Abstract
Fusarium graminearum is a devasting mycotoxin-producing pathogen of grain crops. F. graminearum has been extensively studied to understand its pathogenicity and virulence factors. These studies gained momentum with the advent of next-generation sequencing (NGS) technologies and proteomics. NGS and proteomics have enabled the discovery of a multitude of pathogenicity and virulence factors of F. graminearum. This current review aimed to trace progress made in discovering F. graminearum pathogenicity and virulence factors in general, as well as pathogenicity and virulence factors discovered using NGS, and to some extent, using proteomics. We present more than 100 discovered pathogenicity or virulence factors and conclude that although a multitude of pathogenicity and virulence factors have already been discovered, more work needs to be done to take advantage of NGS and its companion applications of proteomics.
1. Introduction
Plant pathogens have developed sophisticated penetration, infection, and colonization strategies to suppress plant defense mechanisms of susceptible hosts and cause disease. The display of minor symptoms by the plant is of least concern but excessive tissue damage and crop loss result in serious economic losses with adverse implications for society, especially in poor countries. To gain insights into how these plant pathogens develop the sophisticated strategies of attack, platforms such as next-generation sequencing (NGS), and its companion applications like proteomics, are currently being exploited. Pertinent scientific inquiry of pathogenicity and virulence processes of pathogens, comprehensive and integrated investigations remain imperative and must take advantage of such high-tech platforms as NGS to generate multitudes of useful datasets. Among microbes associated with the plant are fungi which encounter plants, penetrate and colonize plant tissue to either cause disease or live within the colonized plant in a symbiotic relationship with the host. Fungi which cause diseases are called fungal pathogens and their adverse effects can range from tiny spots on the plant surface to significant symptom expression and tissue collapse. In causing diseases, fungi employ various strategies. They can penetrate the host using penetration structures and develop a large network of hyphae within the intercellular spaces. Through the hyphae, these fungi draw nutrients from the living plant cells. This mode of attack is called biotrophy. Alternatively, fungal pathogens kill the host tissue and derive nutrients from the dead tissue. This mode of attack is called necrotrophy. In the hemi-biotrophic mode of attack, the fungus successively adopts a necrotrophic lifestyle after biotrophy. The invasion of the plant by the pathogen is usually carried out through the production of factors used for plant tissue manipulation to gain physical access into the tissues as well as draw nutrients. These factors enable the plant to cause disease as well as advance the infection within the interior of the tissue, ultimately worsening the disease condition. The ability of a fungus to cause disease is termed pathogenicity, and the ability to worsen the disease is called virulence. The terms pathogenicity factors and virulence factors are loosely used to refer to any substance a pathogen uses to parasitize the plant. We will also use these concepts jointly and will not attempt to classify the various disease-causing factors as either pathogenicity or virulence factors. As a necrotrophic fungus, the wheat pathogen Fusarium graminearum Schwabe (teleomorph Gibberellae zeae) produces, among other things, cell wall-degrading enzymes (CWDEs) and toxins. Plant tissue exposed to the CWDEs and the toxins loses firmness, macerates and cell contents leak. As the cell contents provide nourishment to the fungus, it grows and advances into the inner tissues. The disease spreads and eventually, depending on other factors, the affected part dies. With its ability to produce toxins, F. graminearum, a homothallic (self-fertile) ascomycetous fungus, of the Order: Sordariomycetes and Family: Nectriaceae causes Fusarium head blight (FHB), one of the most economically important diseases of wheat, barley, rice and other grain crops worldwide [1,2,3,4,5]. Also known as scab, this devastating disease was first described in 1884 in England and since then, its prominence has increased worldwide with outbreaks reported in temperate and semitropical areas globally [1,6,7,8,9]. Under favorable conditions, FHB can advance from initial infection to the destruction of the entire crop within a few weeks [1]. A combination of factors, global warming, increase in wheat production under irrigation and the concomitant increase in no-till practices is the likely cause of the recent resurgence of FHB. F. graminearum is favored by high temperatures and irrigation splash disseminates its propagules. Wheat spikes infected by F. graminearum have a bleached appearance and grain infected with this fungus is shriveled with pale grey color and an occasional pinkish discoloration. Over and above these undesirable qualities, ingestion of significant amounts of mycotoxin-contaminated grain may cause vomiting, headache, and dizziness in humans. Animals may lose weight and suffer anorexia. Serious effects of ingestion of large amounts of mycotoxins include leukoencephalomalacia in horses, pulmonary edema in swine, and kidney and liver cancers in mice (mentioned in Proctor et al. [10]), and a plethora of records link mycotoxin consumption to cancer in humans. However, some of these sicknesses are linked to mycotoxins produced by other Fusarium species, and not F. graminearum. The primary mycotoxin produced by F. graminearum is the trichothecene deoxynivalenol (DON) (see Figure 1 for the classification of DON) and other toxins produced include zearalenone, nivalenol (NIV), 4-acetylnivalenol (4-ANIV), and DON derivatives 3- and 15-acetyldeoxynivalenol (3-ADON and 15-ADON) [2,11,12,13,14]. The serious effects of crop infection by F. graminearum require in-depth studies on the pathogen, primarily its pathogenicity and virulence factors which can be comprehensively studied using, among other techniques, whole-genome sequencing, transcriptomics using the convenient, reliable, and large data-generating tools of NGS and proteomics. Recently, a multitude of data has been generated for F. graminearum either to understand its genome organization or its gene composition as well as genes involved in plant attack. Although more work is still required, the pace at which this information is generated is too high to allow sufficient time for synthesis, organization, and communication. Meanwhile, with the dynamics of climate change, the relations between the pathogen and the plant require active and rapid utilization of the data which is generated for use to ensure plant health and ultimately good human livelihood. Given this background, it remains important to reflect on the work which has been undertaken in F. graminearum research and collate it as a useful resource for various F. graminearum workers. Furthermore, the work to understand the genome of F. graminearum and its pathogenicity and virulence factors requires acceleration taking advantage of NGS technologies and its supplementing proteomics applications. This is the purpose of this current review, which aimed to trace progress made in discovering pathogenicity and virulence factors, in general, as well as using NGS, and to some extent, proteomics. We present more than 100 F. graminearum factors which either directly perform pathogenicity and virulence functions or are indirectly linked to pathogenicity and virulence. We conclude that a multitude of pathogenicity and virulence factors have been discovered, however, more work needs to be done taking advantage of NGS and its companion applications of proteomics. This review article is organized as follows: We provide a narrative background of pathogenicity and virulence, information on F. graminearum and information on NGS. We then discuss in detail the various pathogenicity and virulence factors of F. graminearum and elaborate on the pathogenicity and virulence factors discovered using NGS (see Figure 2 for the thought process and article organization) and, to some extent, those discovered using proteomics. We collated information from various recent scientific publications to uniquely reflect the pathogenicity and virulence factors. We conclude that a multitude of pathogenicity and virulence factors have been discovered, however, more work needs to be done taking advantage of NGS and its companion applications of proteomics.
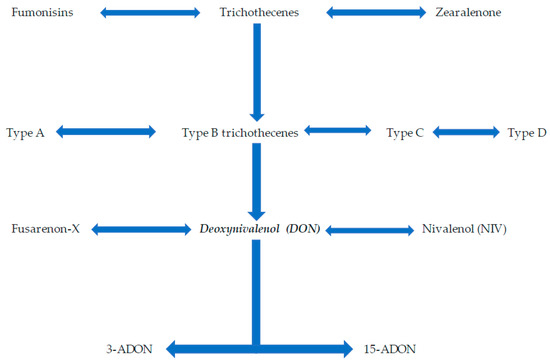
Figure 1.
The classification of the major toxin produced by Fusarium graminearum, DON. The major toxins produced by Fusarium species are fumonisins, trichothecenes, and zearalenone. DON is a type B trichothecene which has derivatives 3-ADON and 15-ADON.
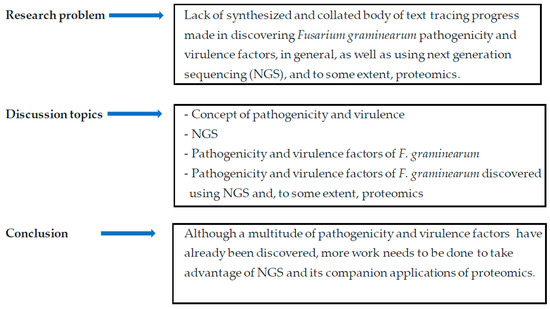
Figure 2.
An illustration of concept development and the organization of the article.
2. Next-Generation Sequencing, Its Relevance in Studying Plant Pathogenic Fungi and Fusarium graminearum
Nucleic acid sequencing has evolved from determining a few nucleotides of a nucleic acid fragment to its current form of generating large datasets of millions of reads which cover significant genomes of organisms. The generation of these large genomic and RNA datasets has been exploited by researchers to unravel various complexities of genome organization and function in many organisms. Nucleic acid sequencing is the establishment of the order of nucleotides of a given DNA or RNA fragment [15,16]. Earlier efforts to sequence nucleic acids, such as Sanger sequencing, would determine a stretch of a few nucleotides. These earlier sequencing efforts were slow and costly and the need for cheaper and faster sequencing methods grew. This demand drove the development of NGS. NGS, also termed high-throughput or massively parallel sequencing, is a genre of technologies that allow for thousands to billions of DNA fragments to be sequenced simultaneously and independently [17,18]. Although different platforms are commercially available, the NGS workflow features are generally similar, with four major steps: (i) DNA library preparation, (ii) clonal amplification, (iii) massive parallel sequencing, and (iv) data analysis [19,20]. In just the past five years, a number of instruments have been invented. The NGS platform Illumina NextSeq was invented in 2014, followed by the Ion Torrent S5/S5XL 540 in 2015. These were followed by Pacific Biosciences PacBio Sequel in 2016 [21]. These are among the instruments which have been recently invented and utilized to generate large datasets for organisms. One of the first inventions was the 454 GS FLX (454 Life Sciences, Branford, CT, USA). It utilizes an amplicon-based technology to create libraries from genetic regions selected with pairs of multiple primers [18,22,23]. On the contrary, Ion Torrent utilizes the pH variation sequencing method [18,24], whereas the modern platforms, such as Illumina MiSeq and Gene Reader System, utilize the fluorescence emission sequencing method [24,25]. NGS technologies have impressively accelerated research and are becoming employed routinely to seek solutions in different areas of research, as well as provide insights into pathogenicity and virulence of various plant pathogens [26,27,28,29,30,31,32,33,34]. Although NGS is more affordable than first-generation sequencing (considering time and money), it is still not within the reach of many laboratories in undeveloped countries [16]. However, sequencing services are offered by many sequencing companies at relatively low prices, and therefore, the need to buy the sequencers is less compelling for many. Despite the many challenges associated with NGS, such as inaccurate sequencing of homopolymer regions on certain NGS platforms which can lead to sequencing errors [16], as well as the lack of expertise in downstream processes such as data analysis, there is still success in the generation of sequences. NGS has been exploited to uncover genome organization and sequence for various plant pathogenic fungi. Plant pathogenic fungi were identified as important in some of the ambitious earlier projects in the area of characterization of fungi, such as the Fungal Genome Initiative [35,36], the Fungal Genomics Program [37,38] and its extension, the 1000 Fungal Genomes (1KFG) Project [39]. By extension, the plant pathogenic fungus F. graminearum has benefited tremendously and still does so from the various sequencing initiatives. The genome of F. graminearum has been sequenced completely and published for the benefit of Fusarium workers worldwide. This is in addition to the numerous gene sequences available in various databases. The complete genome and sequence resources have provided useful information on the biology, pathogenicity, and virulence of F. graminearum. A multitude of genes coding for pathogenicity and virulence factors of F. graminearum has been uncovered and the valuable information analyzed by researchers to deduce the much-needed insight into this devastating plant pathogen. We trace the important pathogenicity and virulence factors discovered and focus our attention on those which were discovered using NGS, and to some extent those which were discovered using proteomics. Within the context of this review article, pathogenicity factors and virulence factors are any substances produced by the fungus to gain access into the plant, manipulate it for its benefit, thus causing disease, as well as advance pathogenicity, thus causing the severity of the disease. Genes encoding these pathogenicity and virulence factors are themselves called pathogenicity and virulence factors. The terms pathogenicity and virulence are used jointly in this article. There does not seem to be clarity on the distinction between pathogenicity factors and virulence factors, and therefore, many publications seem to utilize these terms loosely. There is great justification to generate genome sequence information on F. graminearum. In order to offset the serious pathogenic problems caused by F. graminearum and to develop insights into the virulence and antagonistic defense mechanisms in host plants, it appears imperative to undertake the identification of the pathogenicity and virulence factors which make up the attack arsenal of F. graminearum. For this reason, the MIPS Fusarium graminearum Genome Database (FGDB) with an updated set with an estimated 14,000 genes and downstream analysis in a live gene validation process was established to provide a comprehensive genomic and molecular analysis [40]. Because Fusarium species, including F. graminearum, are among the most important phytopathogenic and toxigenic fungi, it is essential to understand the molecular underpinnings of their pathogenicity and virulence. Based on the comparative genomic analysis conducted on three phenotypically diverse species that include F. graminearum, it was revealed that among others, this particular homothallic fungal species, F. graminearum, has shown a relatively narrow host range that includes important crops like wheat (mentioned in Ma et al. [41]). Before discussing pathogenicity and virulence factors of F. graminearum, we look at pathogenicity and virulence factors of plant pathogens generally.
3. Pathogenicity and Virulence Factors of Plant Pathogens
Pathogenicity is the ability of an organism to cause disease, virulence, on the other hand, refers to the magnitude of the disease caused. There are three modes of attack by fungi. One mode requires ‘cooperation’ from the plant and living plant tissues. The fungus forms structures for penetration and a network of hyphae intertwined within the colonized plant tissues. Nutrients in the plants are extracted by this mass of hyphal structures formed in the intercellular spaces of the plant. This mode is called biotrophy (Figure 3). Other fungi release a barrage of degradative enzymes and toxins to lyse plant tissues and in so doing feed on the leaking cell contents and degraded tissue. This is called necrotrophy (Figure 4). Hemi-biotrophy is a hybrid mode between biotrophy and necrotrophy. Biotrophs produce proteins which circumvent the defense responses of the plant. These secreted proteins may have a counteracting effect on the plant’s defense proteins. This protein interaction in a match-match situation plays out as gene-for-gene interaction. This was found to be the case with flax rust of wheat caused by Melampsora lini and wheat rusts caused by the Puccinias. This gene-for-gene interaction is evidence of co-evolution between the plant and its pathogen. The mechanism of attack is less systematic in necrotrophs in which toxin production can be unspecific. Necrotrophs flood the plant with CWDEs and toxins which “chew up” the tissue. The hard physical defense structures of the plant, such as pectin, lignin, and glucans, crumble and the cells collapse. Pathogenicity and virulence-associated with plant pathogens are made possible by the secretion of pathogenicity and virulence factors which are molecules that help the pathogen colonize the plant [42,43]. For the purpose of this review, genes which are involved in the production of pathogenicity and virulence factors produced are themselves called pathogenicity and virulence factors.
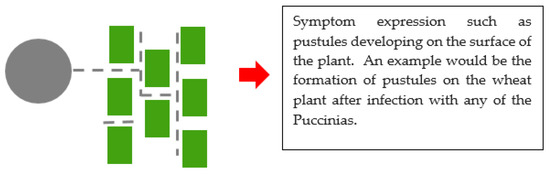
Figure 3.
An illustration of biotrophy between a plant pathogenic fungus and a plant. The grey circle represents a fungal spore. Dotted lines represent the germ tube which has developed to form a network of hyphae, and the green rectangles represent plant cells. Finally, symptoms are expressed.
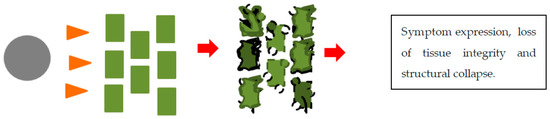
Figure 4.
An illustration of necrotrophy between a plant pathogenic fungus and a plant. The grey circle represents a fungal spore, the orange arrows represent secreted pathogenicity and virulence factors. The green rectangles represent plant cells, the plant cells become deformed, ripped, and cell contents leak. Finally, symptoms are expressed.
Decades of research have disclosed that plant pathogens have used a notable assortment of proteins and toxins as virulence factors [44]. Several categories of pathogenicity genes, namely (i) gene signals, (ii) genes generating or detoxifying toxins, (iii) metabolic enzymes, (iv) genes involved in the generation of particular infection structures, and (v) CWDEs, can be identified (mentioned in Duyvesteijn et al. [45]). Vadlapudi and Naidu [46] have identified several fungal processes and molecules which contribute to fungal pathogenicity or virulence which have the ability to harm the plants, and these include cell-wall-degrading proteins and toxins. Reception and transduction of signals in the early phase of plant infection play a vital role in triggering development and morphogenesis mechanisms involved in the penetration of the plant host. Several plant pathogens produce toxins that can harm the tissues of plants. Toxins may be either non-specific or host selective. The link between host selective toxin (HST) secretion and pathogenicity in plant pathogens and between sensitivity to toxins and susceptibility to disease in plants provides convincing proof that HSTs can be accountable for host-selective infection and disease growth [47]. Non-specific toxins harm phylogenetically unrelated plant cells, while HSTs are both pathogenicity determinants on various hosts and their genetic varieties. Loculoascomycetes produce the most recognized fungal HSTs. Trichothecenes are also compounds that are toxic to plants, and they are believed to be pathogenicity and/or virulence factors. The protein region of the oomycete Phytophthora family of the phytotoxin-like scr74 gene, the C-terminal of Phytophthora RXLR effector paralogues and a single amino acid polymorphism in the Phytophthora EPIC1 effector were related to the capacity to specialize in a fresh host [48,49,50]. Stukenbrock and McDonald [51] also reported polymorphism data showing the spread of two codons in the host-specific necrotrophic effector ToxA produced by Pyrenophora tritici-repentis and Stagonospora nodorum. Various fungi are models for studying plant-directed toxins. These include Alternaria alternata, Sclerotinia sclerotiorum, Botrytis cineria, and Fusarium. Certain plant species can also be used as models to study the effect of toxins on plants as it was demonstrated with trichothecenes on the unicellular plant Chlamydomonas reinhardtii [52]. The production of CWDEs (e.g., cellulases, hemicellulases, xylanases, and pectinases) is one of the processes used by fungi to invade plants. CWDEs degrade the complex polymers, cellulose, hemicellulose, xylan, and pectin to gain access to the contents of the plant cell. The enzymatic action of the CWDEs crumbles the cell wall, its integrity is weakened and eventually it collapses. Coupled with toxin production, the production of CWDEs is part of the infection process of F. graminearum when it attacks wheat spikelets. These compounds have been reported to help the infection spread to other tissues [53]. Several pathogenicity factors identified in Fusarium are also part of preserved complexes or pathways, such as mitogen-activated protein kinases. However, specific genes of Fusarium involved in host-pathogen interactions such as HST, elicitors, or Avr genes are mainly undefined, apart from the effectors secreted in xylem. For the purposes of this review article, attention is focused on pathogenicity and virulence factors which are produced by F. graminearum when it infects its various hosts which are primarily grain crops.
3.1. Pathogenicity and Virulence Factors of Fusarium graminearum
F. graminearum causes FHB, a devastating disease of wheat and other crops which include maize (Zea mays L.) and barley (Hordeum vulgare L.) [1,54] (mentioned in Desjardins et al. [54]). The rate at which this fungus attacks and infects its host plant is dependent on different mechanisms involving the secretion of extracellular enzymes and mycotoxins. Wanjiru and colleagues [53] show that the pathogenicity of F. graminearum is dependent on the extracellular enzymes secreted. The fungus enters the host via the epidermal cell walls with the help of infection-hyphae, leading to the reduction of the host plant’s cellulose, pectin, and xylan. Upon gaining access into the plant, F. graminearum produces various depolymerase enzymes such as CWDEs that macerate plant tissue. Jenczmionka and Schäfer [55] showed that the production of CWDEs such as pectinases, amylases, cellulases, xylanases, proteases, and lipases by F. graminearum enhances its penetration and proliferation in the attacked plant. The production of CWDEs is systematic, regulated, and programmed by the pathogen during attack. To prove regulation and co-ordination, Jenczmionka and Schäfer [55] showed that the F. graminearum Gpmk1 MAP kinase regulates the ability of the fungus to induce extracellular endoglucanase, xylanolytic as well as proteolytic activities.
With the disruption of the cell wall, F. graminearum uses the polysaccharides in the plant cell wall as a source of nutrients when advancing infection into the deeper underlying tissue [56]. These CWDEs have also been reported to elicit primary immune responses in plants [57,58,59,60,61]. According to Kikot and colleagues [62], various studies have shown a link between the presence of pectic enzymes, disease symptoms, and virulence, indicating CWDEs as decisive factors in the process of phytopathogenic fungi. Plant defense requires the pathogen to engage in a more sophisticated attack mode to overcome this defense. The production of CWDEs alone does not suffice, and therefore, other pathogenicity and virulence factors must come into play. Over and above CWDEs, F. graminearum attacks host plants through the release of mycotoxins which include primarily the trichothecene DON, also known as vomitoxin [63]. Sella and colleagues [64] reported that F. graminearum secretes lipase, DON, OS-2 (a stress-activated kinase) and Gpmk1 MAP kinases as essential pathogenicity and virulent factors necessary for the development of full disease symptoms in soybean seedlings, and OS-2 is involved in overcoming the resistance possessed by the soybean phytoalexin. Voigt and colleagues [65] observed that the lipase secreted by F. graminearum encoded by the FGL1 gene is a virulent factor that enhances the fungal pathogenicity against wheat and maize. Through functional annotation by gene deletion, Zhang and colleagues [66] also discovered the involvement of FgNoxR in conidiation, germination, sexual development and pathogenicity of F. graminearum. In another study, Jia and colleagues [67], through gene deletion and mutant analysis, identified a putative secondary metabolite biosynthesis gene cluster fg3_54 responsible for cell-to-cell penetration. The invasiveness of the fg3_54-deleted F. graminearum strains is restored by fusaoctaxin A (an octapeptide), leading to the conclusion that fusaoctaxin A is a virulence factor necessary for cell-to-cell invasion of wheat by F. graminearum. Gardiner and colleagues [68] point out that DON is associated with fungal virulence in wheat and barley and plays a crucial role in the spread of FHB. Moreover, DON elicits various defense responses in wheat which include hydrogen peroxide production, programmed cell death [69]. Trichothecenes are secondary metabolites secreted by many fungi belonging to the genera Fusarium, Myrothecium, Stachybotrys, and Trichoderma. Ueno [70] classifies trichothecenes into four types based on the unique characteristics of their chemical structures. Fusarium spp. secrete mostly type A and type B trichothecenes and not type C and type D [71]. These two types are characterized by the presence of a carbonyl group attached to C-8 of the sequiterpenoid backbone of trichothecenes. The presence of additional 7, 8 epoxides differentiates type C from the other types. Type D contains a macrocyclic ring that connects C-4 and C-15 of the sequiterpenoid backbone. Not a matter of discussion in the current review is the formation of toxin glycosides, also called masked mycotoxins, which are formed by cereal crops which are infected with mycotoxins. Toxin glycosides may be converted to toxins during digestion by the consumers of grain. Of great concern is the fact that toxin glycosides may escape detection by routine laboratory procedures (taken from McCormick et al. [72]. F. graminearum secretes type B trichothecenes DON, 15-ADON and 3-ADON or NIV. Lanoseth and Elen [73] reported the presence of DON, 3-ADON and NIV in infected oats. In F. graminearum-infected barley, wheat and corn, DON is the most common toxin produced by the fungus [74]. Ueno [70] indicates that the mechanism of action of trichothecenes involves binding the toxin to the ribosomes and inhibiting the synthesis of proteins.
A wide variety of proteins plays essential roles in the synthesis of trichothecenes, and most are encoded by genes within a trichothecene biosynthetic cluster (Tri cluster) [75,76]. The Tri cluster encodes 13 proteins that are responsible for the secretion of toxins in F. graminearum. Jenczmionka and colleagues [77] reported the role of MAP kinase Gpmk1/MAP1 in the production of F. graminearum conidia, which implies that the removal of the enzyme could result in a reduction in conidia production and eventually disease spread and pathogenicity. Urban and colleagues [78] observed that in their study the mutants of Δgpmk1/map1 did not cause any disease to wheat, although they were able to produce DON in the infected plant. It was further discovered that Gpmk1 was involved in the regulation of CWDEs (endo-1,4-b-glucanase and other proteolytic, xylanolytic, and lipolytic enzymes) expression [55]. Ochiai and colleagues [79] reported the activities of a protein kinase cascade in trichothecene biosynthesis. According to the authors, the protein kinases are actively involved in histidine kinase signal transduction. Alteration of the genes leads to the reduction in the expression of Tri6, Tri4 and NIV production in rice cultures. Perithecia, ascospore, conidia formation and spore germination are impacted by the activities of the serine/threonine-protein kinase gene (GzSNF1) [80]. GzSNF1 is also involved in the expression of F. graminearum genes which encode for endo-1,4-ß-xylanase 1 precursor (GzXYL1), an endo-1,4-ß-xylanase 2 precursor (GzXYL2) and an extracellular ß-xylosidase (GzXLP) which are involved in depolymerization and plant cell wall degradation. Lee and colleagues [80] also report a reduction in the virulence of Δgzsnf1 mutants on barley, proving the importance of the GzSNF1 gene in pathogenicity and virulence. The docking and fusion of secretory vesicles are required for vegetative and sexual growth in fungi. In the process of docking and fusion, v-SNARE proteins are fused with cargo proteins into vesicles. SNARE proteins are soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptors.
Hong [81] reports that the fusion of the apposing membranes which belong to the transport intermediate together with the target compartment is sped up by the interaction between the membrane-integrated t-SNARE protein and v-SNARE protein. In F. graminearum, syntaxin-like t-SNARE proteins are encoded by GzSYN1 and GzSYN2 and the deletion of these genes results in decreased virulence of the fungus on barley [82]. Syntaxin a is a multidomain protein with a globular amino terminal domain, a SNARE domain, and a carboxyl terminal transmembrane domain. In Δgzsyn1 mutants, perithecia were found to be unevenly distributed despite retaining reproductive ability with a decrease in the radial hyphal growth in culture. The Δgzsyn2 mutants are sterile females that could act like males in outcrosses with Δmat1-2 strains, resulting in the normal formation of perithecia. Radial hyphal growth of Δgzsyn2 mutants looks like the wild-type strains although the mycelia of both Δgzsyn1 and Δgzsyn2 mutants are generally thinner compared to those of wild-type strains [82].
Secreted lipase in F. graminearum is encoded by Fgl1 genes and the alteration of these genes in the fungus can reduce the virulence of the fungus in wheat with symptoms not spreading beyond the spikelets adjacent to inoculated spikelets [65]. Voigt and colleagues [65] further reported that in their study the maize plant infected with Δfgl1 mutants showed slight symptoms and the disease severity estimation was lower in the cobs infected with the mutant compared to the wild-type. The ability of fungi to colonize plants and initiate disease formation is dependent on the transmembrane transport proteins. These protein transporters can also exhibit virulence activities via the provision of protection from poisonous secondary metabolites secreted by host plants or even the fungus itself [83]. Many studies have indicated the roles of multiple ATP-binding cassette transporters or major facilitator superfamily (MFS) proteins in virulence activities of the fungi such as Botrytis cinerea, Cochliobolus carbonum, Cercospora kikuchii, Fusaria, Magnaporthe grisea, Mycosphaerella graminicola and Penicillium digitatum [83,84,85,86,87,88,89,90,91,92]. The same applies to F. graminearum, ATP-Binding Cassette Transporter Gene FgABCC9 was found to be involved in the fungus’s pathogenicity towards wheat [93]. F. graminearum attacks its host plant through the secretion of mycotoxins characterized as virulence factors. These secreted virulence factors and the level of secretion depend on the presence of the relevant genes that facilitate their secretion. Presently, trichothecene mycotoxins have been identified as the major virulence factors associated with F. graminearum [94,95]. The trichothecene gene cluster has diverse allelic genes with some of the genes found outside of the main cluster [95]. The allelic variations in fgTri8 are responsible for producing the DON derivatives [96]. Furthermore, allelic differences in fgTri1 are linked to the formation of the alternate trichothecene NX-2, whereas fgTri13 and fgTri7 are responsible for the trichothecene NIV [97,98]. The genes associated with the trichothecene production are not the only genes discovered to be responsible for the pathogenicity and virulence of F. graminearum. FGSG_04694, a gene-encoding polyketide synthase (PKS2) responsible for mycelial growth and fungi virulence, was found in F. graminearum [99]. Gaffoor and colleagues [100] confirm the function of PKS2 as they observed decreased mycelial growth and virulence in a mutant PKS2 found in F. graminearum. PKS2 is an accessory gene with irregular patterns of conservation that is also found in F. graminearum responsible for secondary metabolism and virulence [101,102]. The genes within these accessory gene clusters include terpene synthase-encoding genes FGSG_08181, FGSG_08182, which encode a putative transcription factor and three putative cytochrome P450 genes FGSG_17088, FGSG_08183 and FGSG_08187 [101,102]. Harris and colleagues [103] observed the expression of these accessory genes in cereals when infected by the fungus, which implies that they are involved in the virulence of the fungus.
ZIF1 that is conserved in filamentous ascomycetes encodes a bZIP (basic leucine zipper) transcription factor. The absence of FgZIF1 affects the virulence and reproduction ability of F. graminearum. Expression of FgZIF1 in mozif1 of M. oryzae mutants shows clearly that this transcription factor is functionally conserved in F. graminearum and M. oryzae [104]. Topoisomerase I encoded by TOP1 is another enzyme involved in the pathogenicity and conidia formation of fungi. The removal of TOP1 in F. graminearum was found to reduce disease expression in infected spikelets and abolition of spore formation [105]. FGSG_10057 encodes a Zn(II)2Cys6-type transcription factor in F. graminearum that enhances radial growth and virulence activity of the fungus in wheat [106]. Most of these factors that enhance virulence are proteins and these proteins need to be transported to the areas in the host plant where disease symptoms are expressed. Most recently, it was found that F. graminearum FgCWM1 encodes a cell wall mannoprotein which plays a role in pathogenicity. FgCWM1 mutants exhibit reduced pathogenicity in wheat [107]. Another recent discovery is that of a SNARE gene FgSec22 which was found to be required for vegetative growth, pathogenesis and DON biosynthesis in F. graminearum [108]. Similarly, an F. graminearum mitochondrial gene mitochondrial gene FgEch1 was found to be important for conidiation, DON production, and plant infection [109]. Another recently discovered pathogenicity and virulence factor is FgPEX4, which is important for development, cell wall integrity and pathogenicity [110]. Elucidation and complete delineation of the pathogenicity and virulence factors of F. graminearum must still be done by increasing efforts in whole-genome sequencing of strains from various parts of the world and by conducting in planta gene expression studies to assess genes which are differently expressed during the infection of a host plant by F. graminearum. Nowadays these can be conveniently done using NGS. The pathogenicity and virulence factors of F. graminearum can be conveniently summed under CWDEs, toxins, toxin biosynthesis genes, and other pathogenicity and virulence determinants. In this section, an elaborate explanation of the different pathogenicity and virulence factors was provided. Below we provide a convenient list of these pathogenicity and virulence factors under different topics, namely, CWDEs, toxins, toxin biosynthesis genes and other pathogenicity and virulence determinants (Table 1). The pathogenicity and virulence factors are therefore summarized under their respective headings. In trying to categorize pathogenicity and virulence factors there may be some ambiguities which may lead to unclear categorizations.

Table 1.
List of pathogenicity and virulence factors of Fusarium graminearum.
3.2. Comparative Genomics and Molecular Basis of Pathogenicity and Virulence in Fusarium graminearum
In order to offset the serious pathogenic problems caused by F. graminearum and to develop insights into the virulence and antagonistic defense mechanisms in host plants, it appears imperative to undertake the identification of fungal pathogenicity and virulence factors which make up the arsenal of this fungal plant pathogen. Presently this is conveniently done using the high-throughput genome sequencing technologies which generate large datasets to reveal genome organization and the various genes present in F. graminearum. The information generated from genome sequencing and RNA sequencing projects can be utilized to understand the mechanisms employed by F. graminearum to gain entry into plant tissue as well as advance with the plant. Various commendable projects have been launched in the previous years and have advanced our understanding of pathogenicity and virulence of F. graminearum. Among these significant projects was the MIPS Fusarium graminearum Genome Database (FGDB) with an estimated 14,000 genes and downstream analysis in a live gene validation process was established to provide a comprehensive genomic and molecular analysis [40]. Because Fusarium species are among the most important phytopathogenic and toxigenic fungi, it is essential to understand the molecular underpinnings of their pathogenicity. In addition, the production of mycotoxins by these fungi put animals and humans who consume the crop product at risk. Given this alarming situation, aggressive research in F. graminearum is imperative and it must take advantage of the presence of convenient tools to generate a multitude of data to elucidate the various pathogenicity and virulence processes of F. graminearum. Based on the comparative genomic analysis conducted on three phenotypically diverse species that include F. graminearum, it was revealed that among others this particular homothallic fungal species, F. graminearum, has shown a relatively narrow host range which includes important cereals [41]. It is therefore important to note that this pathogen is particularly notorious on wheat, barley, rice, oats by causing head blight or ‘scab’ and on maize causing mainly stalk and ear rot disease [2]. However, genomic analysis shows that this fungus may also infect other plant species without causing disease symptoms. Further genome analysis revealed that 67 gene clusters with significant enrichment of predicted secondary metabolites and with functional enzymes were shown to be expressed among which 30% with gene overexpression were likely in virulence [102]. While the exchange of genes between the core and supernumerary genomes bestows significant opportunities for adaptation and evolution on the organism, it appears reminiscent in F. graminearum, to the compartmentalization of genetic material where non-conserved regions are found at various places on the four core chromosomes [111]. Studies from comparative genomics indicate that these mobile pathogenicity chromosomes exist in most Fusarium species with lineage-specific genomic regions [41], nevertheless, the molecular foundation of pathogenicity in F. graminearum was shown to be closely associated with the MAP1 gene which is also responsible for the development of perithecia in the same fungal species [78]. Furthermore, 29 F. graminearum genes are rapidly evolving, in planta-induced and encode secreted proteins, strongly pointing toward effector function [112], implicating genomic footprints that can be used in predicting gene sets likely to be involved in host–pathogen interactions. In association with this, as forward and reverse genetics have improved our understanding of molecular mechanisms involved in pathogenesis, it was revealed that mitogen-activated protein kinase and cyclic AMP-protein kinase A cascades both regulate virulence in Fusarium species and it has been postulated that cell wall integrity might be necessary for invasive growth and/or resistance to plant defense compounds [113]. These snippets which have been discovered to give clues on the pathogenicity and virulence of F. graminearum necessitated a deeper and comprehensive interrogation of the genome of this fungal pathogen to uncover all pathogenicity and virulence genes. NGS was conveniently used to unravel various pathogenicity and virulence factors of F. graminearum to the benefit of F. graminearum researchers worldwide.
3.3. Pathogenicity and Virulence Factors of Fusarium graminearum Discovered Using NGS Technologies
Understanding the molecular mechanisms involved in fungal pathogens in plants has been accelerated over the past decade. Notably, NGS has contributed immensely towards the generation of vast datasets of genomes and transcriptomes. The availability of fungal genome sequences from a majority of plant fungal pathogens has contributed to these discoveries [28,29,114,115]. Genomics, transcriptomics, proteomics, and metabolomics approaches were introduced, allowing possible identification of genes, proteins, and metabolites of fungi in various artificial cultures and during infection of plants under different experimental conditions [116,117]. Fungal pathogens cause diseases in plants, resulting in tissue damage and disease due to pathogenicity and virulence factors which assist fungal pathogen survival and persistence [118]. The pathogens affect their host by adapting to their environment and secreting/producing pathogenicity-related toxins, pectic enzymes, and hormone-like compounds. These products can have devastating effects on the quality and yield of crops in the field and can also cause postharvest diseases [119]. The mechanisms involved in fungal pathogenesis in plants are therefore being studied broadly, to protect plants against diseases of economic importance. F. graminearum is one of the plant pathogens that affect grain cereal crops globally, causing different diseases in different crops [80,120,121]. The pathogen produces metabolites that are toxic or non-toxic, which enables it to manipulate the plant to acquire nutrition. Inhibition of pathogenicity and virulence factors enhanced by the pathogenic fungus results in the development of diseases in plants. Pathogenicity factors involved in plant-pathogen interactions have been investigated extensively in different plants, and genes, proteins, and metabolites have been identified [81,97,122,123,124,125], including those involved in response to F. graminearum infections [55,104,113,120,126,127]. Pathogenicity and virulence genes that have been largely identified belong to the trichothecene biosynthesis gene cluster, as described by Proctor and colleagues [95]. For F. graminearum, it is largely known that pathogenicity and virulence follow a path of germ tube emergence from conidia, production of cell wall-degrading enzymes and the production of trichothecene mycotoxins [53,94,95,128].
However, complete delineation of the infection process of F. graminearum requires sequencing and analysis of the entire fungal genome, conveniently and preferably using NGS. From the various efforts to study pathogenicity and virulence factors of F. graminearum using NGS, a few studies are worth noting. The first is the study of King and colleagues [115] (sequencing was done using the Illumina HiSeq 2000 sequencing platform) which provided a complete genome sequence from a combined genome analysis from various sources which had F. graminearum genome sequence information. Through the modification the gene model set, the FGRRES_17235_M gene was identified to be of particular interest because it is a virulence factor, it encodes for a cysteine-rich secretory protein, allergen V5/Tpx-1-related with CAP and signal peptide domains, with a previous link with plant pathogenesis proteins of the PR-1 family [129] and had been identified in the highly virulent F. graminearum strain CS3005 (gene ID: FG05_09548). Another gene 15917_M was identified as an endo-1,4-beta-xylanase enzyme, which hydrolyzes (1- > 4)-beta-D-xylosidic linkages in xylans, of the cell walls. The second study by Wang and colleagues [130] identified eight genes responsible for F. graminearum-wheat interactions. Three of the genes had already been identified in various studies [112,131]. Their gene annotation revealed largely polymer degrading function i.e., xylanase, catalase, protease and lipase. The third study was by Cuomo and colleagues [114], who identified a variety of pathogenicity and virulence factors belonging to the gene classes cutinases, pectate lyases, pectin lyases and other genes encoding secreted proteins. The fourth study reports, among the findings, the presence of 616 potential effector genes, 126 of which are expressed in a host-specific manner. This same study by Laurent and colleagues [132] which utilized the Illumina HiSeq 2000 identified 252 variants within the genic sequences and the intergenic sequences of Tri genes. Tri genes are involved in the production of type B trichothecenes. Given the mammoth task of elucidating pathogenicity and virulence factors of F. graminearum, increasing efforts in whole-genome sequencing of strains from various parts of the world is necessary. These efforts must be coupled with in planta gene expression studies to assess genes which are differently expressed during the infection of a host plant by F. graminearum. Traditionally, common techniques for gene expression studies included northern blotting, real-time PCR and microarrays. Nowadays most studies which could be done using northern blotting, real-time PCR and microarrays can be conducted conveniently using NGS. However, these relatively old techniques paved the way for NGS.
3.3.1. Notable Studies Which Paved Way for NGS
Before NGS technologies were commonplace, Northern blotting, real-time PCR, and microarrays were used to discover pathogenicity and virulence factors of fungi including F. graminearum. These studies paved the way for studies based on NGS. Some of the studies (reviewed in this section, none of which were performed in the past two years) utilized proteomic approaches and metabolomics. F. graminearum genome analysis reports that the pathogen consists of 1250 genes that encode secreted protein effectors [133]. These genes associated with fungal pathogenesis in vitro and in planta have been identified, and a large number are activated during infection. The F. graminearum classes of genes overlap with other plant-microbe interaction studies. These genes include the trichothecene gene cluster [95], bZIP transcription factor [104], syntaxin-like t-SNARE proteins [81], PKS [134], lipase [65], EBR1 [127], FgATG15 [135], among others. These genes play different roles in fungal pathogenesis. Among these types of genes, the Tri gene cluster has been characterized in the F. graminearum species complex, with the type B cluster being the most studied, due to their ability to cause diseases in both animals and humans [136]. In addition, proteins such as the five TRI proteins TRI1, FG00071; TRI3, FG03534; TRI4, FG03535; TRI14, FG03543; TRI101, FG07896 [137]; kinases [138] and FG00028, metallopeptidase MEP1; FG00060, KP4 killer toxin; FG00150, NADP-dependent oxidoreductase (COG2130); FG00192, peptidase S8 (pfam00082); FG00237, O-acyltransferase (pfam02458), among others [139], were found to be involved in F. graminearum pathogenesis [124,140,141]. The study by Dhokane et al. [124] demonstrates the interface between metabolomics and NGS. The role of the genes found to be involved in F. graminearum pathogenesis [124,140,141] has been identified and characterized using high throughput sequencing approaches, confirming their roles in fungal pathogenesis. NGS studies can employ either studying the genome or gene expression by means of transcriptomics. Both have been instrumental in discovering pathogenicity and virulence factors of F. graminearum.
3.3.2. Fusarium graminearum Pathogenesis-Related Genes Discovered Using RNA-Seq Transcriptomics
Many technologies have been employed for the detection, identification, and quantification of mycotoxins secreted by F. graminearum infections in grain cereals. A few studies, including those by Pasquali and Migheli [136], report the most important fungal mycotoxins belonging to the type B trichothecenes that are produced by the Fusarium spp. Identification of differentially expressed genes regulated by mycotoxins using transcriptomics is one of the approaches to identify and catalog pathogenicity and virulence factors in response to F. graminearum in grain cereals. Using comparative transcriptomic analysis, Walkowiak and colleagues [142] identified 1500 differentially expressed genes of two F. graminearum strains with 3-ADON and 15-ADON trichothecene toxin chemotypes. Furthermore, a whole-genome sequencing and comparative genomics study investigated four Fusarium strains and reported few pathogenicity and virulence genes [99]. These included the g8968 gene, which was predicted to contain the Tri5 domain. The Tri5 is a terpene synthase gene that catalyzes the first step of trichothecene biosynthesis in F. graminearum [95]. Furthermore, Tri8 was also identified in this study, and was reported to have exhibited a high frequency of SNPs and indels. The importance of Tri5 in pathogenicity and virulence was also supported in non-NGS studies. Boddu and colleagues [143] report Tri5 encoding a DON enzyme and revealed that loss-of-function F. graminearum tri5 mutants were unable to produce DON in wheat and barley. Similar results of the importance of Tri5 were also observed in a study by Jonkers and colleagues [144] whereby the Wor1-like Protein Fgp1 regulated pathogenicity, toxin synthesis and reproduction in F. graminearum. The study predicted that the loss of mycotoxin accumulation alone may be enough to explain the associated loss of pathogenicity to wheat.
Using transcriptomic analyses, differentially expressed genes (DEGs) were identified in infected spikelets and rachis wheat samples following F. graminearum infections [145]. From the list of the DEGs identified, a few trichothecene biosynthesis genes of the F. graminearum Tri cluster were mostly upregulated in the pathogen when infecting the resistant near isogeneic lines (NILs). Interestingly, another transcriptomic study was conducted between three host plants infected with F. graminearum strain to identify DEGs during colonization [103]. The study discovered that some genes were only expressed in a specific host, and there was also a difference in the genes’ functional categories identified in each host. In summary, the pathogenicity and virulence factors (listed in Section 3.3) of F. graminearum discovered using NGS technologies are provided in Table 2 below:

Table 2.
List of pathogenicity and virulence factors of Fusarium graminearum, including genes discovered using comparative genomics methods.
3.4. Fusarium Graminearum Pathogenesis Proteins Discovered Using Proteomics Approaches
Proteins are macromolecular machines which undertake various biochemical functions either building blocks, transporters, enzymes, and other functions. Proteins functions are coordinated, they are intertwined with other constituents of organisms like genes, RNA, and metabolites.
Proteomics is a large-scale study of sets of proteins produced by organisms. The set of total proteins produced by organisms is termed the proteome. The proteome varies across cells, and to some extent, it is defined by the underlying transcriptome. Traditionally, proteins were studied using low-throughput methods which focus on a relatively small set of proteins and provide qualitative data on structure, function, and interaction which other cell constituents. The small windows of knowledge opened by these traditional techniques denied biochemists of a broader bigger of the entire proteome of a cell. From the traditional methods of studying proteins emerged proteomics, a large-scale study of the proteome which is able to provide a snapshot of total proteins in an organism. As opposed to gel-based and antibody-based methods of studying proteins, mass spectrometry (MS) has been utilized to produce large datasets on the proteome. The basic workflow followed in proteomics is the extraction from the tissue of total proteins, followed by trypsin digestion, separation chromatography of short peptides from the digestion, and mass analysis by MS. From then onwards is the identification of proteins in the studied sample and the generation of the protein list. Similar to NGS, proteomic studies have been accelerated by the invention of various instruments which perform both the separation of the digested peptides, mass analysis, and other downstream applications. The instruments had to meet a number of qualities which include high throughput and high confidence in the identification of peptides, notably Orbitrap and time-of-flight mass analyzers [147,148,149,150]. The common trend in improving the performance of the MSs was to create hybrid systems. The hybrid systems make use of different ion analyzers or separators to enhance the capability, quality and usefulness of the results obtained. The advent of a triple quadrupole instrument enhanced MS capability over a single quadrupole. With the triple quadrupole, data on m/z values are combined with data on molecule fragmentation patterns to improve accuracy. The fragmentation pattern data is made possible by the presence of the second quadrupole which acts as a collision cell.
Fungi produce proteins for pathogenicity and virulence. Usually, some of these proteins are secreted into the intercellular spaces of plant and may either degrade the cell wall or act as effectors to perform various other pathogenicity and virulence functions. The secreted proteins are part of what is called the secretome. For comprehensive studies of pathogenicity and virulence proteins, studying the whole-organism proteome becomes necessary and it is made possible using high-throughput proteomics instruments. Yang and colleagues [138] pointed out that the invention of “omics” and bioinformatics tools has enhanced the proteome analysis of phytopathogenic fungi and their host interactions. Paper and colleagues [139] identified 120 fungal proteins of F. graminearum which include CWDEs from infected wheat heads through vacuum filtration. Among the identified proteins, about 56% controlled putative secretion signals. Transcriptomics data can be complemented with other omics approaches such as proteomics. The functions of proteins expressed at a given time can be identified and understood using proteomics approaches [138,151]. Although high throughput sequencing technologies have been available for over a decade, identifying differentially expressed proteins involved in fungal pathogenicity in cereals has not been widely investigated. Several proteins have been identified and characterized in F. graminearum and associated infections. However, they focused on the secretome and the impact of DON [137,139,152]. When the expression of F. graminearum proteins was investigated in response to in vitro stimulation of biosynthesis of the mycotoxin, trichothecene, 130 F. graminearum proteins that showed changes in expression were reported [137]. Many of the proteins identified were involved in fungal virulence. Moreover, investigation of a secretome of F. graminearum annotated secreted pathogenesis proteins related to the KP4 killer toxin and gEgh 16 proteins, among others, which were associated with pathogenicity [146].
Recently, a study by Lu and Edwards [153] reported about 190 small secreted cysteine-rich proteins (SSCPs) found in the genome of F. graminearum using genome-wide analysis. From the list of the SSCPs reported, five belonging to the cysteine-rich secretory proteins, Antigen 5 and pathogenesis-related 1 proteins were established. These SSCPs were observed to contain homologies to proteins that have established crystal structures. The authors also maintained that previous studies had not reported these SSCP associations with pathogenicity or virulence in plants. Moreover, in planta expression patterns showed upregulation of nine proteins associated with pathogenesis. These proteins contain conserved domains of Ecp-2-like panels 1 and 4, CFEM-like panel 3, Kp4-like panels 10, 14 and 9, PR-1-like panel 11, hydrophobin-like panel 12 and glycol_61 family panel 13, which are linked to fungal pathogenicity. This is in line with a study by Paper and colleagues [137], who identified 229 in vitro and 120 in planta proteins secreted by F. graminearum during infection of a wheat head using a high-throughput MS/MS comparative study. The study reported that 49 in planta proteins were not present in vitro, indicating that fungal lysis occurred during pathogenesis. Rampitsch and colleagues [154], on the other hand, reported 29 proteins whose relative abundance was affected in their secretome following infection by F. graminearum using a comparative secretome analysis. These proteins included metabolic enzymes, proteins of unknown function and pathogenesis-related proteins. Other studies involved in the identification of F. graminearum pathogenesis-related proteins in vitro and in planta also include the PR-3 and PR-5 proteins [155]. Various forms of proteomics are significant in studying plant-pathogen relations and other factors which include elicitors. An example of this is phosphoproteomics which has to some extent been studied in necrotrophic pathogens like B. cinerea [156,157,158], Septoria tritici [159]. These studies need to be extended to F. graminearum to deepen the understanding of its pathogenicity and virulence.
4. Summary and Conclusions
The infection mechanism of the plant pathogenic fungus F. graminearum is complex and intricate. It involves the production of a germ tube from conidia, the production of CWDEs, and eventually, the production of mycotoxins, chiefly type B trichothecene mycotoxins, DON, and NIV. Within the context of this review article, we regarded the CWDEs and the mycotoxins as pathogenicity and virulence factors which enable F. graminearum to gain entry into the plant and advance within the interior of the infected tissue. Our focus was on pathogenicity and virulence factors so far discovered, and we also devoted attention to those discovered using NGS and, to a limited extent, proteomics. We conclude that a multitude of pathogenicity and virulence factors have been discovered, however, more work needs to be done taking advantage of NGS and its companion applications of proteomics. Discovery of more pathogenicity and virulence and factors may facilitate the newer methods of control of F. graminearum infection of wheat and DON accumulation, for instance, as it has been shown by Machado et al. [160] using RNAi. Progress on the use of RNAi may depend greatly on the discovery of more pathogenicity and virulence factors of F. graminearum.
Author Contributions
M.E.R. wrote parts on the use of next generation sequencing in discovering F. graminearum pathogenicity and virulence factors. U.V.O. wrote about pathogenicity and virulence factors. C.M.K. wrote about pathogenicity and virulence factors of F. graminearum. L.K.L. wrote about next generation sequencing in general. A.A.W. wrote about the significance of NGS in studying fungal pathogenicity and K.N. wrote the remaining parts including the abstract, the introduction and the conclusions, and organized the manuscript to be a coherent article. All the authors participated in the formulation of the concept. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McMullen, M.P.; Jones, R.; Gallenberg, D. Scab of wheat and barley: Are-emerging disease of devastating impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Chang, I.; Kim, H.; Yun, S.; Leslie, J.F.; Lee, Y. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl. Environ. Microbiol. 2009, 84, 3285–3295. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top-10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Transcriptomics of cereal-Fusarium graminearum interactions: What we have learned so far. Mol. Plant Pathol. 2018, 19, 764–778. [Google Scholar] [CrossRef]
- Stack, R.W. Return of an old problem: Fusarium head blight of small grains. Plant Health Prog. 2000, 1, 19. [Google Scholar] [CrossRef]
- Muriuki, J.G. Deoxynivalenol and Nivalenol in Pathogenesis of Fusarium Head Blight in Wheat. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2001. [Google Scholar]
- Schmale, D.G.; Bergstrom, G.C. Fusarium Head Blight in Wheat. The Plant Health Instructor. Available online: http://www.apsnet.org/edcenter/intropp/lessons/fungi/ascomycetes/Pages/Fusarium.aspx (accessed on 1 October 2019).
- Stack, R.W. History of Fusarium head blight with emphasis on North America. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; APS Press: St. Paul, MN, USA, 2003; pp. 1–34. [Google Scholar]
- Proctor, R.H.; Desjardins, A.E.; Plattner, R.D.; Hohn, T.M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 1999, 27, 100–112. [Google Scholar] [CrossRef]
- Tralamazza, S.M.; Bemvenuti, R.H.; Zorzete, P.; de Souza Garcia, F.; Corrêa, B. Fungal diversity and natural occurrence of deoxynivalenol and zearalenone in freshly harvested wheat grains from Brazil. Food Chem. 2016, 196, 445–450. [Google Scholar] [CrossRef]
- Lee, T.; Lee, S.H.; Shin, J.Y.; Kim, H.K.; Yun, S.H.; Kim, H.Y.; Lee, S.; Ryu, J.G. Comparison of trichothecene biosynthetic gene expression between Fusarium graminearum and Fusarium asiaticum. Plant Pathol. J. 2014, 30, 33–42. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Hohn, T.M.; McCormick, S.P. Trichothecene biosynthesis in Fusarium species: Chemistry, genetics, and significance. Microbiol. Mol. Biol. Rev. 1993, 157, 595–604. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Hohn, T.M. Mycotoxins in plant pathogenesis. Mol. Plant Microbe Interact. 1997, 10, 147–152. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Grada, A.; Weinbrecht, K. Next-generation sequencing: Methodology and application. J. Investig. Dematol. 2013, 133, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 47, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Vigliar, E.; Malapelle, U.; de Luca, C.; Bellevicine, C.; Troncone, G. Challenges and opportunities of next-generation sequencing: A cytopathologist’s perspective. Cytopathology 2015, 26, 271–283. [Google Scholar] [CrossRef]
- Li, J.; Batcha, A.M.; Grüning, B.; Mansmann, U.R. An NGS workflow blueprint for DNA sequencing data and its application in individualized molecular oncology. Cancer Inform. 2016, 14, 87–107. [Google Scholar] [CrossRef]
- Kchouk, M.; Gibrat, J.F.; Elloumi, M. Generations of sequencing technologies: From first to next generation. Biol. Med. 2017, 9, 1–8. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Leamon, J.H. The development and impact of 454 sequencing. Nat. Biotechnol. 2008, 26, 1117–1124. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Passiglia, F.; Dendooven, A.; Pauwels, P.; Salgado, R.; Rolfo, C. Next–generation sequencing in clinical practice. In Oncogenomics; Dammacco, F., Silvestris, F., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 4, pp. 57–64. [Google Scholar]
- Koitzsch, U.; Heydt, C.; Attig, H.; Immerschitt, I.; Merkelbach-Bruse, S.; Fammartino, A.; Büttner, R.H.; Kong, Y.; Odenthal, M. Use of the GeneReader NGS System in a clinical pathology laboratory: A comparative study. J. Clin. Pathol. 2017, 70, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Salanoubat, M.; Genin, S.; Artiguenave, F.; Gousy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J.C.; Cattolico, L.; et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Tabolt, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.Q.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ding, S.W.; Zhang, Y.; Zhu, S. Identification of viruses and viroids by Next-Generation Sequencing and homology-dependent and homology-independent algorithms. Annu. Rev. Phytopathol. 2015, 53, 425–444. [Google Scholar] [CrossRef]
- Aylward, J.; Steenkamp, E.T.; Dreyer, L.L.; Roets, F.; Wingfield, B.D.; Wingfield, M.J. A plant pathology perspective of fungal genome sequencing. IMA Fungus 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral diagnostics in plants using Next generation sequencing: Computational analysis in practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next generation sequencing for detection and discovery of plant viruses and viroids: Comparison of two approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef]
- Birren, B.; Fink, G.; Lander, E. Fungal Genome Initiative: White Paper Developed by the Fungal Research Community; Whitehead Institute Center for Genome Research: Cambridge, MA, USA, 2002. [Google Scholar]
- The Fungal Genome Initiative Steering Committee. A White Paper for Fungal Comparative Genomics; Whitehead Institute: Cambridge, MA, USA, 2003. [Google Scholar]
- Grigoriev, I.V.; Cullen, D.; Goodwin, S.B.; Hibbett, D.; Jeffries, T.W.; Kubicek, C.P.; Kuske, C.; Magnuson, J.K.; Martin, F.; Spatafora, J.W.; et al. Fuelling the future with fungal genomics. Mycology 2011, 2, 192–209. [Google Scholar]
- Martin, F.; Cullen, D.; Hibbett, D.; Pisabarro, A.; Spatafora, J.W.; Bakerm, S.E.; Grigoriev, I.V. Sequencing the tree of life. New Phytol. 2011, 190, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J. 1000 fungal genomes to be sequenced. IMA Fungus 2011, 2, 41–45. [Google Scholar] [CrossRef]
- Güldener, U.; Mannhaupt, G.; Münsterkötter, M.; Haase, D.; Oesterheld, M.; Stümpflen, V.; Mewes, H.W.; Adam, G. FGDB: A comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2006, 34, 456–458. [Google Scholar] [CrossRef]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Yoder, O.C. Toxins in pathogenesis. Ann. Rev. Phytopathol. 1980, 18, 103–129. [Google Scholar] [CrossRef]
- Sharma, A.K.; Dhasamana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial virulence factors: Secreted for survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef]
- Speth, E.B.; Lee, Y.N.; He, S.Y. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 2007, 10, 580–586. [Google Scholar] [CrossRef]
- Duyvesteijn, R.G.; van Wijk, R.; Boer, Y.; Rep, M.; Cornelissen, B.J.; Haring, M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005, 57, 1051–1063. [Google Scholar] [CrossRef]
- Vadlapudi, V.; Naidu, K.C. Fungal pathogenicity of plants: Molecular approach. Eur. J. Exp. Biol. 2011, 1, 38–42. [Google Scholar]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bos, J.I.; Armstrong, M.; Whisson, S.C.; da Cunha, L.; Torto-Alalibo, T.; Win, J.; Avrova, A.O.; Wright, F.; Birch, P.R.J.; et al. Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol. Biol. Evol. 2005, 22, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Morgan, W.; Bos, J.; Krasileva, K.V.; Cano, L.M.; Chaparro-Garcia, A.; Ammar, R.; Staskawicz, B.J.; Kamoun, S. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 2007, 19, 2349–2369. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Stam, R.; Cano, L.M.; Song, J.; Sklenar, J.; Yoshida, K.; Bozkurt, T.O.; Oliva, R.; Liu, Z.; Tian, M.Y.; et al. Effector specialization in a lineage of the Irish potato famine pathogen. Science 2014, 343, 552–555. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; McDonald, B.A. Geographical variation and positive diversifying selection in the host-specific toxin SnToxA. Mol. Plant Pathol. 2007, 8, 321–332. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Wanjiru, W.M.; Zhensheng, K.; Buchenauer, H. Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 2002, 108, 803–810. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, H.R.; Bairoch, A.; McCormick, S.P.; Shaner, G.; Buechley, G.; Hohne, T.M. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant Microbe Interact. 1996, 9, 775–781. [Google Scholar] [CrossRef]
- Jenczmionka, N.J.; Schäfer, W. The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Curr. Genet. 2005, 47, 29–36. [Google Scholar] [CrossRef]
- Fernandez, J.; Marroquin-Guzman, M.; Wilson, R.A. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 2014, 52, 155–174. [Google Scholar] [CrossRef]
- Ryan, C.A.; Farmer, E.E. Oligosaccharide signals in plants: A current assessment. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 651–674. [Google Scholar] [CrossRef]
- Felix, G.; Regenass, M.; Boller, T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation and establishment of a refractory state. Plant J. 1993, 4, 307–316. [Google Scholar] [CrossRef]
- Zhang, B.; Ramonell, K.; Somerville, S.; Stacey, G. Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 2002, 15, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, D.; Robatzek, S. Pattern recognition receptors: From the cell surface to intracellular dynamics. Mol. Plant Microbe Interact. 2007, 20, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. How plants recognize pathogens and defend themselves. Cell Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: A review. J. Basic Microbiol. 2009, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yazar, S.; Omurtag, G.Z. Fumonisins, trichothecenes and zearalenone in cereals. Int. J. Mol. Sci. 2008, 9, 2062–2090. [Google Scholar] [CrossRef]
- Sella, L.; Gazzetti, K.; Castiglioni, C.; Schäfer, W.; Favaron, F. Fusarium graminearum possesses virulence factors common to Fusarium Head Blight of wheat and seedling rot of soybean but differing in their impact on disease severity. Phytopathology 2014, 104, 1201–1207. [Google Scholar] [CrossRef]
- Voigt, C.A.; Schäfer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, Y.; Wang, J.; Wang, Y.; Chen, M.; Norvienyeku, J.; Li, G.; Yu, W.; Wang, Z. FgNoxR, a regulatory subunit of NADPH oxidases, is required for female fertility and pathogenicity in Fusarium graminearum. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Jia, L.J.; Tang, H.Y.; Wang, W.Q.; Yuan, T.L.; Wei, W.Q.; Pang, B.; Gong, X.M.; Wang, S.F.; Li, Y.J.; Zhang, D.; et al. A linear nonribosomal octapeptide from Fusarium graminearum facilitates cell-to-cell invasion of wheat. Nat. Commun. 2019, 10, 922. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, S.A.; Boddu, J.; Berthiller, F.; Hametner, C.; Stupar, R.M.; Adam, G.; Muehlbauer, G.J. Transcriptome analysis of the barley-deoxynivalenol interaction: Evidence for a role of glutathione in deoxynivalenol detoxification. Mol. Plant Microbe Interact. 2010, 23, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Desmond, O.J.; Manners, J.M.; Stephens, A.E.; Maclean, D.J.; Schenk, P.M.; Gardiner, D.M.; Munn, A.L.; Kazan, K. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol. Plant Pathol. 2008, 9, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y. Toxicological features of T-2 toxin and related trichothecenes. Fundam. Appl. Toxicol. 1984, 4, 124–132. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef]
- McCormick, S.P.; Kato, T.; Maragos, C.M.; Busman, M.; Lattanzio, V.M.; Galaverna, G.; Dall-Asta, C.; Crich, D.; Price, N.P.; Kurtzman, C.P. Anomericity of T-2 toxin-glucoside: Masked mycotoxin in cereal crops. J. Agric. Food Chem. 2015, 63, 731–738. [Google Scholar] [CrossRef]
- Lanoseth, W.; Elen, O. Differences between barley, oats and wheat in the occurrence of deoxynivalenol and other trichothecenes in Norwegian grain. J. Phytopathol. 1996, 144, 113–118. [Google Scholar] [CrossRef]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; O’Donnell, K.; Ward, T.J.; Fujimura, M.; Hamamoto, H.; Shibata, T.; Yamaguchi, I. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 2003, 539, 105–110. [Google Scholar] [CrossRef]
- Wong, P.; Walter, M.; Lee, W.; Mannhaupt, G.; Münsterkötter, M.; Mewes, H.W.; Adam, G.; Güldener, U. FGDB: Revisiting the genome annotation of the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2011, 39, 637–639. [Google Scholar] [CrossRef]
- Jenczmionka, N.J.; Maier, F.J.; Lösch, A.P.; Schäfer, W. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase Gpmk1. Curr. Genet. 2003, 43, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Mott, E.; Farley, T.; Hammond-Kosack, K. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 2003, 4, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.; Lee, S.; Park, E.H.; Kim, K.W.; Kim, M.D.; Yun, S.H.; Lee, Y.W. GzSNF1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot. Cell 2009, 8, 116–127. [Google Scholar] [CrossRef]
- Hong, W. SNAREs and traffic. Biochim. Biophys. Acta BBA Mol. Cell Res. 2005, 1744, 120–144. [Google Scholar] [CrossRef]
- Hong, S.Y.; So, J.; Lee, J.; Min, K.; Son, H.; Park, C.; Yun, S.H.; Lee, Y.W. Functional analyses of two syntaxin-like SNARE genes, GzSYN1 and GzSYN2, in the ascomycete Gibberella zeae. Fungal Genet. Biol. 2010, 47, 364–372. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Zwiers, L.H.; De Waard, M.A. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 2002, 108, 719–734. [Google Scholar] [CrossRef]
- Pitkin, J.W.; Panaccione, D.G.; Walton, J.D. A putative cyclic peptide efflux pump encoded by the ToxA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 1996, 142, 1557–1565. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. Tri12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Gen. Genet. 1999, 261, 977–984. [Google Scholar] [CrossRef]
- Callahan, T.M.; Rose, M.S.; Meade, M.J.; Ehrenshaft, M.; Upchurch, R.G. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild-type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe Interact. 1999, 12, 901–910. [Google Scholar] [CrossRef]
- Urban, M.; Bhargava, T.; Hamer, J.E. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 1999, 18, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Zwiers, L.H.; De Waard, M.A. Characterization of the ABC transporter genes MgAtr1 and MgAtr2 from the wheat pathogen Mycosphaerella graminicola. Fungal Genet. Biol. 2000, 30, 115–125. [Google Scholar] [CrossRef]
- Nakaune, R.; Hamamoto, H.; Imada, J.; Akutsu, K.; Hibi, T. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol. Genet. Genom. 2002, 267, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, A.; Sopalla, C.; Weltring, K.M. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol. Plant Microbe Interact. 2002, 15, 102–108. [Google Scholar] [CrossRef]
- Schoonbeek, H.J.; van Nistelrooy, J.G.M.; de Waard, M.A. Functional analysis of ABC transporter genes from Botrytis cinerea identifies BcatrB as a transporter of eugenol. Eur. J. Plant Pathol. 2003, 109, 1003–1011. [Google Scholar] [CrossRef]
- Skov, J.; Lemmens, M.; Giese, H. Role of a Fusarium culmorum ABC transporter (FcABC1) during infection of wheat and barley. Physiol. Mol. Plant Pathol. 2004, 64, 245–254. [Google Scholar] [CrossRef]
- Qi, P.F.; Zhang, Y.Z.; Liu, C.H.; Zhu, J.; Chen, Q.; Guo, Z.R.; Wang, Y.; Xu, B.J.; Zheng, T.; Jiang, Y.F.; et al. Fusarium graminearum ATP-binding cassette transporter gene FgABCC9 is required for its transportation of salicylic acid, fungicide resistance, mycelial growth and pathogenicity towards wheat. Int. J. Mol. Sci. 2018, 19, 2351. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes with wildtype and trichodiene synthase gene disrupted Fusarium graminearum expressing a green fluorescence protein marker. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Alexander, N.J.; Desjardins, A.E. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 2009, 74, 1128–1142. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 2002, 68, 2959–2964. [Google Scholar] [CrossRef]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; McCormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Han, Y.K.; Kim, K.H.; Yun, S.H.; Lee, Y.W. Tri13 and Tri7 determine deoxynivalenol-and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 2002, 68, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Rowland, O.; Rodrigue, N.; Subramaniam, R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016, 17, 1014. [Google Scholar] [CrossRef] [PubMed]
- Gaffoor, I.; Brown, D.W.; Trail, F. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 2005, 4, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.T.; Gardiner, D.M.; Lysøe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Lee, W.; Wong, P.; Münsterkötter, M.; Mewes, H.W.; Schmeitzl, C.; Varga, E.; Berthiller, F.; Adam, G.; Güldener, U. The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS ONE 2014, 9, e0110311. [Google Scholar] [CrossRef]
- Harris, L.J.; Balcerzak, M.; Johnston, A.; Schneiderman, D.; Ouellet, T. Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016, 120, 111–123. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Hou, Z.; Wang, C.; Zhou, X.; Jonkers, W.; Ding, S.; Kistler, H.C.; Xu, J.R. A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2011, 24, 118–128. [Google Scholar] [CrossRef]
- Baldwin, T.K.; Urban, M.; Brown, N.; Hammond-Kosack, K.E. A role for topoisomerase in Fusarium graminearum and F. culmorum pathogenesis and sporulation. Mol. Plant Microbe Interact. 2010, 23, 566–577. [Google Scholar] [CrossRef]
- Dufresne, M.; van der Lee, T.; Ben M’Barek, S.; Xu, X.D.; Zhang, X.; Liu, T.G.; Waalwijk, C.; Zhang, W.; Kema, G.H.; Daboussi, M.J. Transposon-tagging identifies novel pathogenicity genes in Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 1552–1561. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Chen, Q.; Liu, C.H.; Lei, L.; Li, Y.; Zhao, K.; Wei, M.Q.; Guo, Z.R.; Wang, Y.; Xu, B.J.; et al. Fusarium graminearum FgCWM1 Encodes a Cell Wall Mannoprotein Conferring Sensitivity to Salicylic Acid and Virulence to Wheat. Toxins 2019, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G.D. R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr. Genet. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yu, X.; Zhang, L.; Zhang, L.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Mitochondrial FgEch1 is responsible for conidiation and full virulence in Fusarium graminearum. Curr. Genet. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, L.; Liang, Y.; Yu, J. FgPEX4 is involved in development, pathogenicity, and cell wall integrity in Fusarium graminearum. Curr. Genet. 2019, 65, 747–758. [Google Scholar] [CrossRef]
- Waalwijk, C.; Vanheule, A.; Audenaert, K.; Zhang, H.; Warris, S.; van de Geest, H.; van der Lee, T. Fusarium in the age of genomics. Trop. Plant Pathol. 2017, 42, 184–189. [Google Scholar] [CrossRef]
- Sperschneider, J.; Gardiner, D.M.; Thatcher, L.F.; Lyons, R.; Singh, K.B.; Manners, J.M.; Taylor, J.M. Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol. Evol. 2015, 7, 1613–1627. [Google Scholar] [CrossRef]
- Michielse, C.B.; van Wijk, R.; Reijnen, L.; Cornelissen, B.J.; Rep, M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 2009, 10. [Google Scholar] [CrossRef]
- Cuomo, C.A.; Guldener, U.; Xu, J.R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.J.; Baker, S.E.; Rep, M.; et al. Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef]
- King, R.; Urban, M.; Hammond-Kosack, M.C.; Hassani-Pak, K.; Hammond-Kosack, K.E. The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genom. 2015, 16, 544–564. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, J.; Song, X.; Yang, J.; Li, H.; Tang, H.; Liao, Y.C. Combined metabolomic and quantitative real-time PCR analyses reveal systems metabolic changes of Fusarium graminearum induced by Tri5 gene deletion. J. Proteome Res. 2011, 10, 2273–2285. [Google Scholar] [CrossRef]
- Fall, L.A.; Salazar, M.M.; Drnevich, J.; Holmes, J.R.; Tseng, M.C.; Kolb, F.L.; Mideros, S.X. Field pathogenomics of Fusarium head blight reveals pathogen transcriptome differences due to host resistance. Mycologia 2019, 111, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Cumagun, C.J.R.; Bowden, R.L.; Jurgenson, J.E.; Leslie, J.F.; Miedaner, T. Genetic mapping of pathogenicity and aggressiveness of Gibberella zeae (Fusarium graminearum) toward wheat. Phytopathology 2004, 94, 520–526. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Dhokane, D.; Karre, S.; Kushalappa, A.C.; McCartney, C. Integrated metabolo-transcriptomics reveals Fusarium Head Blight candidate resistance genes in wheat QTL-Fhb2. PLoS ONE 2016, 11, e0155851. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Sondergaard, T.E.; Covarelli, L.; Fuertes, P.R.; Hansen, F.T.; Frandsen, R.J.; Saei, W.; Lukassen, M.B.; Wimmer, R.; Nielsen, K.F.; et al. Identification of the biosynthetic gene clusters for the lipopeptides fusaristatin A and W493 B in Fusarium graminearum and F. pseudograminearum. J. Nat. Prod. 2014, 77, 2619–2625. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome dynamics associated with resistance and susceptibility against Fusarium head blight in four wheat genotypes. BMC Genom. 2018, 19, 642. [Google Scholar] [CrossRef]
- Zhao, C.; Waalwijk, C.; De Wit, P.J.; van der Lee, T.; Tang, D. EBR1, a novel Zn2Cys6 transcription factor, affects virulence and apical dominance of hyphal tip in Fusarium graminearum. Mol. Plant Microbe Interact. 2011, 24, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.Y.; Zhao, X.; Xu, J.R.; Güldener, U.; Kistler, H.C. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.C.; Cutt, J.R.; Klessig, D.F. Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 1991, 10, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, C.; Wang, C.; Chen, C.; Xu, J.R.; Liu, H. Characterization of the two-speed subgenomes of Fusarium graminearum reveals the fast-speed subgenome specialized for adaption and infection. Front. Plant Sci. 2017, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Gardiner, D.M.; Taylor, J.M.; Hane, J.K.; Singh, K.B.; Manners, J.M.A. Comparative hidden Markov model analysis pipeline identifies proteins characteristic of cereal-infecting fungi. BMC Genom. 2013, 14, 807–829. [Google Scholar] [CrossRef] [PubMed]
- Laurent, B.; Moinard, M.; Spataro, C.; Ponts, N.; Barreau, C.; Foulongne-Oriol, M. Landscape of genomic diversity and host adaptation in Fusarium graminearum. BMC Genom. 2017, 18, 203–229. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Panstruga, R. Pathogenomics of fungal plant parasites: What have we learnt about pathogenesis? Curr. Opin. Plant Biol. 2011, 14, 392–399. [Google Scholar] [CrossRef]
- Garvey, G.S.; McCormick, S.P.; Alexander, N.J.; Rayment, I. Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides. Protein Sci. 2009, 18, 747–761. [Google Scholar]
- Nguyen, L.N.; Bormann, J.; Le, G.T.; Stärkel, C.; Olsson, S.; Nosanchuk, J.D.; Giese, H.; Schäfer, W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet. Biol. 2011, 48, 217–224. [Google Scholar] [CrossRef]
- Pasquali, M.; Migheli, Q. Genetic approaches to chemotype determination in type B-trichothecene producing Fusaria. Int. J. Food Microbiol. 2014, 189, 164–182. [Google Scholar] [CrossRef]
- Taylor, R.D.; Saparno, A.; Blackwell, B.; Anoop, V.; Gleddie, S.; Tinker, N.A.; Harris, L.J. Proteomic analyses of Fusarium graminearum grown under mycotoxin-inducing conditions. Proteomics 2008, 8, 2256–2265. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jacobsen, S.; Jørgensen, H.J.; Collinge, D.B.; Svensson, B.; Finnie, C. Fusarium graminearum and its interactions with cereal heads: Studies in the proteomics era. Front. Plant Sci. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Paper, J.M.; Scott-Craig, J.S.; Adhikari, N.D.; Cuomo, C.A.; Walton, J.D. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 2007, 7, 3171–3183. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Faubert, D.; Hamzehzarghani, H. Mass spectrometry-based metabolomics application to identify quantitative resistance-related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 2010, 11, 769–782. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K.; Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Mamer, O.; Faubert, D. Metabolomics technology to phenotype resistance in barley against Gibberella zeae. Eur. J. Plant Pathol. 2011, 130, 29–43. [Google Scholar] [CrossRef]
- Walkowiak, S.; Bonner, C.T.; Wang, L.; Blackwell, B.; Rowland, O.; Subramaniam, R. Intraspecies interaction of Fusarium graminearum contributes to reduced toxin production and virulence. Mol. Plant Microbe Interact. 2015, 28, 1256–1267. [Google Scholar] [CrossRef]
- Boddu, J.; Cho, S.; Muehlbauer, G.J. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant Microbe Interact. 2007, 20, 1364–1375. [Google Scholar] [CrossRef]
- Jonkers, W.; Dong, Y.; Broz, K.; Kistler, H.C. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog. 2012, 8, e1002724. [Google Scholar] [CrossRef]
- Hofstad, A.N.; Nussbaumer, T.; Akhunov, E.; Shin, S.; Kugler, K.G.; Kistler, H.C.; Mayer, K.F.; Muehlbauer, G.J. Examining the transcriptional response in wheat Fhb1 near-isogenic lines to Fusarium graminearum infection and deoxynivalenol treatment. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Brown, N.A.; Antoniw, J.; Hammond-Kosack, K.E. The predicted secretome of the plant pathogenic fungus Fusarium graminearum: A refined comparative analysis. PLoS ONE 2012, 7, e33731. [Google Scholar] [CrossRef]
- Marshall, A.G.; Hendrickson, C.L. High-resolution mass spectrometers. Annu. Rev. Anal. Chem. 2008, 1, 579–599. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R.; Cociorva, D.; Liao, L.J.; Zabrouskov, V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal. Chem. 2006, 78, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Makarov, A.; Denisov, E.; Lange, O.; Horning, S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Noll, R.J.; Li, H.; Makarov, A.; Hardman, M.; Graham Cooks, R. The Orbitrap: A new mass spectrometer. J. Mass Spectrom. 2005, 40, 430–443. [Google Scholar] [CrossRef]
- Mehta, A.; Brasileiro, A.C.; Souza, D.S.; Romano, E.; Campos, M.A.; Grossi-de-Sá, M.F.; Silva, M.S.; Franco, O.L.; Fragoso, R.R.; Bevitori, R.; et al. Plant–pathogen interactions: What is proteomics telling us? FEBS J. 2008, 275, 3731–3746. [Google Scholar] [CrossRef]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef]
- Lu, S.; Edwards, M.C. Genome-wide analysis of small secreted cysteine-rich proteins identifies candidate effector proteins potentially involved in Fusarium graminearum−wheat interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef]
- Rampitsch, C.; Day, J.; Subramaniam, R.; Walkowiak, S. Comparative secretome analysis of Fusarium graminearum and two of its non-pathogenic mutants upon deoxynivalenol induction in vitro. Proteomics 2013, 13, 1913–1921. [Google Scholar] [CrossRef]
- Geddes, J.; Eudes, F.; Laroche, A.; Selinger, L.B. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics 2008, 8, 545–554. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Phosphoproteome profile of B. cinerea under different pathogenicity stages by plant based elicitors. J. Proteom. 2016, 139, 84–94. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Modifications of fungal membrane proteins profile under pathogenicity induction: A proteomic analysis of Botrytis cinerea membranome. Proteomics 2016, 16, 2363–2376. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Dataset of the Botrytis cinerea phosphoproteome induced by different plant-based elicitors. J. Proteom. 2016, 7, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Melo-Braga, M.N.; Larsen, M.R.; Jørgensen, H.J.; Palmisano, G. Battle through signaling between wheat and the fungal pathogen Septoria tritici revealed by proteomics and phosphoproteomics. Mol. Cell. Proteom. 2013, 12, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.K.; Brown, N.A.; Urban, M.; Kanyuka, K.; Hammond-Kosack, K.E. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 2018, 74, 790–799. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).