Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Extraction/Preparation of Garlic Samples
2.2. Strains and Culturing Conditions
2.3. Antimicrobial Assay
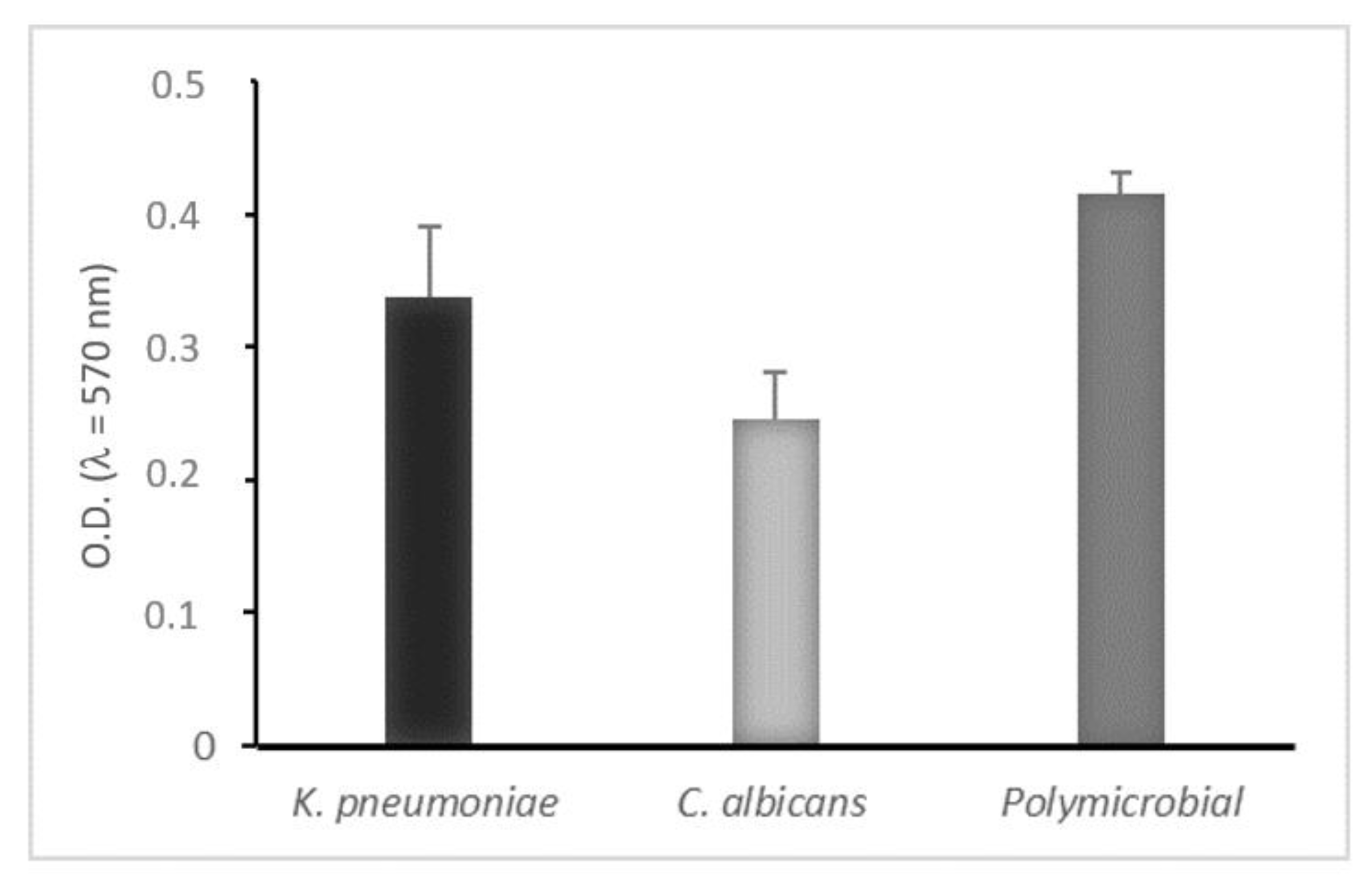
2.4. Mono- and Dual-Species Biofilm: In Vitro Static Model
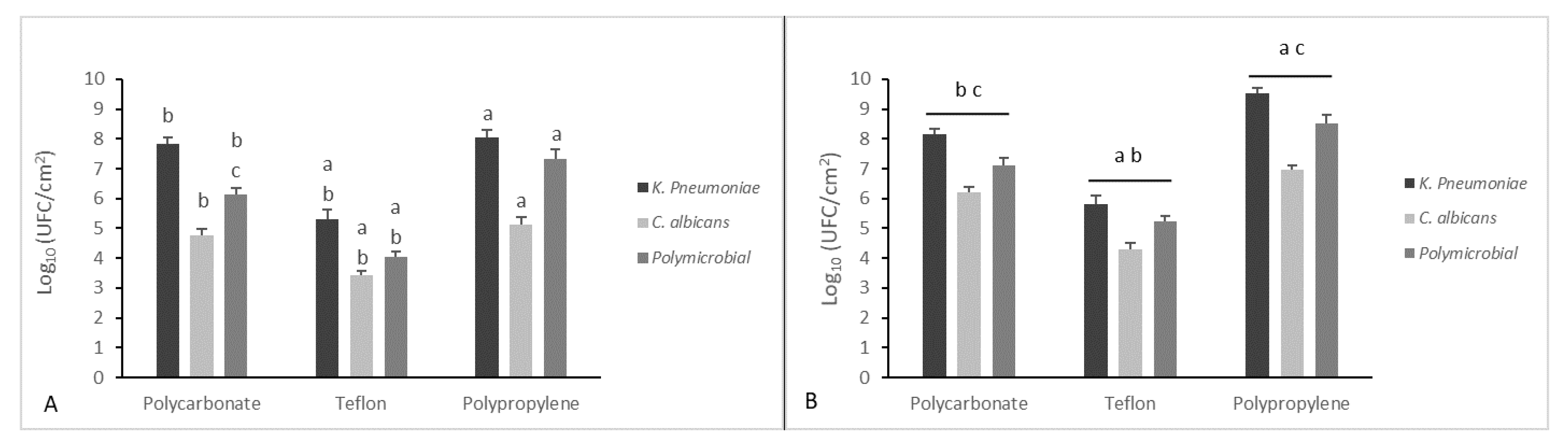
2.5. Mono- and Dual-Species Biofilm: In Vitro Dynamic Biofilm Model
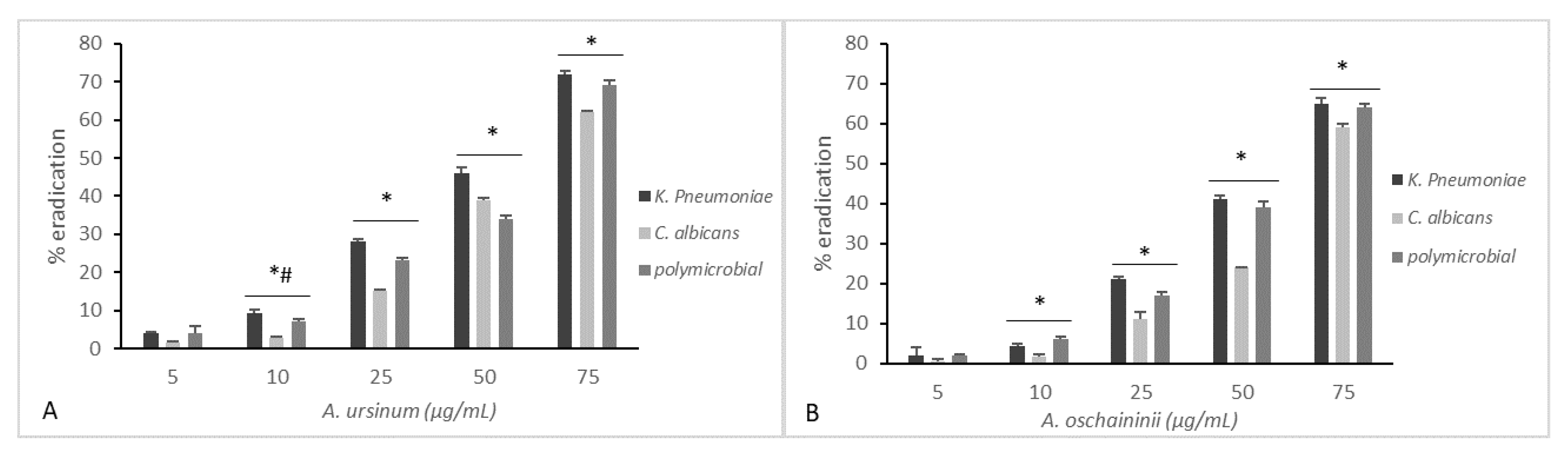
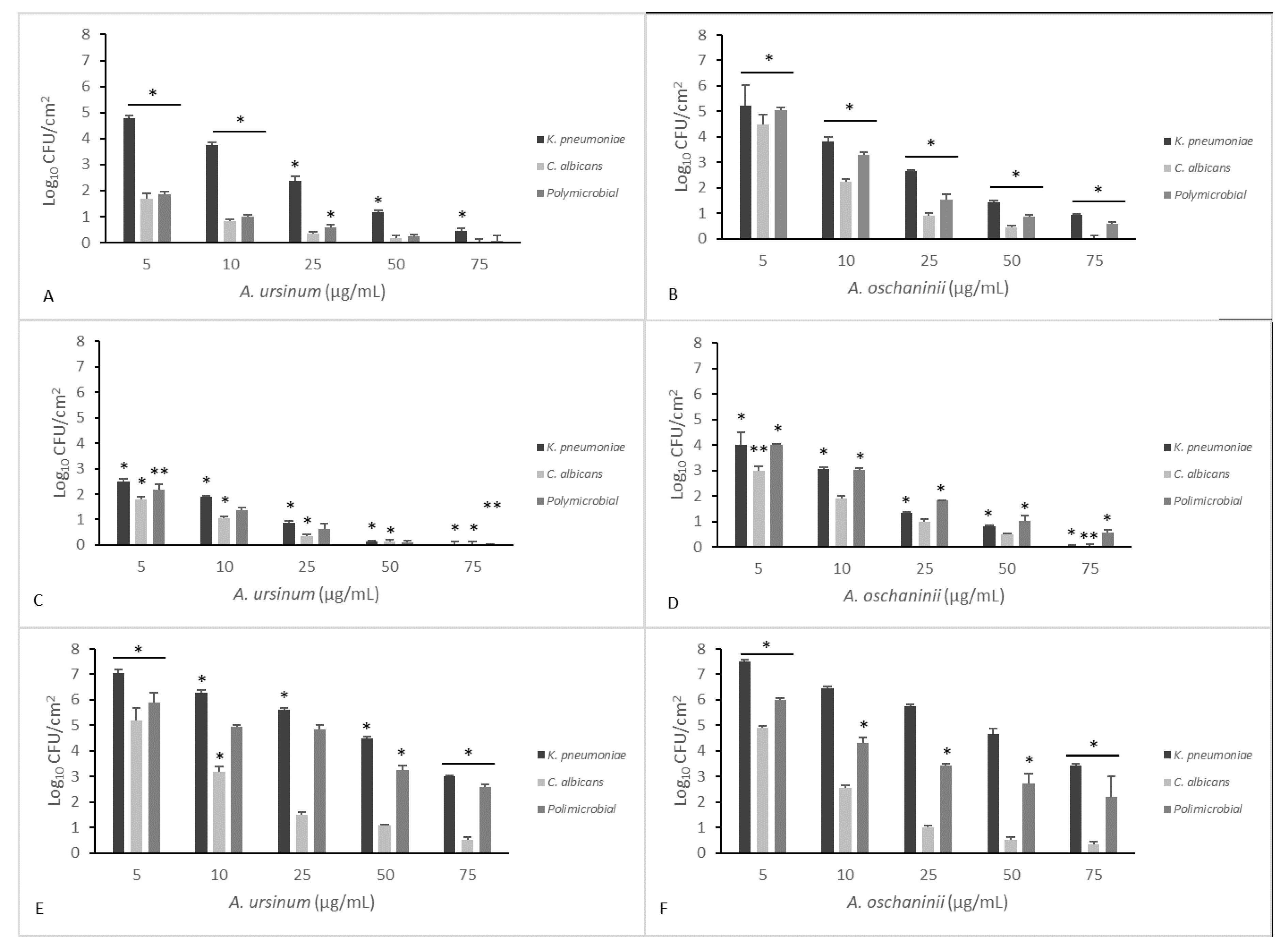
2.6. Eradication of Established Biofilm
2.7. Statistical Analysis
3. Results and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ribeiro, S.M.; Felício, M.R.; Boas, E.V.; Gonçalves, S.; Costa, F.F.; Samy, R.P.; Santos, N.C.; Franco, O.L. New frontiers for anti-biofilm drug development. Pharmacol. Ther. 2016, 160, 133–144. [Google Scholar] [CrossRef]
- Rahdar, H.A.; Malekabad, E.S.; Dadashi, A.R.; Takei, E.; Keikha, M.; Kazemian, H.; Karami-Zarandi, M. Correlation between biofilm formation and carbapenem resistance among clinical isolates of Klebsiella pneumoniae. Ethiop. J. Health Sci. 2019, 29, 745–750. [Google Scholar] [PubMed]
- Ciofu, O.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A. Antibiotic treatment of biofilm infections. APMIS 2017, 125, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Coombs, G.; Ling, T.; Balaji, V.; Rodrigues, C.; Mikamo, H.; Kim, M.J.; Rajasekaram, D.G.; Mendoza, M.; Tan, T.Y.; et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010–2013. Int. J. Antimicrob. Agents 2016, 47, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Liao, C.H.; Teng, L.J.; Hsu, H.S.; Hsueh, P.R. Reinfection and relapse of recurrent bacteremia caused by Klebsiella pneumoniae in a medical center in Taiwan. Future Microbiol. 2016, 11, 1157–1165. [Google Scholar] [CrossRef]
- Al-Mousa, H.H.; Omar, A.A.; Rosenthal, V.D.; Salama, M.F.; Aly, N.Y.; El-Dossoky Noweir, M.; Rebello, F.M.; Narciso, D.M.; Sayed, A.F.; Kurian, A.; et al. Device-associated infection rates, bacterial resistance, length of stay, and mortality in Kuwait: International Nosocomial Infection Consortium findings. Am. J. Infect. Control 2016, 44, 444–449. [Google Scholar] [CrossRef]
- Guo, S.; Xu, J.; Wei, Y.; Xu, J.; Li, Y.; Xue, R. Clinical and molecular characteristics of Klebsiella pneumoniae ventilator-associated pneumonia in mainland China. BMC Infect. Dis. 2016, 16, 608. [Google Scholar] [CrossRef]
- de Campos, P.A.; Royer, S.; Batistão, D.W.; Araújo, B.F.; Queiroz, L.L.; de Brito, C.S.; Gontijo-Filho, P.P.; Ribas, R.M. Multidrug Resistance Related to Biofilm Formation in Acinetobacter baumannii and Klebsiella pneumoniae Clinical Strains from Different Pulsotypes. Curr. Microbiol. 2016, 72, 617–627. [Google Scholar] [CrossRef]
- Dibartola, A.C.; Swearingen, M.C.; Granger, J.F.; Stoodley, P.; Dusane, D.H. Biofilms in orthopedic infections: A review of laboratory methods. APMIS 2017, 125, 418–428. [Google Scholar] [CrossRef]
- Hawser, S.P.; Douglas, L.J. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect. Immun. 1994, 62, 915–921. [Google Scholar] [CrossRef]
- Busscher, H.J.; Geertsema-Doornbusch, G.I.; van der Mei, H.C. Adhesion to silicone rubber of yeasts and bacteria isolated from voice prostheses: Influence of salivary conditioning films. J. Biomed. Mater. Res. 1997, 34, 201–209. [Google Scholar] [CrossRef]
- Bauters, T.G.M.; Moerman, M.; Vermeersch, H.; Nelis, H.J. Colonization of Voice Prostheses by Albicans and Non-Albicans Candida Species. Laryngoscope 2002, 112, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Ranjan, A.; Dongari-Bagtzoglou, A. Tipping the Balance: C. albicans Adaptation in Polymicrobial Environments. J. Fungi 2018, 4, 112. [Google Scholar]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Farrag, H.A.; Hosny, A.E.M.S.; Hawas, A.M.; Hagras, S.A.A.; Helmy, O.M. Potential efficacy of garlic lock therapy in combating biofilm and catheter-associated infections; experimental studies on an animal model with focus on toxicological aspects. Saudi Pharm. J. 2019, 27, 830–840. [Google Scholar] [CrossRef]
- Bhatwalkar, S.B.; Gound, S.S.; Mondal, R.; Srivastava, R.K.; Anupam, R. Anti-biofilm and Antibacterial Activity of Allium sativum Against Drug Resistant Shiga-Toxin Producing Escherichia coli (STEC) Isolates from Patient Samples and Food Sources. Indian, J. Microbiol. 2019, 59, 171–179. [Google Scholar] [CrossRef]
- Moodley, K.; Joseph, K.; Naidoo, Y.; Islam, S.; Mackraj, I. Antioxidant, antidiabetic and hypolipidemic effects of Tulbaghia violacea Harv. (wild garlic) rhizome methanolic extract in a diabetic rat model. BMC Complementary Altern. Med. 2015, 15, 408. [Google Scholar] [CrossRef]
- Nok, A.J.; Williams, S.; Onyenekwe, P.C. Allium sativum-induced death of African trypanosomes. Parasitol. Res. 1996, 82, 634–637. [Google Scholar] [CrossRef]
- Sendl, A.; Elbl, G.; Steinke, B.; Redl, K.; Breu, W.; Wagner, H. Comparative Pharmacological Investigations of Allium ursinum and Allium sativum. Planta Med. 1992, 58, 1–7. [Google Scholar] [CrossRef]
- Van Wyk, B.-E.; Wink, M. Medicinal Plants of the World; Briza Publications: Pretoria, South Africa, 2017. [Google Scholar]
- Van Wyk, B.E.; Wink, M. Phytomedicines, Herbal Drugs, and Poisons; University of Chicago Press: Chicago, IL, USA, 2015. [Google Scholar]
- Bagiu, R.V.; Vlaicu, B.; Butnariu, M. Chemical composition and in vitro antifungal activity screening of the Allium ursinum L. (Liliaceae). Int. J. Mol. Sci. 2012, 13, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Motsei, M.L.; Lindsey, K.L.; van Staden, J.; Jäger, A.K. Screening of traditionally used South African plants for antifungal activity against Candida albicans. J. Ethnopharmacol. 2003, 86, 235–241. [Google Scholar] [CrossRef]
- Singh, G.; Wulansari, S.G. Pattern of bacterial and fungal pathogen in patients with high risk for invasive fungal disease in an indonesian tertiary care hospital: An observational study. Pan Afr. Med. J. 2018, 29, 60. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed]
- Buckingham-Meyer, K.; Goeres, D.M.; Hamilton, M.A. Comparative evaluation of biofilm disinfectant efficacy tests. J. Microbiol. Methods 2007, 70, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Naviglio, D.; Scarano, P.; Ciaravolo, M.; Gallo, M. Rapid Solid-Liquid Dynamic Extraction (RSLDE): A Powerful and Greener Alternative to the Latest Solid-Liquid Extraction Techniques. Foods 2019, 8, 245. [Google Scholar] [CrossRef]
- Majewski, M. Allium sativum: Facts and myths regarding human health. Rocz Panstw Zakl Hig 2014, 65, 1–8. [Google Scholar]
- Gallo, M.; Formato, A.; Formato, G.; Naviglio, D. Comparison between Two Solid-Liquid Extraction Methods for the Recovery of Steviol Glycosides from Dried Stevia Leaves Applying a Numerical Approach. Processes 2018, 6, 105. [Google Scholar] [CrossRef]
- Gallo, M.; Vitulano, M.; Andolfi, A.; Della Greca, M.; Conte, E.; Ciaravolo, M.; Naviglio, D. Rapid Solid-Liquid Dynamic Extraction (RSLDE): A New Rapid and Greener Method for Extracting Two Steviol Glycosides (Stevioside and Rebaudioside A) from Stevia Leaves. Plant Foods Human Nutr. 2017, 72, 141–148. [Google Scholar] [CrossRef]
- CLSI. Methods for diluition antimicrobial susceptibility tests for bacteria that grow aerobically; approved standards-6. In Document m7-a6 Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne PA, USA, 2006. [Google Scholar]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Galdiero, E.; Siciliano, A.; Gesuele, R.; Di Onofrio, V.; Falanga, A.; Maione, A.; Liguori, R.; Libralato, G.; Guida, M. Melittin Inhibition and Eradication Activity for Resistant Polymicrobial Biofilm Isolated from a Dairy Industry after Disinfection. Int. J. Microbiol. 2019, 2019, 4012394. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Banerjee, G.; Garg, R.; Singh, M. Comparative Study of Biofilm Formation in Pseudomonas aeruginosa Isolates from Patients of Lower Respiratory Tract Infection. J. Clin. Diagn. Res. 2014, 8, DC09–DC11. [Google Scholar] [PubMed]
- ASTM International. Standard Test Method for Quantification of Pseudomonas Aeruginosa Biofilm Grown with High Shear and Continuous Flow Using CDC Biofilm Reactor; ASTM International: West Conshohocken, PA, USA, 2007. [Google Scholar]
- Gîtin, L.; Dinică, R.; Neagu, C.; Dumitrascu, L. Sulfur compounds identification and quantification from Allium spp. fresh leaves. J. Food Drug Anal. 2014, 22, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.H.; Slusarenko, A.J. A Comparison of the Antibacterial and Antifungal Activities of Thiosulfinate Analogues of Allicin. Sci. Rep. 2018, 8, 6763. [Google Scholar] [CrossRef]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef]
- James, G.A.; Boegli, L.; Hancock, J.; Bowersock, L.; Parker, A.; Kinney, B.M. Bacterial Adhesion and Biofilm Formation on Textured Breast Implant Shell Materials. Aesthetic Plast. Surg. 2019, 43, 490–497. [Google Scholar] [CrossRef]
- Teughels, W.; Van Assche, N.; Sliepen, I.; Quirynen, M. Effect of material characteristics and/or surface topography on biofilm development. Clin. Oral. Implant. Res. 2006, 17, 68–81. [Google Scholar] [CrossRef]
- Nagant, C.; Pitts, B.; Nazmi, K.; Vandenbranden, M.; Bolscher, J.G.; Stewart, P.S.; Dehaye, J.P. Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments. Antimicrob. Agents Chemother. 2012, 56, 5698–5708. [Google Scholar] [CrossRef]
- Nagant, C.; Pitts, B.; Stewart, P.S.; Feng, Y.; Savage, P.B.; Dehaye, J.P. Study of the effect of antimicrobial peptide mimic, CSA-13, on an established biofilm formed by Pseudomonas aeruginosa. MicrobiologyOpen 2013, 2, 318–325. [Google Scholar] [CrossRef]
- Ribeiro, M.M.; Graziano, K.U.; Olson, N.; França, R.; Alfa, M.J. The polytetrafluoroethylene (PTFE) channel model of cyclic-buildup biofilm and traditional biofilm: The impact of friction, and detergent on cleaning and subsequent high-level disinfection. Infect. Control Hosp. Epidemiol. 2020, 41, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Manner, S.; Goeres, D.M.; Skogman, M.; Vuorela, P.; Fallarero, A. Prevention of Staphylococcus aureus biofilm formation by antibiotics in 96-Microtiter Well Plates and Drip Flow Reactors: Critical factors influencing outcomes. Sci. Rep. 2017, 7, 43854. [Google Scholar] [CrossRef] [PubMed]
- Bandeira, M.; Borges, V.; Gomes, J.P.; Duarte, A.; Jordao, L. Insights on Klebsiella pneumoniae Biofilms Assembled on Different Surfaces Using Phenotypic and Genotypic Approaches. Microorganisms 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef]
- Macià, M.D.; del Pozo, J.L.; Díez-Aguilar, M.; Guinea, J. Diagnóstico microbiológico de las infecciones relacionadas con la formación de biopelículas. Enfermedades Infecciosas y Microbiología Clínica 2018, 36, 375–381. [Google Scholar] [CrossRef]
- Di Onofrio, V.; Gesuele, R.; Maione, A.; Liguori, G.; Liguori, R.; Guida, M.; Nigro, R.; Galdiero, E. Prevention of Pseudomonas aeruginosa Biofilm Formation on Soft Contact Lenses by Allium sativum Fermented Extract (BGE) and Cannabinol Oil Extract (CBD). Antibiotics 2019, 8, 258. [Google Scholar] [CrossRef]
| A. ursinum | A. oschaninii | |||||
|---|---|---|---|---|---|---|
| MIC (µg/mL) | S.D. | MBC/MFC (µg/mL) | MIC (µg/mL) | S.D. | MBC/MFC (µg/mL) | |
| Klebsiella pneumoniae ATCC 10031 | 150 | 0.05 | 200 | 200 | 0.06 | 250 |
| Candida albicans ATCC 90028 | 75 | 0.07 | 150 | 80 | 0.03 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galdiero, E.; Di Onofrio, V.; Maione, A.; Gambino, E.; Gesuele, R.; Menale, B.; Ciaravolo, M.; Carraturo, F.; Guida, M. Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models. Microorganisms 2020, 8, 336. https://doi.org/10.3390/microorganisms8030336
Galdiero E, Di Onofrio V, Maione A, Gambino E, Gesuele R, Menale B, Ciaravolo M, Carraturo F, Guida M. Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models. Microorganisms. 2020; 8(3):336. https://doi.org/10.3390/microorganisms8030336
Chicago/Turabian StyleGaldiero, Emilia, Valeria Di Onofrio, Angela Maione, Edvige Gambino, Renato Gesuele, Bruno Menale, Martina Ciaravolo, Federica Carraturo, and Marco Guida. 2020. "Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models" Microorganisms 8, no. 3: 336. https://doi.org/10.3390/microorganisms8030336
APA StyleGaldiero, E., Di Onofrio, V., Maione, A., Gambino, E., Gesuele, R., Menale, B., Ciaravolo, M., Carraturo, F., & Guida, M. (2020). Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models. Microorganisms, 8(3), 336. https://doi.org/10.3390/microorganisms8030336







