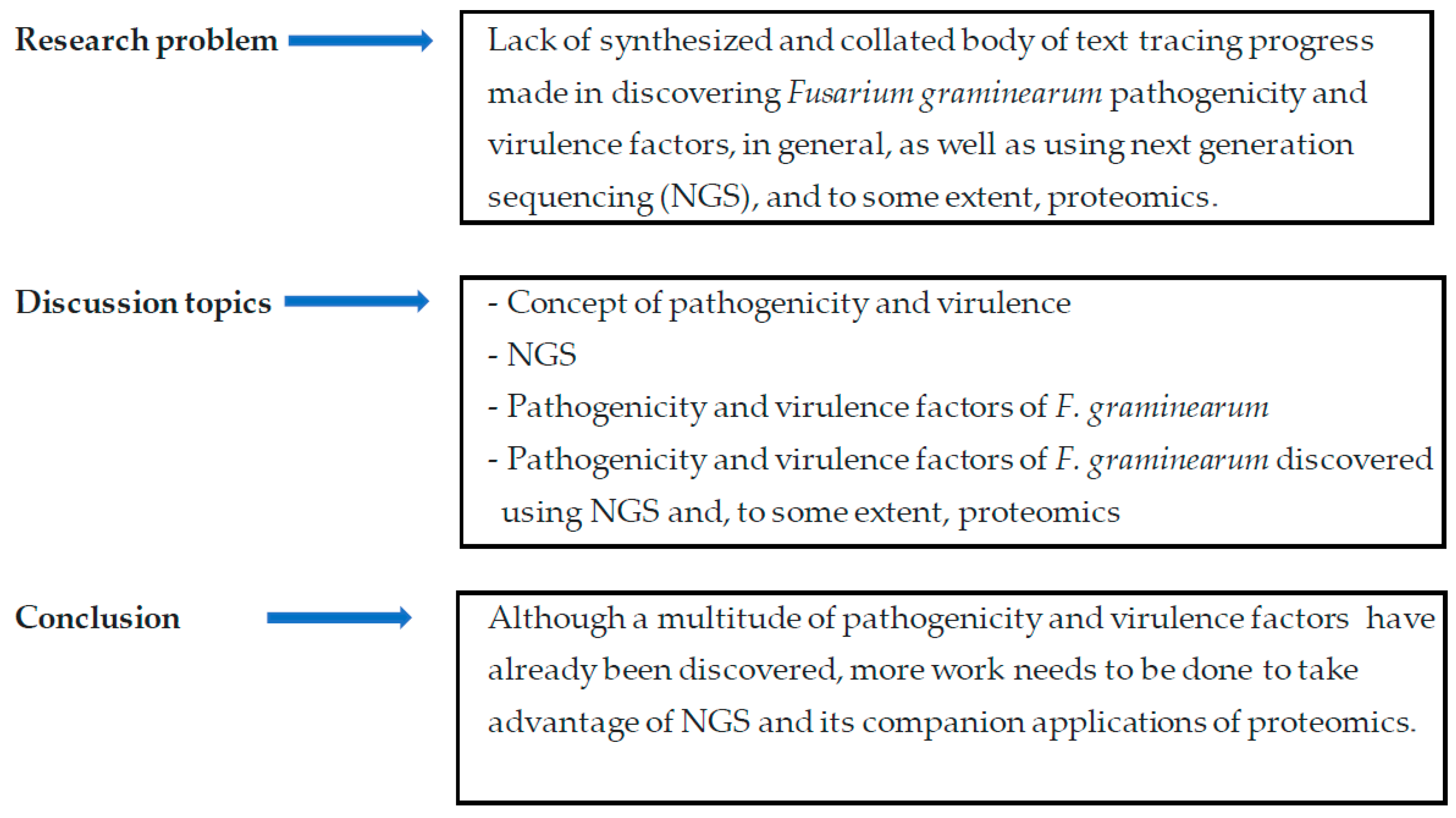
Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics
Abstract
1. Introduction
2. Next-Generation Sequencing, Its Relevance in Studying Plant Pathogenic Fungi and Fusarium graminearum
3. Pathogenicity and Virulence Factors of Plant Pathogens
3.1. Pathogenicity and Virulence Factors of Fusarium graminearum
3.2. Comparative Genomics and Molecular Basis of Pathogenicity and Virulence in Fusarium graminearum
3.3. Pathogenicity and Virulence Factors of Fusarium graminearum Discovered Using NGS Technologies
3.3.1. Notable Studies Which Paved Way for NGS
3.3.2. Fusarium graminearum Pathogenesis-Related Genes Discovered Using RNA-Seq Transcriptomics
3.4. Fusarium Graminearum Pathogenesis Proteins Discovered Using Proteomics Approaches
4. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- McMullen, M.P.; Jones, R.; Gallenberg, D. Scab of wheat and barley: Are-emerging disease of devastating impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Chang, I.; Kim, H.; Yun, S.; Leslie, J.F.; Lee, Y. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl. Environ. Microbiol. 2009, 84, 3285–3295. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top-10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Transcriptomics of cereal-Fusarium graminearum interactions: What we have learned so far. Mol. Plant Pathol. 2018, 19, 764–778. [Google Scholar] [CrossRef]
- Stack, R.W. Return of an old problem: Fusarium head blight of small grains. Plant Health Prog. 2000, 1, 19. [Google Scholar] [CrossRef]
- Muriuki, J.G. Deoxynivalenol and Nivalenol in Pathogenesis of Fusarium Head Blight in Wheat. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2001. [Google Scholar]
- Schmale, D.G.; Bergstrom, G.C. Fusarium Head Blight in Wheat. The Plant Health Instructor. Available online: http://www.apsnet.org/edcenter/intropp/lessons/fungi/ascomycetes/Pages/Fusarium.aspx (accessed on 1 October 2019).
- Stack, R.W. History of Fusarium head blight with emphasis on North America. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; APS Press: St. Paul, MN, USA, 2003; pp. 1–34. [Google Scholar]
- Proctor, R.H.; Desjardins, A.E.; Plattner, R.D.; Hohn, T.M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 1999, 27, 100–112. [Google Scholar] [CrossRef]
- Tralamazza, S.M.; Bemvenuti, R.H.; Zorzete, P.; de Souza Garcia, F.; Corrêa, B. Fungal diversity and natural occurrence of deoxynivalenol and zearalenone in freshly harvested wheat grains from Brazil. Food Chem. 2016, 196, 445–450. [Google Scholar] [CrossRef]
- Lee, T.; Lee, S.H.; Shin, J.Y.; Kim, H.K.; Yun, S.H.; Kim, H.Y.; Lee, S.; Ryu, J.G. Comparison of trichothecene biosynthetic gene expression between Fusarium graminearum and Fusarium asiaticum. Plant Pathol. J. 2014, 30, 33–42. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Hohn, T.M.; McCormick, S.P. Trichothecene biosynthesis in Fusarium species: Chemistry, genetics, and significance. Microbiol. Mol. Biol. Rev. 1993, 157, 595–604. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Hohn, T.M. Mycotoxins in plant pathogenesis. Mol. Plant Microbe Interact. 1997, 10, 147–152. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Grada, A.; Weinbrecht, K. Next-generation sequencing: Methodology and application. J. Investig. Dematol. 2013, 133, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 47, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Vigliar, E.; Malapelle, U.; de Luca, C.; Bellevicine, C.; Troncone, G. Challenges and opportunities of next-generation sequencing: A cytopathologist’s perspective. Cytopathology 2015, 26, 271–283. [Google Scholar] [CrossRef]
- Li, J.; Batcha, A.M.; Grüning, B.; Mansmann, U.R. An NGS workflow blueprint for DNA sequencing data and its application in individualized molecular oncology. Cancer Inform. 2016, 14, 87–107. [Google Scholar] [CrossRef]
- Kchouk, M.; Gibrat, J.F.; Elloumi, M. Generations of sequencing technologies: From first to next generation. Biol. Med. 2017, 9, 1–8. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Leamon, J.H. The development and impact of 454 sequencing. Nat. Biotechnol. 2008, 26, 1117–1124. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Passiglia, F.; Dendooven, A.; Pauwels, P.; Salgado, R.; Rolfo, C. Next–generation sequencing in clinical practice. In Oncogenomics; Dammacco, F., Silvestris, F., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 4, pp. 57–64. [Google Scholar]
- Koitzsch, U.; Heydt, C.; Attig, H.; Immerschitt, I.; Merkelbach-Bruse, S.; Fammartino, A.; Büttner, R.H.; Kong, Y.; Odenthal, M. Use of the GeneReader NGS System in a clinical pathology laboratory: A comparative study. J. Clin. Pathol. 2017, 70, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Salanoubat, M.; Genin, S.; Artiguenave, F.; Gousy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J.C.; Cattolico, L.; et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Tabolt, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.Q.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ding, S.W.; Zhang, Y.; Zhu, S. Identification of viruses and viroids by Next-Generation Sequencing and homology-dependent and homology-independent algorithms. Annu. Rev. Phytopathol. 2015, 53, 425–444. [Google Scholar] [CrossRef]
- Aylward, J.; Steenkamp, E.T.; Dreyer, L.L.; Roets, F.; Wingfield, B.D.; Wingfield, M.J. A plant pathology perspective of fungal genome sequencing. IMA Fungus 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral diagnostics in plants using Next generation sequencing: Computational analysis in practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next generation sequencing for detection and discovery of plant viruses and viroids: Comparison of two approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef]
- Birren, B.; Fink, G.; Lander, E. Fungal Genome Initiative: White Paper Developed by the Fungal Research Community; Whitehead Institute Center for Genome Research: Cambridge, MA, USA, 2002. [Google Scholar]
- The Fungal Genome Initiative Steering Committee. A White Paper for Fungal Comparative Genomics; Whitehead Institute: Cambridge, MA, USA, 2003. [Google Scholar]
- Grigoriev, I.V.; Cullen, D.; Goodwin, S.B.; Hibbett, D.; Jeffries, T.W.; Kubicek, C.P.; Kuske, C.; Magnuson, J.K.; Martin, F.; Spatafora, J.W.; et al. Fuelling the future with fungal genomics. Mycology 2011, 2, 192–209. [Google Scholar]
- Martin, F.; Cullen, D.; Hibbett, D.; Pisabarro, A.; Spatafora, J.W.; Bakerm, S.E.; Grigoriev, I.V. Sequencing the tree of life. New Phytol. 2011, 190, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J. 1000 fungal genomes to be sequenced. IMA Fungus 2011, 2, 41–45. [Google Scholar] [CrossRef][Green Version]
- Güldener, U.; Mannhaupt, G.; Münsterkötter, M.; Haase, D.; Oesterheld, M.; Stümpflen, V.; Mewes, H.W.; Adam, G. FGDB: A comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2006, 34, 456–458. [Google Scholar] [CrossRef]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Yoder, O.C. Toxins in pathogenesis. Ann. Rev. Phytopathol. 1980, 18, 103–129. [Google Scholar] [CrossRef]
- Sharma, A.K.; Dhasamana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial virulence factors: Secreted for survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef]
- Speth, E.B.; Lee, Y.N.; He, S.Y. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 2007, 10, 580–586. [Google Scholar] [CrossRef]
- Duyvesteijn, R.G.; van Wijk, R.; Boer, Y.; Rep, M.; Cornelissen, B.J.; Haring, M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005, 57, 1051–1063. [Google Scholar] [CrossRef]
- Vadlapudi, V.; Naidu, K.C. Fungal pathogenicity of plants: Molecular approach. Eur. J. Exp. Biol. 2011, 1, 38–42. [Google Scholar]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bos, J.I.; Armstrong, M.; Whisson, S.C.; da Cunha, L.; Torto-Alalibo, T.; Win, J.; Avrova, A.O.; Wright, F.; Birch, P.R.J.; et al. Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol. Biol. Evol. 2005, 22, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Morgan, W.; Bos, J.; Krasileva, K.V.; Cano, L.M.; Chaparro-Garcia, A.; Ammar, R.; Staskawicz, B.J.; Kamoun, S. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 2007, 19, 2349–2369. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Stam, R.; Cano, L.M.; Song, J.; Sklenar, J.; Yoshida, K.; Bozkurt, T.O.; Oliva, R.; Liu, Z.; Tian, M.Y.; et al. Effector specialization in a lineage of the Irish potato famine pathogen. Science 2014, 343, 552–555. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; McDonald, B.A. Geographical variation and positive diversifying selection in the host-specific toxin SnToxA. Mol. Plant Pathol. 2007, 8, 321–332. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Wanjiru, W.M.; Zhensheng, K.; Buchenauer, H. Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 2002, 108, 803–810. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, H.R.; Bairoch, A.; McCormick, S.P.; Shaner, G.; Buechley, G.; Hohne, T.M. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant Microbe Interact. 1996, 9, 775–781. [Google Scholar] [CrossRef]
- Jenczmionka, N.J.; Schäfer, W. The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Curr. Genet. 2005, 47, 29–36. [Google Scholar] [CrossRef]
- Fernandez, J.; Marroquin-Guzman, M.; Wilson, R.A. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 2014, 52, 155–174. [Google Scholar] [CrossRef]
- Ryan, C.A.; Farmer, E.E. Oligosaccharide signals in plants: A current assessment. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 651–674. [Google Scholar] [CrossRef]
- Felix, G.; Regenass, M.; Boller, T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation and establishment of a refractory state. Plant J. 1993, 4, 307–316. [Google Scholar] [CrossRef]
- Zhang, B.; Ramonell, K.; Somerville, S.; Stacey, G. Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 2002, 15, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, D.; Robatzek, S. Pattern recognition receptors: From the cell surface to intracellular dynamics. Mol. Plant Microbe Interact. 2007, 20, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. How plants recognize pathogens and defend themselves. Cell Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: A review. J. Basic Microbiol. 2009, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yazar, S.; Omurtag, G.Z. Fumonisins, trichothecenes and zearalenone in cereals. Int. J. Mol. Sci. 2008, 9, 2062–2090. [Google Scholar] [CrossRef]
- Sella, L.; Gazzetti, K.; Castiglioni, C.; Schäfer, W.; Favaron, F. Fusarium graminearum possesses virulence factors common to Fusarium Head Blight of wheat and seedling rot of soybean but differing in their impact on disease severity. Phytopathology 2014, 104, 1201–1207. [Google Scholar] [CrossRef]
- Voigt, C.A.; Schäfer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, Y.; Wang, J.; Wang, Y.; Chen, M.; Norvienyeku, J.; Li, G.; Yu, W.; Wang, Z. FgNoxR, a regulatory subunit of NADPH oxidases, is required for female fertility and pathogenicity in Fusarium graminearum. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef][Green Version]
- Jia, L.J.; Tang, H.Y.; Wang, W.Q.; Yuan, T.L.; Wei, W.Q.; Pang, B.; Gong, X.M.; Wang, S.F.; Li, Y.J.; Zhang, D.; et al. A linear nonribosomal octapeptide from Fusarium graminearum facilitates cell-to-cell invasion of wheat. Nat. Commun. 2019, 10, 922. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, S.A.; Boddu, J.; Berthiller, F.; Hametner, C.; Stupar, R.M.; Adam, G.; Muehlbauer, G.J. Transcriptome analysis of the barley-deoxynivalenol interaction: Evidence for a role of glutathione in deoxynivalenol detoxification. Mol. Plant Microbe Interact. 2010, 23, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Desmond, O.J.; Manners, J.M.; Stephens, A.E.; Maclean, D.J.; Schenk, P.M.; Gardiner, D.M.; Munn, A.L.; Kazan, K. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol. Plant Pathol. 2008, 9, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y. Toxicological features of T-2 toxin and related trichothecenes. Fundam. Appl. Toxicol. 1984, 4, 124–132. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef]
- McCormick, S.P.; Kato, T.; Maragos, C.M.; Busman, M.; Lattanzio, V.M.; Galaverna, G.; Dall-Asta, C.; Crich, D.; Price, N.P.; Kurtzman, C.P. Anomericity of T-2 toxin-glucoside: Masked mycotoxin in cereal crops. J. Agric. Food Chem. 2015, 63, 731–738. [Google Scholar] [CrossRef]
- Lanoseth, W.; Elen, O. Differences between barley, oats and wheat in the occurrence of deoxynivalenol and other trichothecenes in Norwegian grain. J. Phytopathol. 1996, 144, 113–118. [Google Scholar] [CrossRef]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; O’Donnell, K.; Ward, T.J.; Fujimura, M.; Hamamoto, H.; Shibata, T.; Yamaguchi, I. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 2003, 539, 105–110. [Google Scholar] [CrossRef]
- Wong, P.; Walter, M.; Lee, W.; Mannhaupt, G.; Münsterkötter, M.; Mewes, H.W.; Adam, G.; Güldener, U. FGDB: Revisiting the genome annotation of the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2011, 39, 637–639. [Google Scholar] [CrossRef]
- Jenczmionka, N.J.; Maier, F.J.; Lösch, A.P.; Schäfer, W. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase Gpmk1. Curr. Genet. 2003, 43, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Mott, E.; Farley, T.; Hammond-Kosack, K. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 2003, 4, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.; Lee, S.; Park, E.H.; Kim, K.W.; Kim, M.D.; Yun, S.H.; Lee, Y.W. GzSNF1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot. Cell 2009, 8, 116–127. [Google Scholar] [CrossRef]
- Hong, W. SNAREs and traffic. Biochim. Biophys. Acta BBA Mol. Cell Res. 2005, 1744, 120–144. [Google Scholar] [CrossRef]
- Hong, S.Y.; So, J.; Lee, J.; Min, K.; Son, H.; Park, C.; Yun, S.H.; Lee, Y.W. Functional analyses of two syntaxin-like SNARE genes, GzSYN1 and GzSYN2, in the ascomycete Gibberella zeae. Fungal Genet. Biol. 2010, 47, 364–372. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Zwiers, L.H.; De Waard, M.A. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 2002, 108, 719–734. [Google Scholar] [CrossRef]
- Pitkin, J.W.; Panaccione, D.G.; Walton, J.D. A putative cyclic peptide efflux pump encoded by the ToxA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 1996, 142, 1557–1565. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. Tri12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Gen. Genet. 1999, 261, 977–984. [Google Scholar] [CrossRef]
- Callahan, T.M.; Rose, M.S.; Meade, M.J.; Ehrenshaft, M.; Upchurch, R.G. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild-type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe Interact. 1999, 12, 901–910. [Google Scholar] [CrossRef]
- Urban, M.; Bhargava, T.; Hamer, J.E. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 1999, 18, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Zwiers, L.H.; De Waard, M.A. Characterization of the ABC transporter genes MgAtr1 and MgAtr2 from the wheat pathogen Mycosphaerella graminicola. Fungal Genet. Biol. 2000, 30, 115–125. [Google Scholar] [CrossRef]
- Nakaune, R.; Hamamoto, H.; Imada, J.; Akutsu, K.; Hibi, T. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol. Genet. Genom. 2002, 267, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, A.; Sopalla, C.; Weltring, K.M. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol. Plant Microbe Interact. 2002, 15, 102–108. [Google Scholar] [CrossRef]
- Schoonbeek, H.J.; van Nistelrooy, J.G.M.; de Waard, M.A. Functional analysis of ABC transporter genes from Botrytis cinerea identifies BcatrB as a transporter of eugenol. Eur. J. Plant Pathol. 2003, 109, 1003–1011. [Google Scholar] [CrossRef]
- Skov, J.; Lemmens, M.; Giese, H. Role of a Fusarium culmorum ABC transporter (FcABC1) during infection of wheat and barley. Physiol. Mol. Plant Pathol. 2004, 64, 245–254. [Google Scholar] [CrossRef]
- Qi, P.F.; Zhang, Y.Z.; Liu, C.H.; Zhu, J.; Chen, Q.; Guo, Z.R.; Wang, Y.; Xu, B.J.; Zheng, T.; Jiang, Y.F.; et al. Fusarium graminearum ATP-binding cassette transporter gene FgABCC9 is required for its transportation of salicylic acid, fungicide resistance, mycelial growth and pathogenicity towards wheat. Int. J. Mol. Sci. 2018, 19, 2351. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes with wildtype and trichodiene synthase gene disrupted Fusarium graminearum expressing a green fluorescence protein marker. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Alexander, N.J.; Desjardins, A.E. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 2009, 74, 1128–1142. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 2002, 68, 2959–2964. [Google Scholar] [CrossRef]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; McCormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Han, Y.K.; Kim, K.H.; Yun, S.H.; Lee, Y.W. Tri13 and Tri7 determine deoxynivalenol-and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 2002, 68, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Rowland, O.; Rodrigue, N.; Subramaniam, R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016, 17, 1014. [Google Scholar] [CrossRef] [PubMed]
- Gaffoor, I.; Brown, D.W.; Trail, F. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 2005, 4, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.T.; Gardiner, D.M.; Lysøe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Lee, W.; Wong, P.; Münsterkötter, M.; Mewes, H.W.; Schmeitzl, C.; Varga, E.; Berthiller, F.; Adam, G.; Güldener, U. The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS ONE 2014, 9, e0110311. [Google Scholar] [CrossRef]
- Harris, L.J.; Balcerzak, M.; Johnston, A.; Schneiderman, D.; Ouellet, T. Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016, 120, 111–123. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Hou, Z.; Wang, C.; Zhou, X.; Jonkers, W.; Ding, S.; Kistler, H.C.; Xu, J.R. A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2011, 24, 118–128. [Google Scholar] [CrossRef]
- Baldwin, T.K.; Urban, M.; Brown, N.; Hammond-Kosack, K.E. A role for topoisomerase in Fusarium graminearum and F. culmorum pathogenesis and sporulation. Mol. Plant Microbe Interact. 2010, 23, 566–577. [Google Scholar] [CrossRef][Green Version]
- Dufresne, M.; van der Lee, T.; Ben M’Barek, S.; Xu, X.D.; Zhang, X.; Liu, T.G.; Waalwijk, C.; Zhang, W.; Kema, G.H.; Daboussi, M.J. Transposon-tagging identifies novel pathogenicity genes in Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 1552–1561. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Chen, Q.; Liu, C.H.; Lei, L.; Li, Y.; Zhao, K.; Wei, M.Q.; Guo, Z.R.; Wang, Y.; Xu, B.J.; et al. Fusarium graminearum FgCWM1 Encodes a Cell Wall Mannoprotein Conferring Sensitivity to Salicylic Acid and Virulence to Wheat. Toxins 2019, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G.D. R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr. Genet. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yu, X.; Zhang, L.; Zhang, L.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Mitochondrial FgEch1 is responsible for conidiation and full virulence in Fusarium graminearum. Curr. Genet. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, L.; Liang, Y.; Yu, J. FgPEX4 is involved in development, pathogenicity, and cell wall integrity in Fusarium graminearum. Curr. Genet. 2019, 65, 747–758. [Google Scholar] [CrossRef]
- Waalwijk, C.; Vanheule, A.; Audenaert, K.; Zhang, H.; Warris, S.; van de Geest, H.; van der Lee, T. Fusarium in the age of genomics. Trop. Plant Pathol. 2017, 42, 184–189. [Google Scholar] [CrossRef]
- Sperschneider, J.; Gardiner, D.M.; Thatcher, L.F.; Lyons, R.; Singh, K.B.; Manners, J.M.; Taylor, J.M. Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol. Evol. 2015, 7, 1613–1627. [Google Scholar] [CrossRef]
- Michielse, C.B.; van Wijk, R.; Reijnen, L.; Cornelissen, B.J.; Rep, M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 2009, 10. [Google Scholar] [CrossRef]
- Cuomo, C.A.; Guldener, U.; Xu, J.R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.J.; Baker, S.E.; Rep, M.; et al. Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef]
- King, R.; Urban, M.; Hammond-Kosack, M.C.; Hassani-Pak, K.; Hammond-Kosack, K.E. The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genom. 2015, 16, 544–564. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, J.; Song, X.; Yang, J.; Li, H.; Tang, H.; Liao, Y.C. Combined metabolomic and quantitative real-time PCR analyses reveal systems metabolic changes of Fusarium graminearum induced by Tri5 gene deletion. J. Proteome Res. 2011, 10, 2273–2285. [Google Scholar] [CrossRef]
- Fall, L.A.; Salazar, M.M.; Drnevich, J.; Holmes, J.R.; Tseng, M.C.; Kolb, F.L.; Mideros, S.X. Field pathogenomics of Fusarium head blight reveals pathogen transcriptome differences due to host resistance. Mycologia 2019, 111, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Cumagun, C.J.R.; Bowden, R.L.; Jurgenson, J.E.; Leslie, J.F.; Miedaner, T. Genetic mapping of pathogenicity and aggressiveness of Gibberella zeae (Fusarium graminearum) toward wheat. Phytopathology 2004, 94, 520–526. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Dhokane, D.; Karre, S.; Kushalappa, A.C.; McCartney, C. Integrated metabolo-transcriptomics reveals Fusarium Head Blight candidate resistance genes in wheat QTL-Fhb2. PLoS ONE 2016, 11, e0155851. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Sondergaard, T.E.; Covarelli, L.; Fuertes, P.R.; Hansen, F.T.; Frandsen, R.J.; Saei, W.; Lukassen, M.B.; Wimmer, R.; Nielsen, K.F.; et al. Identification of the biosynthetic gene clusters for the lipopeptides fusaristatin A and W493 B in Fusarium graminearum and F. pseudograminearum. J. Nat. Prod. 2014, 77, 2619–2625. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome dynamics associated with resistance and susceptibility against Fusarium head blight in four wheat genotypes. BMC Genom. 2018, 19, 642. [Google Scholar] [CrossRef]
- Zhao, C.; Waalwijk, C.; De Wit, P.J.; van der Lee, T.; Tang, D. EBR1, a novel Zn2Cys6 transcription factor, affects virulence and apical dominance of hyphal tip in Fusarium graminearum. Mol. Plant Microbe Interact. 2011, 24, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.Y.; Zhao, X.; Xu, J.R.; Güldener, U.; Kistler, H.C. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.C.; Cutt, J.R.; Klessig, D.F. Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 1991, 10, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, C.; Wang, C.; Chen, C.; Xu, J.R.; Liu, H. Characterization of the two-speed subgenomes of Fusarium graminearum reveals the fast-speed subgenome specialized for adaption and infection. Front. Plant Sci. 2017, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Gardiner, D.M.; Taylor, J.M.; Hane, J.K.; Singh, K.B.; Manners, J.M.A. Comparative hidden Markov model analysis pipeline identifies proteins characteristic of cereal-infecting fungi. BMC Genom. 2013, 14, 807–829. [Google Scholar] [CrossRef] [PubMed]
- Laurent, B.; Moinard, M.; Spataro, C.; Ponts, N.; Barreau, C.; Foulongne-Oriol, M. Landscape of genomic diversity and host adaptation in Fusarium graminearum. BMC Genom. 2017, 18, 203–229. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Panstruga, R. Pathogenomics of fungal plant parasites: What have we learnt about pathogenesis? Curr. Opin. Plant Biol. 2011, 14, 392–399. [Google Scholar] [CrossRef]
- Garvey, G.S.; McCormick, S.P.; Alexander, N.J.; Rayment, I. Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides. Protein Sci. 2009, 18, 747–761. [Google Scholar]
- Nguyen, L.N.; Bormann, J.; Le, G.T.; Stärkel, C.; Olsson, S.; Nosanchuk, J.D.; Giese, H.; Schäfer, W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet. Biol. 2011, 48, 217–224. [Google Scholar] [CrossRef]
- Pasquali, M.; Migheli, Q. Genetic approaches to chemotype determination in type B-trichothecene producing Fusaria. Int. J. Food Microbiol. 2014, 189, 164–182. [Google Scholar] [CrossRef]
- Taylor, R.D.; Saparno, A.; Blackwell, B.; Anoop, V.; Gleddie, S.; Tinker, N.A.; Harris, L.J. Proteomic analyses of Fusarium graminearum grown under mycotoxin-inducing conditions. Proteomics 2008, 8, 2256–2265. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jacobsen, S.; Jørgensen, H.J.; Collinge, D.B.; Svensson, B.; Finnie, C. Fusarium graminearum and its interactions with cereal heads: Studies in the proteomics era. Front. Plant Sci. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Paper, J.M.; Scott-Craig, J.S.; Adhikari, N.D.; Cuomo, C.A.; Walton, J.D. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 2007, 7, 3171–3183. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Faubert, D.; Hamzehzarghani, H. Mass spectrometry-based metabolomics application to identify quantitative resistance-related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 2010, 11, 769–782. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K.; Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Mamer, O.; Faubert, D. Metabolomics technology to phenotype resistance in barley against Gibberella zeae. Eur. J. Plant Pathol. 2011, 130, 29–43. [Google Scholar] [CrossRef]
- Walkowiak, S.; Bonner, C.T.; Wang, L.; Blackwell, B.; Rowland, O.; Subramaniam, R. Intraspecies interaction of Fusarium graminearum contributes to reduced toxin production and virulence. Mol. Plant Microbe Interact. 2015, 28, 1256–1267. [Google Scholar] [CrossRef]
- Boddu, J.; Cho, S.; Muehlbauer, G.J. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant Microbe Interact. 2007, 20, 1364–1375. [Google Scholar] [CrossRef]
- Jonkers, W.; Dong, Y.; Broz, K.; Kistler, H.C. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog. 2012, 8, e1002724. [Google Scholar] [CrossRef]
- Hofstad, A.N.; Nussbaumer, T.; Akhunov, E.; Shin, S.; Kugler, K.G.; Kistler, H.C.; Mayer, K.F.; Muehlbauer, G.J. Examining the transcriptional response in wheat Fhb1 near-isogenic lines to Fusarium graminearum infection and deoxynivalenol treatment. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Brown, N.A.; Antoniw, J.; Hammond-Kosack, K.E. The predicted secretome of the plant pathogenic fungus Fusarium graminearum: A refined comparative analysis. PLoS ONE 2012, 7, e33731. [Google Scholar] [CrossRef]
- Marshall, A.G.; Hendrickson, C.L. High-resolution mass spectrometers. Annu. Rev. Anal. Chem. 2008, 1, 579–599. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R.; Cociorva, D.; Liao, L.J.; Zabrouskov, V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal. Chem. 2006, 78, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Makarov, A.; Denisov, E.; Lange, O.; Horning, S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Noll, R.J.; Li, H.; Makarov, A.; Hardman, M.; Graham Cooks, R. The Orbitrap: A new mass spectrometer. J. Mass Spectrom. 2005, 40, 430–443. [Google Scholar] [CrossRef]
- Mehta, A.; Brasileiro, A.C.; Souza, D.S.; Romano, E.; Campos, M.A.; Grossi-de-Sá, M.F.; Silva, M.S.; Franco, O.L.; Fragoso, R.R.; Bevitori, R.; et al. Plant–pathogen interactions: What is proteomics telling us? FEBS J. 2008, 275, 3731–3746. [Google Scholar] [CrossRef]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef]
- Lu, S.; Edwards, M.C. Genome-wide analysis of small secreted cysteine-rich proteins identifies candidate effector proteins potentially involved in Fusarium graminearum−wheat interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef]
- Rampitsch, C.; Day, J.; Subramaniam, R.; Walkowiak, S. Comparative secretome analysis of Fusarium graminearum and two of its non-pathogenic mutants upon deoxynivalenol induction in vitro. Proteomics 2013, 13, 1913–1921. [Google Scholar] [CrossRef]
- Geddes, J.; Eudes, F.; Laroche, A.; Selinger, L.B. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics 2008, 8, 545–554. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Phosphoproteome profile of B. cinerea under different pathogenicity stages by plant based elicitors. J. Proteom. 2016, 139, 84–94. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Modifications of fungal membrane proteins profile under pathogenicity induction: A proteomic analysis of Botrytis cinerea membranome. Proteomics 2016, 16, 2363–2376. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Dataset of the Botrytis cinerea phosphoproteome induced by different plant-based elicitors. J. Proteom. 2016, 7, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Melo-Braga, M.N.; Larsen, M.R.; Jørgensen, H.J.; Palmisano, G. Battle through signaling between wheat and the fungal pathogen Septoria tritici revealed by proteomics and phosphoproteomics. Mol. Cell. Proteom. 2013, 12, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.K.; Brown, N.A.; Urban, M.; Kanyuka, K.; Hammond-Kosack, K.E. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 2018, 74, 790–799. [Google Scholar] [CrossRef] [PubMed]
| Cell Wall Degrading Enzymes | |||
|---|---|---|---|
| Category/Type/Classification | Function | At Least One Reference Where Mentioned | |
| Depolymerase enzyme | Degradative enzyme | Catalyses depolymerization reactions | [53] |
| Pectinase | Degradative enzyme | Breaks down pectin | [55] |
| Cellulase | Degradative enzyme | Breaks down cellulose | [55] |
| Extracellular endoglucanase | Degradative enzyme | Breaks down glucan | [55] |
| Endo-1,4-b-glucanase | Degradative enzyme | Breaks down glucan | [55] |
| Proteolytic enzyme | Degradative enzyme | Breaks down proteins | [55] |
| Xylanolytic enzyme | Degradative enzyme | Breaks down xylan | [55] |
| Lipolytic enzyme | Degradative enzyme | Breaks down lipids | [55] |
| Toxins | |||
| Trichothecene NX-2 | Type A trichothecene (toxin) | Toxicity | [97] |
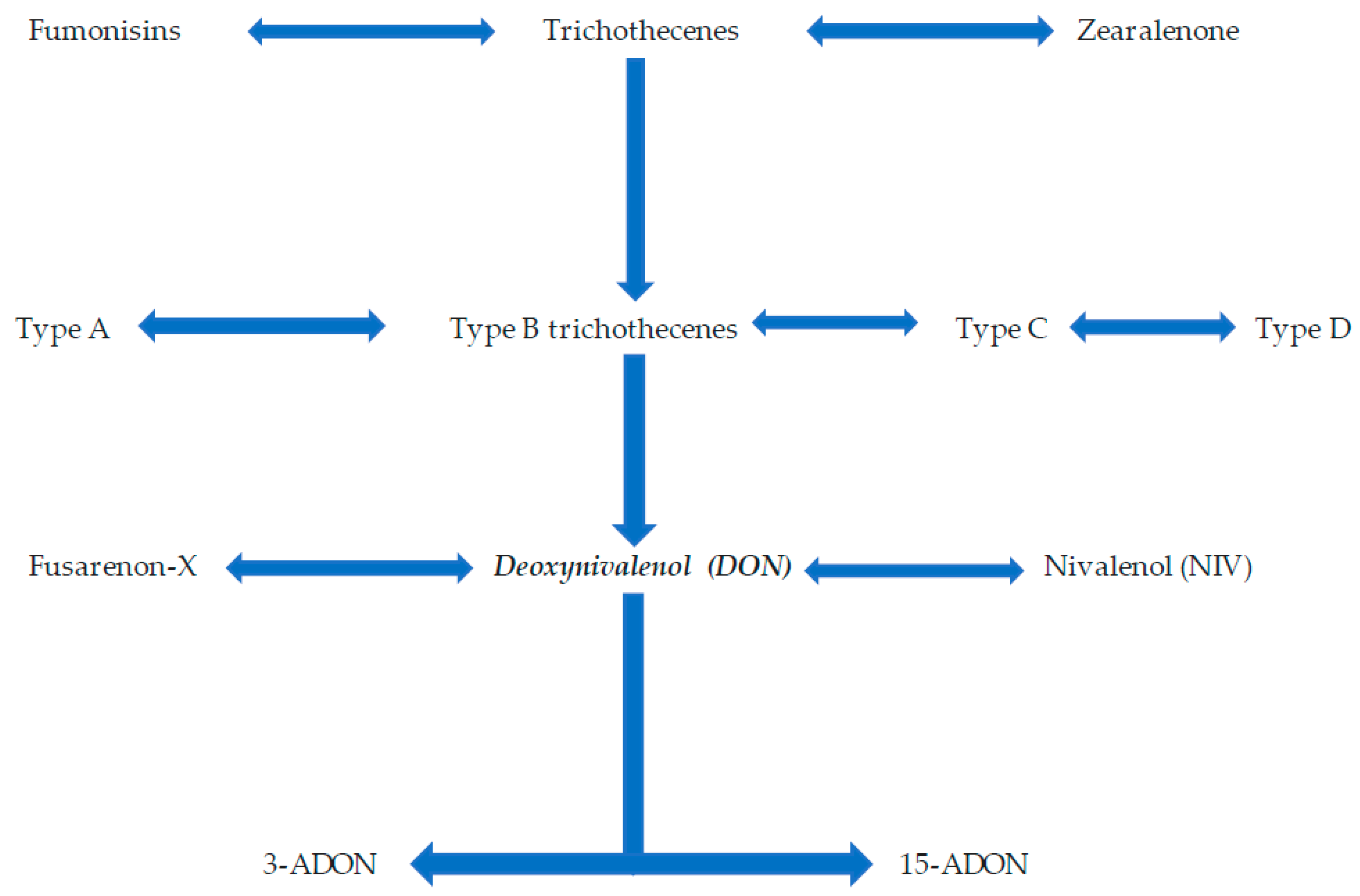
| Deoxynivalenol (DON) | Type B trichothecene (toxin) | Toxicity | [2] |
| 3-acetyldeoxynivalenol (3-ADON) | Type B trichothecene (toxin) | Toxicity | [11] |
| 15-ADON | Type B trichothecene (toxin) | Toxicity | [11] |
| Nivalenol (NIV) | Type B trichothecene (toxin) | Toxicity | [11] |
| Fusaoctaxin A | Toxin | Responsible for cell-to-cell invasion of wheat by F. graminearum | [67] |
| Toxin biosynthesis genes | |||
| Tri cluster genomic region | Trichothecene biosynthesis genes | Involved in the synthesis of trichothecene mycotoxins | [75] |
| Other pathogenicity and virulence determinants | |||
| Gpmk1 MAP kinase | MAP kinase | Involved in mating, conidiation, and pathogenicity | [77] |
| OS-2 | Stress activated kinase | Involved in conferring resistance to a soybean phytoalexin | [64] |
| FGL1 | Lipase gene | Enhances the fungal pathogenicity against wheat and maize | [65] |
| FgNoxR | A regulatory subunit of NADPH oxidase | Involved in conidiation, sexual development and pathogenicity of F. graminearum | [66] |
| fg3_54 | Putative secondary metabolite biosynthesis gene cluster | Responsible for cell-to-cell invasiveness | [67] |
| Protein kinase | Kinase cascade in trichothecene biosynthesis | Involved in trichothecene production | [79] |
| Histidine kinase | Kinase | Involved in trichothecene production | [79] |
| GzSNF1 | Serine/threonine-protein kinase | Responsible sexual, asexual development, and virulence | [80] |
| GzXYL1 | Endo-1,4-ß-xylanase 1 precursor gene | Involved in plant cell wall degradation | [80] |
| GzXYL2 | Endo-1,4-ß-xylanase 2 precursor gene | Involved in plant cell wall degradation | [80] |
| GzXLP | Extracellular ß-xylosidase gene | Involved in depolymerization and plant cell wall degradation | [80] |
| v-SNARE protein | Vesicle related proteins SNARE | Interacts with t-SNARE to catalyze the fusion of the apposing membranes of the transport intermediate and the target compartment) | [82] |
| t-SNARE protein | Target membrane-related SNARE | Interacts with v-SNARE to catalyze the fusion of the apposing membranes of the transport intermediate and the target compartment | [82] |
| Syntaxin-like t-SNARE protein | Syntaxin-like membrane-integrated protein | Required for vegetative growth, sexual reproduction, and virulence in Gibberella zeae. Proteins are also encoded by GzSYN1 and GzSYN2 in F. graminearum that enhanced virulence of the fungus on barley | [82] |
| GzSYN1 | Syntaxin-like SNARE gene | Enhances perithecia and radial hyphal growth | [82] |
| GzSYN2 | Syntaxin-like SNARE gene | Enhances perithecia and radial hyphal growth | [82] |
| Multiple ATP-binding cassette transporter | Major facilitator superfamily of membrane transporter | Involved in virulence | [83] |
| FgABCC9 | ATP-binding cassette transporter gene | involved in the fungal pathogenicity towards wheat | [93] |
| FGSG_04694 | Gene-encoding polyketide synthase PKS2 | Responsible for mycelial growth and fungi virulence | [99] |
| PKS2 | Polyketide synthase gene | Responsible for secondary metabolism and virulence | [101] |
| FGSG_08181 | Terpene synthase-encoding gene | Involved in the virulence of the fungus | [99] |
| FGSG_08182 | Terpene synthase-encoding gene | Involved in the virulence of the fungus | [103] |
| FGSG_17088 | Putative cytochrome P450 gene | Responsible for the expression of disease in fungus-infected cereals | [99] |
| FGSG_08183 | Putative cytochrome P450 gene | Responsible for the expression of disease in fungus-infected cereals | [99] |
| FGSG_08187 | Putative cytochrome P450 gene | Responsible for the expression of disease in fungus-infected cereals | [99] |
| ZIF1 | Encodes bZIP transcription factor | Involved in virulence and reproduction ability of F. graminearum | [104] |
| bZIP transcription factor | Transcription factor | Enhances the virulence of F. graminearum in infected plants | [104] |
| TOP1 I | Enzyme | Involved in sporulation and pathogenicity | [105] |
| FGSG_10057 | Conserved hypothetical protein | Involved in growth and virulence | [106] |
| Zn(II)2Cys6-type transcription factor | Transcription factor | Regulates fungal reproduction and pathogenicity | [106] |
| Cell Wall Degrading Enzymes | |||
|---|---|---|---|
| Category/Type/Classification | Function | At Least One Reference Where Mentioned | |
| 15917_M | Endo-1,4-beta-xylanase enzyme | Hydrolyses (1- > 4)-beta-D-xylosidic linkages in xylans of the cell walls | [115] |
| Xylanase | Degradative enzyme | Bring about the disintegration of xylan and cell wall penetration | [55] |
| Protease | Degradative enzyme | Responsible for the breakdown of protein | [55] |
| Lipase | Degradative enzyme | Responsible for the breakdown of lipids | [55] |
| Cutinases | Degradative enzyme | Plays polymer degrading function | [114] |
| Pectate lyases | Degradative enzyme | Plays polymer degrading function | [114] |
| Pectin lyases | Degradative enzyme | Plays polymer degrading function | [114] |
| β-amylase protein | Degradative enzyme | Involved in F. graminearum pathogenesis | [138] |
| Metallopeptidase | Degradative enzyme | Involved in F. graminearum pathogenesis | [139] |
| Peptidase | Degradative enzyme | Involved in F. graminearum pathogenesis | [139] |
| Toxins | |||
| Type B trichothecenes | Trichothecene mycotoxin | Toxicity | [2] |
| KP4 killer toxin | Toxic polypeptide | Toxicity | [146] |
| Genes for toxin biosynthesis | |||
| TRI1 gene | Tri cluster gene | Involved in F. graminearum pathogenesis | [137] |
| FGRRES_17235_M | Virulence-related gene | Encodes cysteine-rich secretory protein, allergen V5/Tpx-1-related with CAP and PR-1 family | [115] |
| 15917_M | Endo-1,4-beta-xylanase enzyme | Hydrolyses (1- > 4)-beta-D-xylosidic linkages in xylans of the cell walls | [115] |
| g8968 gene (predicted to contain the Tri5 domain) | Pathogenicity and virulence gene | Predicted to contain the Tri5 domain | [99] |
| Tri5 | Tri cluster gene | Involved in trichothecene biosynthesis | [95] |
| Tri8 | Tri cluster gene | Involved Fusarium trichothecene phytotoxicity | [96] |
| Pathogenicity and virulence proteins | |||
| TRI3 | Trichothecene biosynthesis protein | Involved in F. graminearum pathogenesis | [137] |
| TRI4 | Trichothecene biosynthesis protein | Involved in F. graminearum pathogenesis | [137] |
| TRI101 | Trichothecene biosynthesis protein | Involved in F. graminearum pathogenesis | [137] |
| Other pathogenicity and virulence determinants | |||
| Hormone-like compounds | Compounds with hormone-like properties | Enhances the adaptation of the fungi to the host plant environment | [119] |
| PR-1 family proteins | Pathogenicity related protein | Involved in pathogenicity | [129] |
| Basic leucine zipper (bZIP) transcription factor | Transcription factor | Enhances the virulence and reproduction ability of F. graminearum in infected plants | [104] |
| Syntaxin-like t-SNARE proteins | Syntaxin-like membrane-integrated proteins | Required for vegetative growth, sexual reproduction, and virulence in G. zeae. Proteins are also encoded by GzSYN1 and GzSYN2 in F. graminearum that enhanced virulence of the fungus on barley | [82] |
| Polyketide synthase (PKS) gene | Polyketide synthase gene | Responsible for secondary metabolism and virulence | [101] |
| Enhanced branching 1 (EBR1) | Zn(2)Cys(6) transcription factor | Involved in F. graminearum pathogenesis | [127] |
| NADP-dependent oxidoreductase | Oxidoreductase enzyme | Involved in F. graminearum pathogenesis | [139] |
| O-acyltransferase | Transferase enzyme | Involved in F. graminearum pathogenesis | [139] |
| Wor1-like Protein Fgp1 | Regulatory protein | Regulates pathogenicity, toxin synthesis, and reproduction in F. graminearum | [144] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rauwane, M.E.; Ogugua, U.V.; Kalu, C.M.; Ledwaba, L.K.; Woldesemayat, A.A.; Ntushelo, K. Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics. Microorganisms 2020, 8, 305. https://doi.org/10.3390/microorganisms8020305
Rauwane ME, Ogugua UV, Kalu CM, Ledwaba LK, Woldesemayat AA, Ntushelo K. Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics. Microorganisms. 2020; 8(2):305. https://doi.org/10.3390/microorganisms8020305
Chicago/Turabian StyleRauwane, Molemi E., Udoka V. Ogugua, Chimdi M. Kalu, Lesiba K. Ledwaba, Adugna A. Woldesemayat, and Khayalethu Ntushelo. 2020. "Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics" Microorganisms 8, no. 2: 305. https://doi.org/10.3390/microorganisms8020305
APA StyleRauwane, M. E., Ogugua, U. V., Kalu, C. M., Ledwaba, L. K., Woldesemayat, A. A., & Ntushelo, K. (2020). Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics. Microorganisms, 8(2), 305. https://doi.org/10.3390/microorganisms8020305







