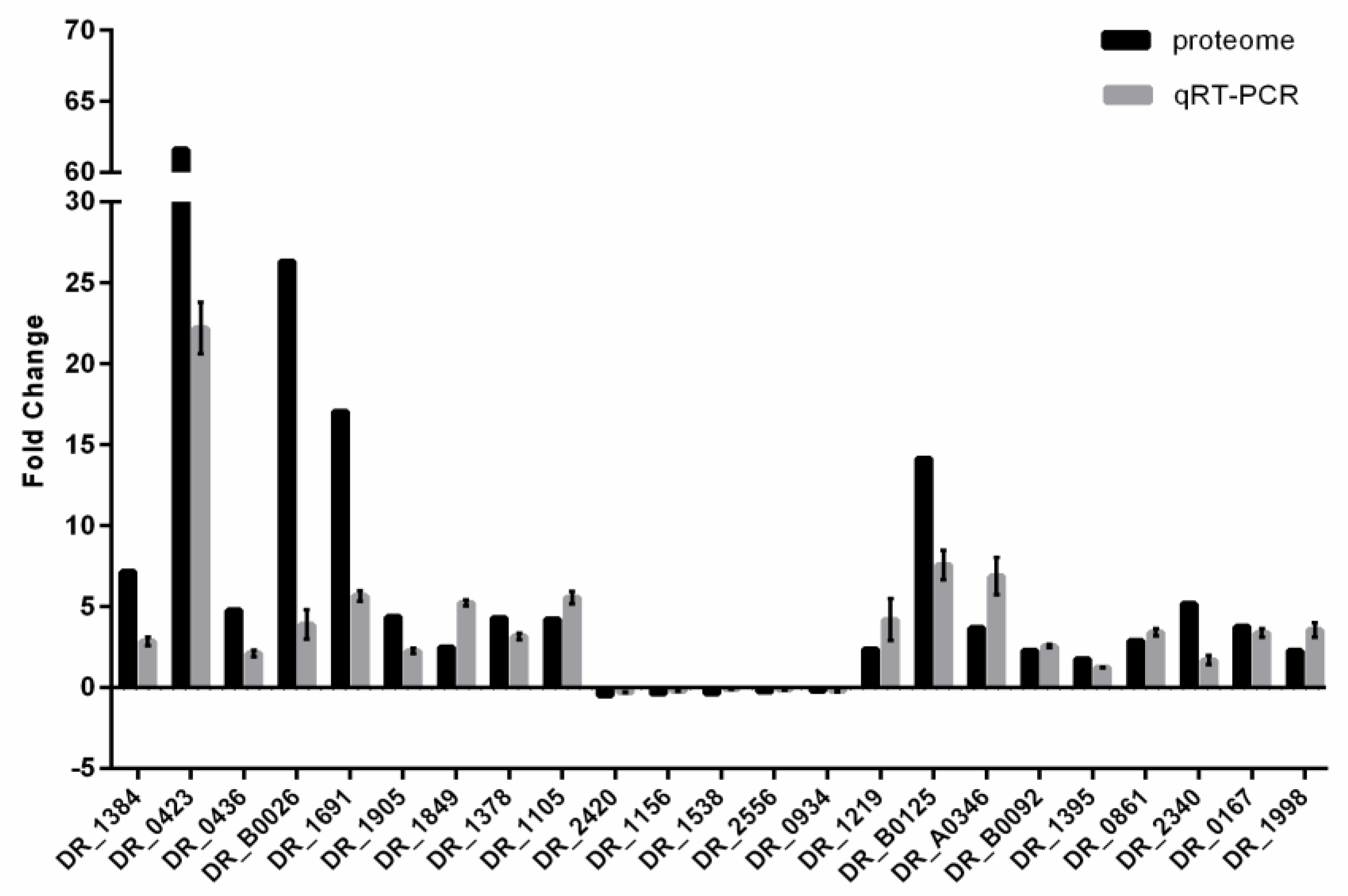
2.8. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Survival of D. radiodurans under Oxidative Stress
3.2. Global Analysis of the Differentially Expressed Proteins (DEPs) in DC and DH
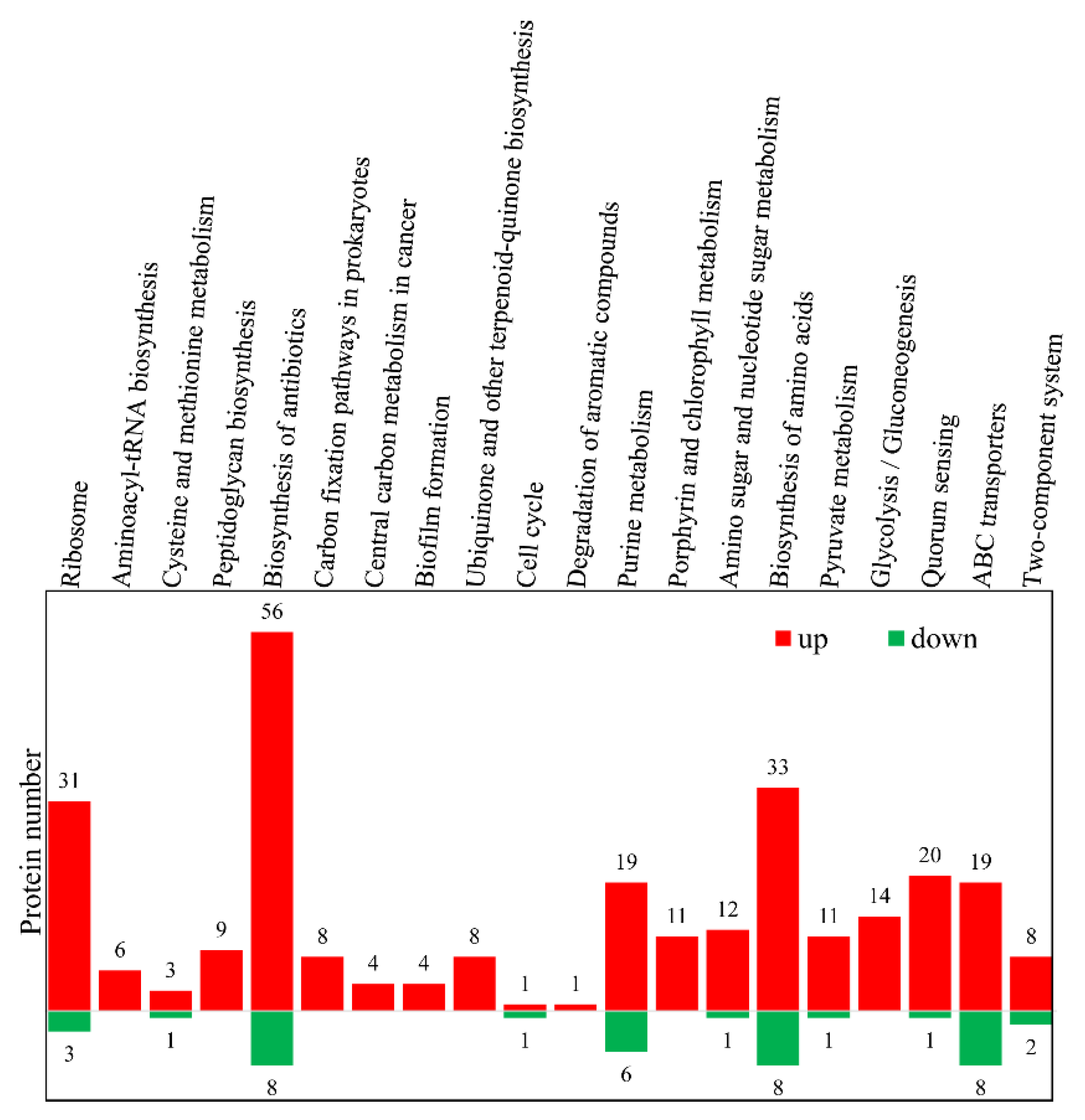
3.3. Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genome (KEGG) Analyses of the DEPs
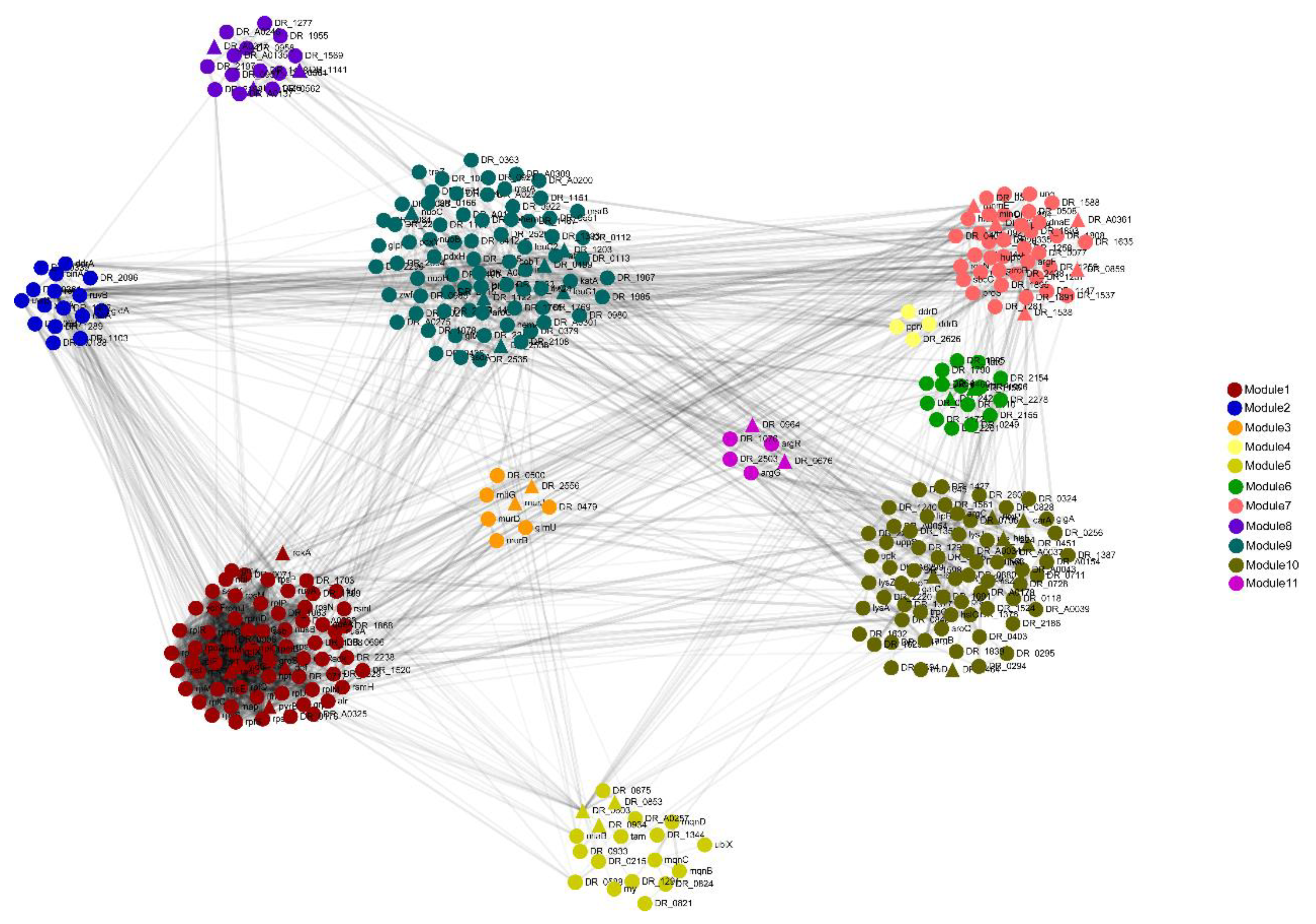
3.4. Protein–Protein Interaction (PPI) Network of DEPs
3.5. Differential Proteins between DC and DH Enriched in Antioxidant Activity
3.5.1. Catalase Detoxification of H2O2
3.5.2. DNA-Binding Proteins during the Stationary Phase (Dps)
3.5.3. Proteins Involved in Carotenoid Synthesis
3.5.4. Mn2+/Fe2+ Homeostasis
3.5.5. The Cyclic AMP (Adenosine MonoPhosphate) Receptor Protein DdrI
3.5.6. Radiation Response Metalloprotease IrrE
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ghosal, D.; Omelchenko, M.V.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Venkateswaran, A.; Zhai, M.; Kostandarithes, H.M.; Brim, H.; Makarova, K.S.; et al. How Radiation Kills Cells: Survival of Deinococcus radiodurans and Shewanella oneidensis under Oxidative Stress. Fems Microbiol. Rev. 2005, 29, 361–375. [Google Scholar]
- Potts, M. Desiccation Tolerance of Prokaryotes. Microbiol. Rev. 1994, 58, 755–805. [Google Scholar] [CrossRef]
- Jurkiewicz, B.A.; Buettner, G.R. Ultraviolet Light-Induced Free Radical Formation in Skin: An Electron Paramagnetic Resonance Study. Photochem. Photobiol. 1994, 59, 1–4. [Google Scholar] [CrossRef]
- Lown, J.W.; Sim, S.K.; Chen, H.H. Hydroxyl Radical Production by Free and DNA-bound Aminoquinone Antibiotics and Its Role in DNA Degradation. Electron Spin Resonance Detection of Hydroxyl Radicals by Spin Trapping. Can. J. Biochem. 1978, 56, 1042–1047. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef]
- Qi, H.Z.; Wang, W.Z.; He, J.Y.; Ma, Y.; Xiao, F.Z.; He, S.Y. Antioxidative system of Deinococcus radiodurans. Res. Microbiol. 2020, 171, 45–54. [Google Scholar] [CrossRef]
- Agapov, A.A.; Kulbachinskiy, A.V. Mechanisms of Stress Resistance and Gene Regulation in the Radioresistant Bacterium Deinococcus radiodurans. Biochemistry 2015, 80, 1201–1216. [Google Scholar] [CrossRef]
- Daly, M.J.; Minton, K.W. Interchromosomal Recombination in the Extremely Radioresistant Bacterium Deinococcus radiodurans. J. Bacteriol. 1995, 177, 5495–5505. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.; Kemner, K.M.; et al. Protein Oxidation Implicated as the Primary Determinant of Bacterial Radioresistance. PLoS Biol. 2007, 5, e92. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Protein Damage and Death by Radiation in Escherichia Coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 2010, 107, 14373–14377. [Google Scholar] [CrossRef]
- Daly, M.J. A New Perspective on Radiation Resistance Based on Deinococcus radiodurans. Nat. Rev. Microbiol. 2009, 7, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Gerard, E.; Jolivet, E.; Prieur, D.; Forterre, P. DNA Protection Mechanisms are not Involved in the Radioresistance of the Hyperthermophilic Archaea Pyrococcus Abyssi and P. Furiosus. Mol. Genet. Genomics 2001, 266, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xue, D.; Sun, W.; Han, J.; Li, J.; Gao, R.; Zhou, Z.; Zhang, W.; Chen, M.; Lin, M.; et al. SRNA OsiA Stabilizes Catalase mRNA during Oxidative Stress Response of Deincoccus radiodurans R1. Microorganisms 2019, 7, e422. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, A.; Uivarosi, V.; Andries, A. Recent Progress in Understanding the Molecular Mechanisms of Radioresistance in Deinococcus Bacteria. Extremophiles 2015, 19, 707–719. [Google Scholar] [CrossRef]
- Gygi, S.P.; Corthals, G.L.; Zhang, Y.; Rochon, Y.; Aebersold, R. Evaluation of Two-Dimensional Gel Electrophoresis-Based Proteome Analysis Technology. Proc. Natl. Acad. Sci. USA 2000, 97, 9390–9395. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation Between Protein and mRNA Abundance in Yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef]
- Pradet-Balade, B.; Boulme, F.; Beug, H.; Mullner, E.W.; Garcia-Sanz, J.A. Translation Control: Bridging the Gap Between Genomics and Proteomics? Trends Biochem. Sci. 2001, 26, 225–229. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Liu, X.; Liu, Y.; Guo, C.; Zhang, L.; Han, J.; Wu, X.; Xue, D.; Gomaa, A.E.; et al. DrwH, a Novel WHy Domain-Containing Hydrophobic LEA5C Protein from Deinococcus radiodurans, Protects Enzymatic Activity under Oxidative Stress. Sci. Rep. 2017, 7, 9281. [Google Scholar] [CrossRef]
- Zhang, H.; Zhan, Y.; Yan, Y.; Liu, Y.; Hu, G.; Wang, S.; Yang, H.; Qiu, X.; Liu, Y.; Li, J.; et al. The Pseudomonas stutzeri-Specific Regulatory Noncoding RNA NfiS Targets katB mRNA Encoding a Catalase Essential for Optimal Oxidative Resistance and Nitrogenase Activity. J. Bacteriol. 2019, 201, e00334-19. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Xu, B.; Gao, Y.; Zhan, S.; Xiong, F.; Qiu, W.; Qian, X.; Wang, T.; Wang, N.; Zhang, D.; Yang, Q.; et al. Quantitative Protein Profiling of Hippocampus during Human Aging. Neurobiol. Aging 2016, 39, 46–56. [Google Scholar] [CrossRef]
- Xue, D.; Liu, W.; Chen, Y.; Liu, Y.; Han, J.; Geng, X.; Li, J.; Jiang, S.; Zhou, Z.; Zhang, W.; et al. RNA-Seq-Based Comparative Transcriptome Analysis Highlights New Features of the Heat-Stress Response in the Extremophilic Bacterium Deinococcus radiodurans. Int. J. Mol. Sci. 2019, 20, e5603. [Google Scholar] [CrossRef]
- Zhan, Y.; Deng, Z.; Yan, Y.; Zhang, H.; Lu, C.; Yang, Z.; Shang, L.; Huang, Y.; Lv, F.; Liu, Y.; et al. NfiR, a New Regulatory Noncoding RNA (ncRNA), is Required in Concert with the NfiS ncRNA for Optimal Expression of Nitrogenase Genes in Pseudomonas stutzeri A1501. Appl. Environ. Microbiol. 2019, 85, e00762-19. [Google Scholar] [CrossRef]
- Brioukhanov, A.L.; Netrusov, A.I.; Eggen, R. The Catalase and Superoxide Dismutase Genes are Transcriptionally up-Regulated upon Oxidative Stress in the Strictly Anaerobic Archaeon Methanosarcina Barkeri. Microbiology 2006, 152, 1671–1677. [Google Scholar] [CrossRef]
- Chelikani, P.; Fita, I.; Loewen, P.C. Diversity of Structures and Properties Among Catalases. Cell. Mol. Life Sci. 2004, 61, 192–208. [Google Scholar] [CrossRef]
- Zamocky, M.; Furtmuller, P.G.; Obinger, C. Evolution of Catalases from Bacteria to Humans. Antioxid. Redox Signal. 2008, 10, 1527–1548. [Google Scholar] [CrossRef]
- Lipton, M.S.; Pasa-Tolic’, L.; Anderson, G.A.; Anderson, D.J.; Auberry, D.L.; Battista, J.R.; Daly, M.J.; Fredrickson, J.; Hixson, K.K.; Kostandarithes, H.; et al. Global Analysis of the Deinococcus radiodurans Proteome by Using Accurate Mass Tags. Proc. Natl. Acad. Sci. USA 2002, 99, 11049–11054. [Google Scholar] [CrossRef]
- Jeong, S.W.; Jung, J.H.; Kim, M.K.; Seo, H.S.; Lim, H.M.; Lim, S. The Three Catalases in Deinococcus radiodurans: Only Two Show Catalase Activity. Biochem. Biophys. Res. Commun. 2016, 469, 443–448. [Google Scholar] [CrossRef]
- Kobayashi, I.; Tamura, T.; Sghaier, H.; Narumi, I.; Yamaguchi, S.; Umeda, K.; Inagaki, K. Characterization of Monofunctional Catalase KatA from Radioresistant Bacterium Deinococcus radiodurans. J. Biosci. Bioeng. 2006, 101, 315–321. [Google Scholar] [CrossRef]
- Zeth, K. Dps Biomineralizing Proteins: Multifunctional Architects of Nature. Biochem. J. 2012, 445, 297–311. [Google Scholar] [CrossRef]
- Wolf, S.G.; Frenkiel, D.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. DNA Protection by Stress-Induced Biocrystallization. Nature 1999, 400, 83–85. [Google Scholar] [CrossRef]
- Zhao, G.; Ceci, P.; Ilari, A.; Giangiacomo, L.; Laue, T.M.; Chiancone, E.; Chasteen, N.D. Iron and Hydrogen Peroxide Detoxification Properties of DNA-binding Protein from Starved Cells. A Ferritin-Like DNA-binding Protein of Escherichia Coli. J. Biol. Chem. 2002, 277, 27689–27696. [Google Scholar] [CrossRef]
- Ceci, P.; Ilari, A.; Falvo, E.; Chiancone, E. The Dps Protein of Agrobacterium Tumefaciens Does Not Bind to DNA but Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties. J. Biol. Chem. 2003, 278, 20319–20326. [Google Scholar] [CrossRef]
- Reon, B.J.; Nguyen, K.H.; Bhattacharyya, G.; Grove, A. Functional Comparison of Deinococcus radiodurans Dps Proteins Suggests Distinct in vivo Roles. Biochem. J. 2012, 447, 381–391. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Smith, L.T.; Xiao, L.; Bhattacharyya, G.; Grove, A. On the Stoichiometry of Deinococcus radiodurans Dps-1 Binding to Duplex DNA. Proteins 2012, 80, 713–721. [Google Scholar] [CrossRef]
- Santos, S.P.; Mitchell, E.P.; Franquelim, H.G.; Castanho, M.A.; Abreu, I.A.; Romao, C.V. Dps from Deinococcus radiodurans: Oligomeric Forms of Dps1 with Distinct Cellular Functions and Dps2 Involved in Metal Storage. FEBS J. 2015, 282, 4307–4327. [Google Scholar] [CrossRef]
- Armstrong, G.A.; Hearst, J.E. Carotenoids 2: Genetics and Molecular Biology of Carotenoid Pigment Biosynthesis. FASEB J. 1996, 10, 228–237. [Google Scholar] [CrossRef]
- Coker, J.A. Extremophiles and Biotechnology: Current Uses and Prospects. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Tian, B.; Sun, Z.; Shen, S.; Wang, H.; Jiao, J.; Wang, L.; Hu, Y.; Hua, Y. Effects of Carotenoids from Deinococcus radiodurans on Protein Oxidation. Lett. Appl. Microbiol. 2009, 49, 689–694. [Google Scholar] [CrossRef]
- Ji, H.F. Insight into the Strong Antioxidant Activity of Deinoxanthin, a Unique Carotenoid in Deinococcus radiodurans. Int. J. Mol. Sci. 2010, 11, 4506–4510. [Google Scholar] [CrossRef]
- Carbonneau, M.A.; Melin, A.M.; Perromat, A.; Clerc, M. The Action of Free Radicals on Deinococcus radiodurans Carotenoids. Arch. Biochem. Biophys. 1989, 275, 244–251. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Venkateswaran, A.; Hess, M.; Omelchenko, M.V.; Kostandarithes, H.M.; Makarova, K.S.; et al. Accumulation of Mn(II) in Deinococcus radiodurans Facilitates Gamma-Radiation Resistance. Science 2004, 306, 1025–1028. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xu, H.; Wang, J.; Liu, C.; Lu, H.; Liu, M.; Zhao, Y.; Tian, B.; Wang, L.; Hua, Y. Cyclic AMP Receptor Protein Acts as a Transcription Regulator in Response to Stresses in Deinococcus radiodurans. PLoS ONE 2016, 11, e155010. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.; Coste, G.; Sommer, S.; Oberto, J.; Confalonieri, F.; Servant, P.; Pasternak, C. DdrI, a cAMP Receptor Protein Family Member, Acts as a Major Regulator for Adaptation of Deinococcus radiodurans to Various Stresses. J. Bacteriol. 2018, 200, e00129-18. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Narumi, I.; Gao, G.; Tian, B.; Satoh, K.; Kitayama, S.; Shen, B. PprI: A General Switch Responsible for Extreme Radioresistance of Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 2003, 306, 354–360. [Google Scholar] [CrossRef]
- Bauermeister, A.; Bentchikou, E.; Moeller, R.; Rettberg, P. Roles of PprA, IrrE, and RecA in the Resistance of Deinococcus radiodurans to Germicidal and Environmentally Relevant UV Radiation. Arch. Microbiol. 2009, 191, 913–918. [Google Scholar] [CrossRef]
- Lu, H.; Gao, G.; Xu, G.; Fan, L.; Yin, L.; Shen, B.; Hua, Y. Deinococcus radiodurans PprI Switches on DNA Damage Response and Cellular Survival Networks after Radiation Damage. Mol. Cell. Proteomics 2009, 8, 481–494. [Google Scholar] [CrossRef]
- Gao, G.; Tian, B.; Liu, L.; Sheng, D.; Shen, B.; Hua, Y. Expression of Deinococcus radiodurans PprI Enhances the Radioresistance of Escherichia Coli. DNA Rep. 2003, 2, 1419–1427. [Google Scholar] [CrossRef]
- Pan, J.; Wang, J.; Zhou, Z.; Yan, Y.; Zhang, W.; Lu, W.; Ping, S.; Dai, Q.; Yuan, M.; Feng, B.; et al. IrrE, a Global Regulator of Extreme Radiation Resistance in Deinococcus radiodurans, Enhances Salt Tolerance in Escherichia Coli and Brassica Napus. PLoS ONE 2009, 4, e4422. [Google Scholar] [CrossRef]
- Zhao, P.; Zhou, Z.; Zhang, W.; Lin, M.; Chen, M.; Wei, G. Global Transcriptional Analysis of Escherichia Coli Expressing IrrE, a Regulator from Deinococcus radiodurans, in Response to NaCl Shock. Mol. Biosyst. 2015, 11, 1165–1171. [Google Scholar] [CrossRef]
- Ma, R.; Zhang, Y.; Hong, H.; Lu, W.; Lin, M.; Chen, M.; Zhang, W. Improved Osmotic Tolerance and Ethanol Production of Ethanologenic Escherichia Coli by IrrE, a Global Regulator of Radiation-Resistance of Deinococcus radiodurans. Curr. Microbiol. 2011, 62, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Ujaoney, A.K.; Padwal, M.K.; Basu, B. Proteome Dynamics during Post-Desiccation Recovery Reveal Convergence of Desiccation and Gamma Radiation Stress Response Pathways in Deinococcus radiodurans. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jung, J.H.; Blanchard, L.; de Groot, A. Conservation and Diversity of Radiation and Oxidative Stress Resistance Mechanisms in Deinococcus Species. FEMS Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; Omelchenko, M.V.; Beliaev, A.S.; Venkateswaran, A.; Stair, J.; Wu, L.; Thompson, D.K.; Xu, D.; Rogozin, I.B.; et al. Transcriptome Dynamics of Deinococcus radiodurans Recovering from Ionizing Radiation. Proc. Natl. Acad. Sci. USA 2003, 100, 4191–4196. [Google Scholar] [CrossRef]
- Zhang, C.; Wei, J.; Zheng, Z.; Ying, N.; Sheng, D.; Hua, Y. Proteomic Analysis of Deinococcus radiodurans Recovering from Gamma-Irradiation. Proteomics 2005, 5, 138–143. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Zhou, Z.; Chen, X.; Zhang, W.; Lin, M.; Chen, M. Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans. Microorganisms 2020, 8, 451. https://doi.org/10.3390/microorganisms8030451
Gao L, Zhou Z, Chen X, Zhang W, Lin M, Chen M. Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans. Microorganisms. 2020; 8(3):451. https://doi.org/10.3390/microorganisms8030451
Chicago/Turabian StyleGao, Lihua, Zhengfu Zhou, Xiaonan Chen, Wei Zhang, Min Lin, and Ming Chen. 2020. "Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans" Microorganisms 8, no. 3: 451. https://doi.org/10.3390/microorganisms8030451
APA StyleGao, L., Zhou, Z., Chen, X., Zhang, W., Lin, M., & Chen, M. (2020). Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans. Microorganisms, 8(3), 451. https://doi.org/10.3390/microorganisms8030451



