1. Introduction
The fungus
Magnaporthiopsis maydis is known to severely affect sensitive maize (
Zea mays) plants at the maturity stage. It has two additional synonyms,
Cephalosporium maydis [
1] and
Harpophora maydis [
2]. The disease, commonly known as “late wilt”, is reported so far in about eight countries but is considered the most harmful maize disease in Egypt [
3] and Israel [
4,
5]. Although some prevention methods can restrict disease outbreak (for example, see [
6]), the use of resistance maize cultivars is still the most common method for minimizing yield losses [
7], and it was shown that plant hormones might be involved in restricting the pathogen [
8]. Until lately, the only familiar and confirmed alternative host for
M. maydis was
Lupinus termis (lupine), which is widely cultivated in Egypt [
9].
M. maydis was recently identified in
Gossypium hirsutum (Pima cotton, Goliath cv.),
Citrullus lanatus (watermelon, Malali cv.), and
Setaria viridis (green foxtail) [
10].
Late wilt is often associated with contamination by secondary plant pathogenic fungi, enhancing the stem symptoms. Indeed, fungi such as
Fusarium verticillioides causing stalk rot,
Macrophomina phaseolina causing charcoal rot, and
M. maydis are grouped in a post-flowering stalk rot complex, which was identified as being one of the most widespread and destructive groups of diseases in maize [
11]. The interactions between
M. maydis and
Fusarium oxysporum in the roots of cotton and maize were previously investigated [
12]. These interactions are associated with an appreciable decrease in the severity of the cotton wilt disease but do not affect the pathogenicity of
M. maydis to maize. The reduction in infection in cotton was more pronounced when the
M. maydis precedes
F. oxysporum in the soil than when they were inoculated simultaneously.
M. maydis exerts little or no such effect when it follows
F. oxysporum in the soil. These interactions are particularly interesting since both pathogens can be found abundantly in Egyptian soils and share these two summer crop hosts (cotton and maize), which are cultivated alternately in two-year rotations [
12]. Interestingly, both
F. oxysporum f. sp. lupini and
M. maydis are also common fungal pathogens in lupine plants in Egypt, causing wilt diseases that result in substantial economic losses. The presence of both pathogens,
M. maydis and
F. oxysporum, in diseased maize, cotton, and lupine plants [
12] evoke the question of how their interactions or cross-influence affect these and other potential host plants.
Despite the pivotal role of
M. maydis, accumulating lines of evidence suggest that it does not act alone; instead, it is part of a more massive complex of maize pathogenic fungi [
11]. Some of these fungi, such as
F. verticillioides, are a secondary invader or opportunist that developed in late wilt-diseased attenuated maize plants [
5]. Hence, the co-influence between
M. maydis and other phytopathogenic fungi has only now begun to be revealed. The current study is an additional step towards uncovering the intriguing relationship between the late wilt pathogen and other phytoparasitic fungi. In particular, the interactions between
M. maydis and
M. phaseolina in maize and cotton plants were investigated.
M. phaseolina (Tassi) Goidanich, the charcoal rot causal agent, is a common pathogen affecting a wide range of cultivated and wild species in temperate, warm and tropical regions around the world [
13]. The fungus has a vast host range and is responsible for causing damage to more than 500 cultivated and wild plants [
14], including maize and cotton. Similar to
M. maydis,
M. phaseolina is a primarily soil-borne and seed-borne pathogen, but unlike
M. maydis, it has a highly competitive saprophytic ability [
15].
Both fungi have a similar pathogenic mode of action and produce similar visible symptoms. These pathogens are a challenge to control because they often survive in the soil for long periods. When a susceptible host plant is seeded, the fungi can penetrate the plants’ roots, causing root necrosis and affecting sprout development [
14,
16]. Usually, the symptoms develop later in the season as plants begin to flower and are enhanced under drought conditions [
17,
18]. When the growth season advances,
M. maydis and
M. phaseolina may spread upwards inside the plant’s vascular system, disrupt the water supply, and lead to dehydration and yield reduction [
19,
20]. While
M. phaseolina is capable of producing phytotoxins [
14], so far, no secreted phytotoxins have been identified in
M. maydis. However, similar to
M. phaseolina,
M. maydis culture filtrate negatively affects maize seed development [
21].
In a survey conducted in 2016 (Roni Cohen, unpublished results),
M. phaseolina was identified at a high frequency in cotton fields scattered throughout the Hula Valley in the Upper Galilee (northern Israel). These fields are also known to be heavily infested with
M. maydis, which causes a severe disease to sensitive maize hybrids. In the summer of 2017, both
M. maydis and
M. phaseolina were identified using quantitative real-time polymerase chain reaction (qPCR) in diseased cotton plants in a commercial field in Yavne (southern coastal plain of Israel) [
10]. These accumulating lines of evidence suggested possible co-interactions between the two pathogens in cotton and maize in commercial fields where they coexist.
To the best of our knowledge, this is the first study of the co-influence of
M. maydis and
M. phaseolina under field conditions with molecular DNA tracking of each pathogen in maize and cotton tissues. This study aims at deepening our understanding of these curious relationships by measuring changes in the plants’ phenological development, health condition, and yield production. Previous reports of antagonisms among plant pathogens are relatively sparse [
22]. The scarcity of reports may belie their frequent occurrence. A recent meta-analysis suggests that these interactions are probably much more prevalent than previously estimated [
23]. Reports on pathogen–pathogen antagonisms encompass diverse plant disease systems, including tree pathogens and wood-rotting fungi [
24], storage-rotting fungi [
25], foliar pathogens [
26], and root rots [
27]. Studies of foliar pathogens are most common, with antagonisms being reported among foliar pathogens of wheat, barley, and peas (see, for example, [
26,
28,
29]). This may be due to their relative ease of observation and not necessarily due to some innate association.
2. Materials and Methods
2.1. Fungal Isolates and Growth Conditions
The
M. maydis isolate called
Hm-2 (CBS 133165, CBS-KNAW Fungal Biodiversity Center, Utrecht, the Netherlands) was recovered from wilting maize plants sampled in Sde Nehemia (the Hula Valley, Upper Galilee, northern Israel) in 2001, and identified using pathogenicity, physiology, colony morphology, and microscopic and molecular traits [
5,
30]. The
M. phaseolina isolate called
Mp-1 was recovered from wilted cotton plants in 2017 (Roni Cohen’s lab, Newe Ya’ar Research Center, northern Israel) and was identified using pathogenicity, physiology, colony morphology, and microscopic characteristics. Final molecular identification of this isolate was accomplished in this study by qPCR with a primer set targeting
M. phaseolina species-specific fragments [
31], and a primer set targeting the Internal transcribed spacers (ITS), ITS1 and ITS4, as we will elaborate below. The qPCR resultant oligonucleotide was identified by sequencing. The fungi were grown on rich potato dextrose agar (PDA) (Difco, Detroit, MI, USA) at 28 ± 1 °C in the dark for 4-7 days.
2.2. Maize and Cotton Cultivars Selected for This Study
The Prelude cv. sweet maize from SRS Snowy River seeds, Australia (supplied by Green 2000 Ltd., Israel) was chosen for this study. The Prelude cv. had been previously tested for susceptibility to late wilt in the field [
6,
20] and proved to be highly sensitive. The Pima cotton, Goliath cv. (extra-long-staple [ELS] cotton) is commonly grown in different parts of Israel (supplied by Isreal Seeds, Kibbutz Shefaim). This cotton cultivar is also traditionally grown on late wilt contaminated fields during crop rotation and was recently reported to be
M. maydis vulnerable [
10]. In an extensive survey conducted by Roni Cohen across Israel (Newe Ya’ar Research Center, northern Israel), the Pima cotton, Goliath cv. was reported to be sensitive to
M. phaseolina charcoal rot disease [
32].
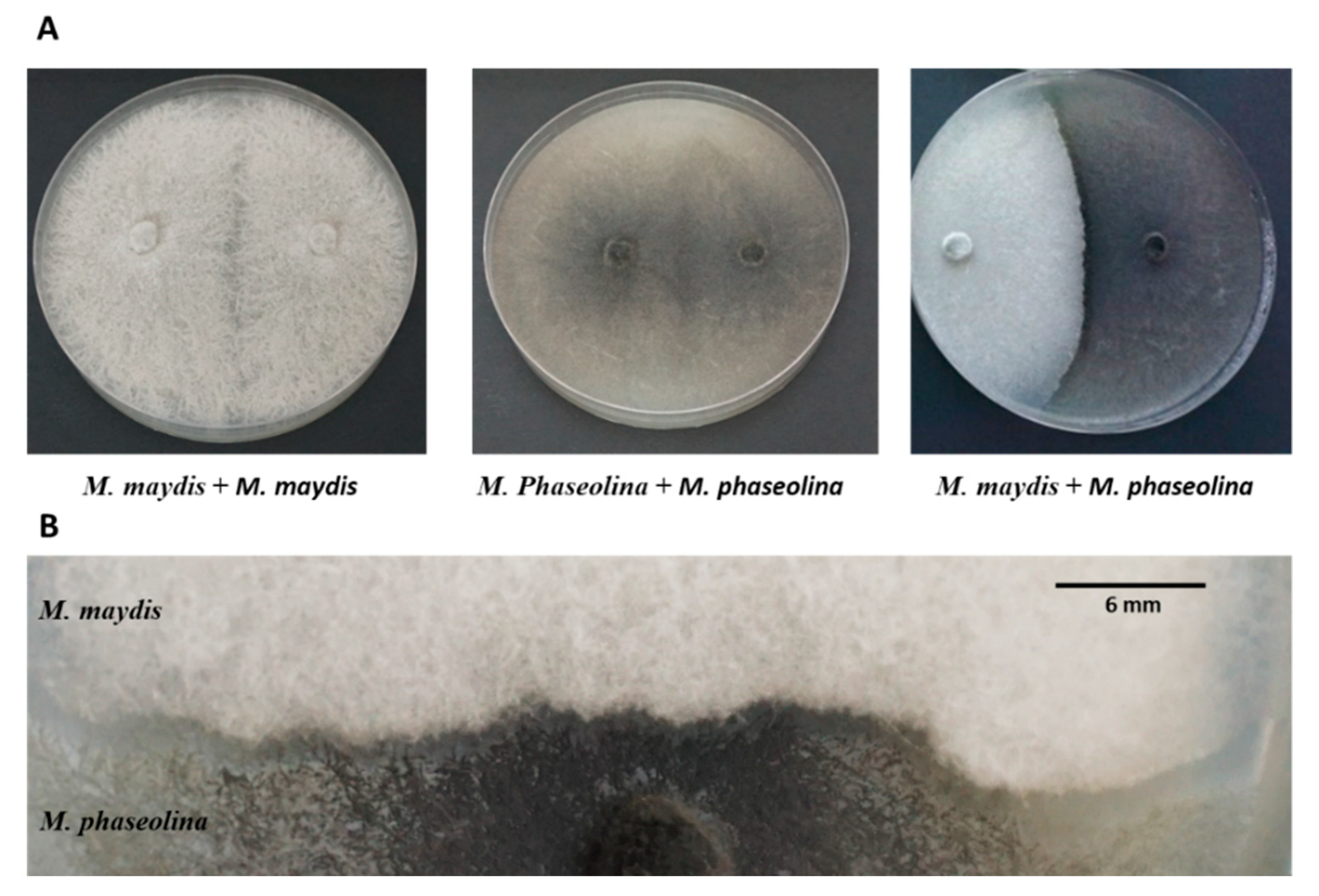
2.3. Plate Confrontation Assay
The interactions between M. maydis and M. phaseolina when both fungi are grown together on the surface of potato dextrose agar can be referred to as antagonism or a mycoparasitism. The plate confrontation assays were performed by positioning a mycelia disk (6 mm in diameter, taken from the margins of a 4-6-day-old colony) of M. maydis at one pole of the culture plate and mycelia disk (at the same size and age) of M. phaseolina on the opposite pole. The M. phaseolina was added to the plate two days after M. maydis since it grows significantly faster. The two fungi were then allowed to grow under optimal conditions (28 ± 1 °C in the dark) for six days until the colonies’ margins met in the area of interaction.
2.4. Full-Growth Season Pot Experiments under Field Conditions
This study examined the combined effect of M. maydis and M. phaseolina on the growth and yield of maize and cotton plants grown in pots in an open-air enclosure under field conditions. The experiments aimed at simulating field conditions and the reason for using pots (positioned in the open-air field) with naturally infested soil instead of sowing the plants directly to the field soil was to allow enhancing the soil inocula in order to achieve, as much as possible, high and equable infection, and for better control of the water regime. To elaborate on this, pathogenicity trials cannot rely on natural soil infestation alone, which can lead to highly variable results. Even in heavily infested fields, the spreading of the pathogen is not uniform. The pathogen is scattered in small quantities in the soil, and the disease spreading is not uniform in the field.
The experiments were conducted in an experimental farm located near Kibbutz Amir (in the Hula Valley Upper Galilee, northern Israel) during a whole growing season and were subsequently repeated twice in the spring and summer of 2018 and 2019. The two subsequent experiments were performed in a completely randomized design. Each treatment included 10 independent replications (pots). Each pot was 10 L in volume. The negative control in the experiments was soil taken from a nearby field that had no history of
M. maydis or
M. phaseolina infestation, and if such an infestation did exist, it was assumed to be very low. All the plants received fertilization and insecticides according to the recommended growth protocol of the Israel Ministry of Agriculture Consultation Service (SAHAM). Each of the pots was seeded with five seeds. The sprouts were diluted to one plant per pot during the seedling growth stage (34 and 23 days after sowing, DAS, in the maize pots, and 28 and 41 DAS in the cotton pots in 2018 and 2019, respectively). Watering was done by drip line irrigation (two droppers per pot) and controlled by a computerized irrigation system. The specific irrigation amount varied for each of the two experiments, as detailed in
Table 1. The 2019 repetition included additional treatments to evaluate the effect of the deficient water regime, as will be detailed below. The average meteorological parameters measured during the 2018 and 2019 experimental periods were similar, with higher soil temperature (in both the maize and cotton growth periods) and radiation (only during the maize growth period) in 2019, as detailed in
Table 2.
The methodology used for plant inoculation and growth was similar to that of Degani et al., 2019 [
20]. The inoculum method involved mixing naturally infested peat soil taken from the Neot Mordechai maize field (Hula Valley, Upper Galilee, northern Israel), which was known to be
M. maydis infested for many years [
33] and was probably also subjected to
M. phaseolina charcoal rot disease [
32], with 30% Perlite No. 4 (to aerate the soil).
Additionally, a complementary inoculation with the
Hm-2 isolate was carried out in two steps. First, 40 g of sterilized infected wheat seeds were added to the top 20 cm of the soil of each pot with the sowing. These seeds were previously incubated for three weeks at 28 °C in the dark with
M. maydis or
M. phaseolina culture agar disks (10 disks per 100 g seeds) and were used here to disperse the pathogen in the soil, as previously described [
5,
34]. Second, with the above-ground appearance, two agar disks (6-mm-diameter) taken from five-day-old
M. maydis or
M. phaseolina colonies (grown for six days at 28 °C in the dark) were added to the upper parts of the roots (4 cm beneath the ground surface). In the maize pots, this procedure was performed 13 or 9 days after the sowing (in the 2018 and 2019 experiments, respectively). In the cotton pots, this procedure was performed 22 or 9 days after the sowing (in the 2018 and 2019 experiments, respectively). In the 2019 repetition, an
M. phaseolina complementary inoculum with
Mp-1 isolate spore suspension was added to enhance the disease severity outcome. The
M. phaseolina spore suspension was prepared by washing and collecting the spores from 10 five-day-old
M. phaseolina colonies grown, as described above, on PDA plates. The spores were then suspended in 1 L sterile double distilled water (DDW). Ten ml of this suspension was sprinkled onto each seed with the sowing.
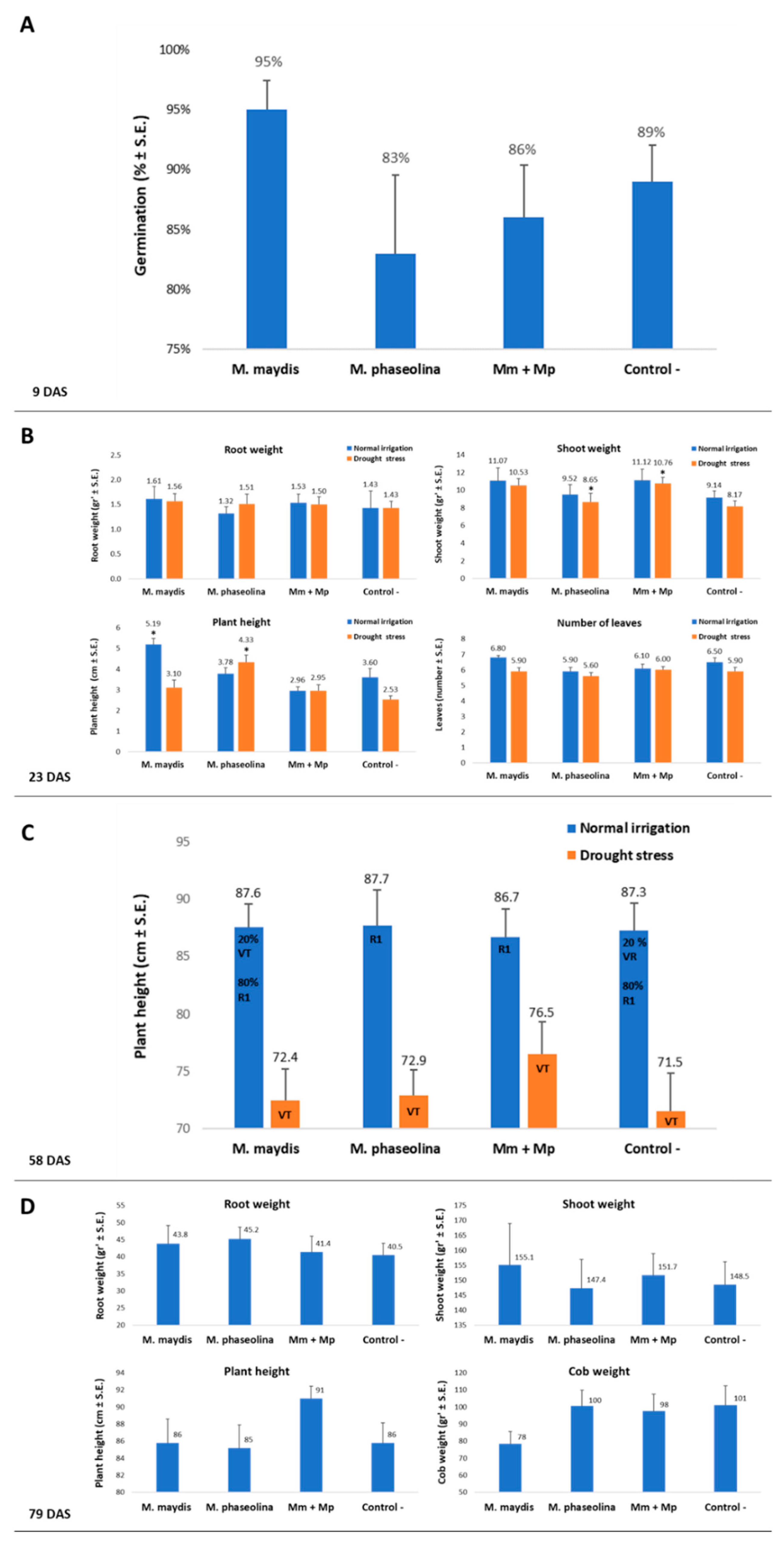
Maize developmental stages are stated according to [
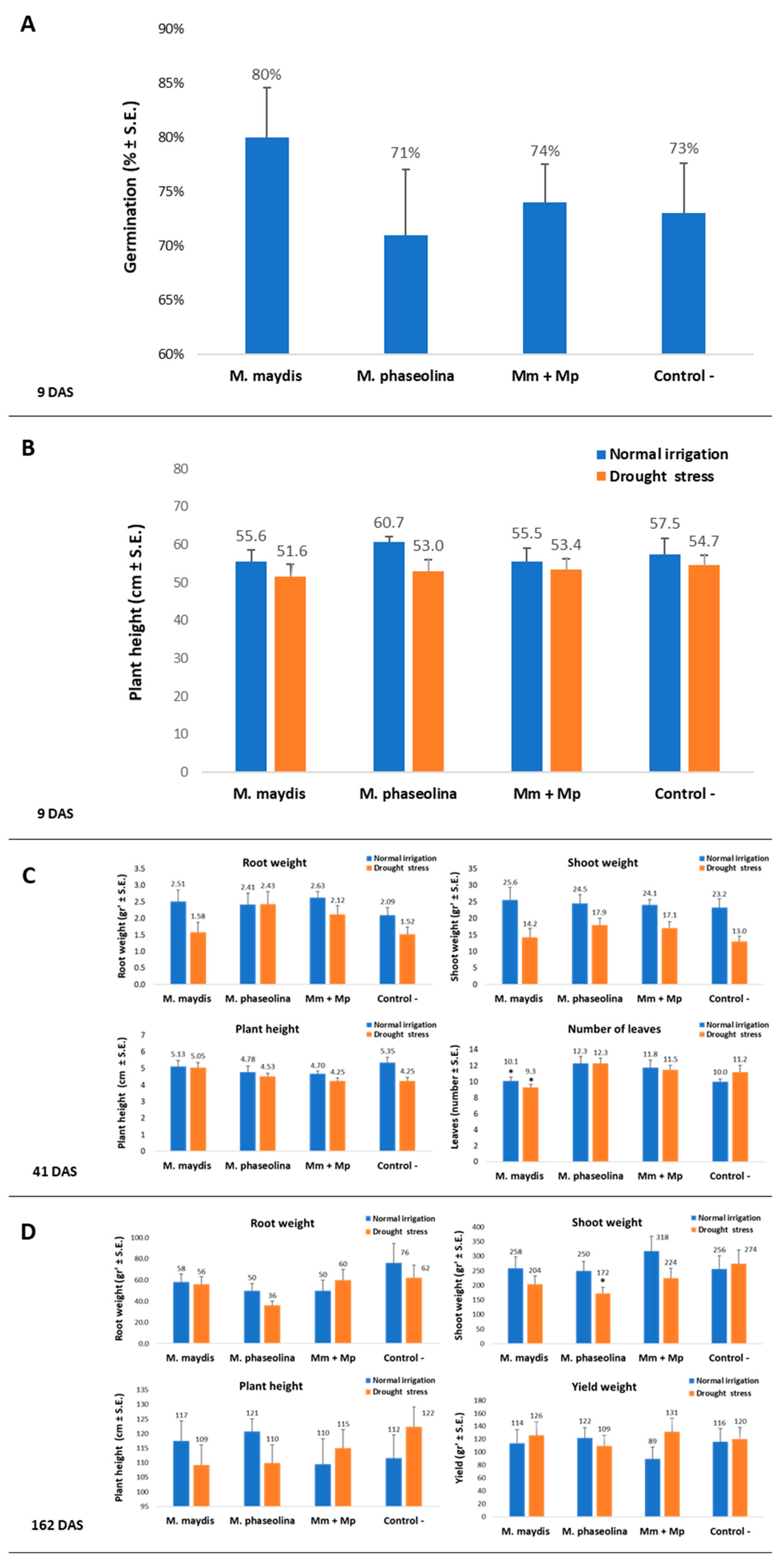
35]. Emergence percentages evaluation conducted 9 DAS for all the plants. The growth parameters were evaluated at the maize sprouting phase (29, 34-37, 54 DAS). Later, at the end of the experiment, the maize harvest day (79-82 DAS), phenological stage evaluation, wilt determination, and yield assessment were performed. A dehydration assessment was done on harvest day by calculating the percentage of plants showing typical maize late wilt dehydration symptoms—the upper leaves’ color alternation to light-silver and then to light-brown and rolling inward from the edges of the leaf. This assessment was done using four categories: 0—the plant is completely dried, 1—the plant has severe dehydration symptoms (over 50% of its part are dehydrated), 2—the plant shows light symptoms and most of its parts are green, 4—the plant is healthy, green, and without visible signs of disease. Thus, each repeat (individual plant) was evaluated according to this scale, and the dehydration proportions are the mean result received from each treatment. A similar symptom evaluation procedure was conducted for the cotton plants 37-41 and 54 DAS, as well as on the harvest day (154 or 167 DAS in 2018 and 2019, respectively), including growth parameter assessment and yield determination. Additionally, all of the plants in each treatment were uprooted, and each plant’s above-ground parts were measured to determine their height and wet weight. The roots’ and shoots’ dry biomass was determined after drying the plants at 65 °C for 62 h. Samples of tissue were taken in order to identify the fungal DNA inside the host tissues using qPCR, as will be described below.
The 2018 experiment. In the first experiment (2018), the sowing of the cotton plants was performed on May 5, 2018, and the sowing of maize was performed on May 24, 2018. Both cotton and maize plants emerged above the ground surface eight days after sowing. The maize fertilization took place 57 DAS (about one week after the male flowering). At that age, most of the plants were at the R1 development stage (silking: plants are at a maximum or near-maximum height and have one or more silks extending outside the husk leaves). The maize plants were harvested on August 14, 2018 (82 DAS, 25 days after fertilization, DAF). The cotton plants flowered on May 30, 2018 (58 DAS) and were harvested on October 3, 2018 (154 DAS).
The 2019 experiment. The field experiment, described above, was repeated in the summer of the following year (2019) at the same location and according to the same design. This repetition included additional treatments to evaluate the effect of water regime on pathogenesis outcome and disease symptoms development. Thus, half of the treatments received reduced irrigation (see
Table 1) in order to produce a restricted, near-drought water regime that may encourage the development of
M. maydis and
M. phaseolina, and to facilitate the disease symptoms outbreak.
The sowing of the maize and cotton plants was performed on May 21, 2019. The maize plants’ fertilization took place 58 DAS (about one week after the male flowering). At that age, in the maize plot that received a regular water supply, most of the plants were at the R1 development stage. At the same age (58 DAS), the maize plots that had received reduced irrigation were at the VT development stage (tasseling: plants with fully visible tassel branches). The low water supply did not allow the plant to continue developing beyond this age (58 DAS), and they suffered from severe drought. Consequently, the reduced water supply plots were harvested one week later, 65 DAS, before the development of cobs. The maize plants that received a regular water regime were harvested on August 08, 2019 (79 DAS, 21 DAF). In the cotton plots, on 58 DAS, the plants had blossom buttons but had not yet flowered. The cotton plants flowered about one week later and were harvested on October 29, 2019 (161 DAS(.
2.5. Molecular Diagnosis
A molecular diagnosis was performed on plants collected from the pots experiment. Roots were washed to remove soil under tap water. The root tissue samples were taken from the uppermost part of the root—from the crown, seminal, and lateral roots (the fibrous root system)—and not from the primary root (the radicle root). The stem tissue samples were taken from the joint section between the aboveground first and second internode. Sampling was done by removing a cross-section of approximately 2 cm in length from each plant stem. The total weight of the samples (roots or stem) was adjusted to 0.7 g and considered to be one repeat. Tissue samples were placed in universal extraction bags (Bioreba, Reinach, Switzerland) with 4 mL of cetyl hexadecyltrimethylammonium bromide (CTAB) buffer and were ground with a manual tissue homogenizer (Bioreba, Reinach, Switzerland) for 5 min until the tissues were completely homogenized. The homogenized samples were treated for DNA purification as previously described [
36]. The DNA was suspended in 100 µL of HPLC-grade water and kept at −20 °C until use in the qPCR analysis.
All of the qPCR reactions were performed as previously described [
36] using the ABI PRISM
® 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) for 384-well plates. The 5 µL of total reaction was used per sample well, including 2 µL of sample DNA extract, 2.5 µL of iTaq™ Universal SYBR
® Green Supermix (Bio-Rad Laboratories Ltd., Rishon Le Zion, Israel), 0.25 µL of forward primer and 0.25 µL of reverse primer (10 µM from each primer to a well). The qPCR program was as follows: precycle activation stage, 1 min at 95 °C; 40 cycles of denaturation (15 s at 95 °C) and annealing and extension (30 s at 60 °C), followed by a melting curve analysis. Plant samples (root and stem tissues) from each experiment were analyzed separately by qPCR. Each sample was tested three times by qPCR to ensure consistency of the results. The A200a primers were utilized for qPCR (sequences in
Table 3). The gene encoding for the last enzyme in the respiratory electron transport chain of the eukaryotic mitochondria-cytochrome c oxidase (
COXI gene) was used as a “housekeeping” reference gene to normalize the amount of DNA [
37]. The
COX gene was amplified using the COX F/R primer set (
Table 3). The amplified gene is from both the plants and the fungi that inoculate it. Relative quantification of the target
M. maydis fungal DNA was calculated according to the ΔCt model [
38]. The efficiency was presumed to be the same for all samples.
2.6. Statistical Analyses
A fully randomized statistical design was used in all experiments. Statistics and data analysis were carried out using the JMP program, 7th Edition, SAS Institute Inc., Cary, NC, USA. For the evaluation of M. maydis and M. phaseolina infection outcome in the experiments, the one-way ANOVA followed by post-hoc multiple comparisons of Student’s t-test for each pair was used. The t-test compared (with a significance threshold of p = 0.05) each treatment to the control.
4. Discussion
In Israel,
M. maydis is considered a major threat to commercial maize fields, while
M. phaseolina is a primary pathogen causing economic damage in cotton fields. Nevertheless, both pathogens can be found in both crops and may contribute to disease outbreak and spread [
10,
41]. The current study reveals a complex and competitive interaction between
M. maydis and
M. phaseolina species that had been previously unknown. The results of this study strongly suggest an antagonistic co-influence of these two vascular wilt pathogens, which results in restricting
M. maydis spreading in maize plants and
M. phaseolina spreading in cotton. This antagonism also has a positive influence on plant growth and yield.
Antagonistic interactions between phytoparasitic fungi have previously been reported in other host-pathogen species pairs [
23], as detailed in the Introduction. In many of those studies, the molecular DNA tracing proved to be an essential research tool [
25,
27,
42]. An example that is very similar to the current research is one of the most documented antagonisms among root pathogens, the suppression of
Cochliobolus sativus by
Fusarium (
roseum) species [
42]. In this recent study, a field trial was conducted that included the monitoring of spring wheat plant health and pathogen populations using qPCR. Across field locations,
C. sativus and
F. pseudograminearum isolates consistently and significantly reduced one another. However, our data suggest more complex interactions than a simple two-direction antagonism.
It is interesting to note that infecting maize plants solely with
M. phaseolina caused a 20% reduction in the number of healthy plants compared to the negative control (from seven plants to five plants,
Table 6). Thus, if both pathogens were acting without interfering with each other, their combined effect (50% in the
M. maydis treatment and 20% in the
M. phaseolina treatment) should have caused the total wilting of the plants. Instead, the dual inoculation led to some recovery of the plants. Indeed,
M. maydis reduced
M. phaseolina DNA in the roots of cotton plants by 4-fold, and
M. phaseolina reduced
M. maydis DNA in maize plants stems by 321-fold. Nevertheless, the mixed inoculation also resulted in higher levels of
M. phaseolina DNA in maize and higher levels of
M. maydis DNA in cotton. This suggests that the suppression effect of the fungal coexistence is of each pathogen in its primary host, while in its secondary host, the interactions with other phytopathogens can occasionally allow it to flourish. The nature of these interesting relationships and their causes will require further work to resolve. Another question triggered by the results presented here is what is the nature of
M. maydis interactions with cotton plants?
It was previously reported that, in cotton, infecting the soil with
M. maydis caused an increased growth of lateral roots and the appearance of local dark red lesions and shallow cracks on young cotton roots (up to 45 DAS) [
12]. Later, as the plants matured, these lesions disappeared, as well as the fungus. Furthermore,
M. maydis was not recovered from these symptomatic cotton plants [
12]. It was recently reported that
M. maydis infection also affected the root biomass and phenological development of young cotton plants (37 DAS) in a growth chamber [
10]. The absence of symptoms in mature cotton plants reported in the above-cited work evokes a new inquisitive question: does this pathogen have a hidden lifestyle as an endophyte in cotton?
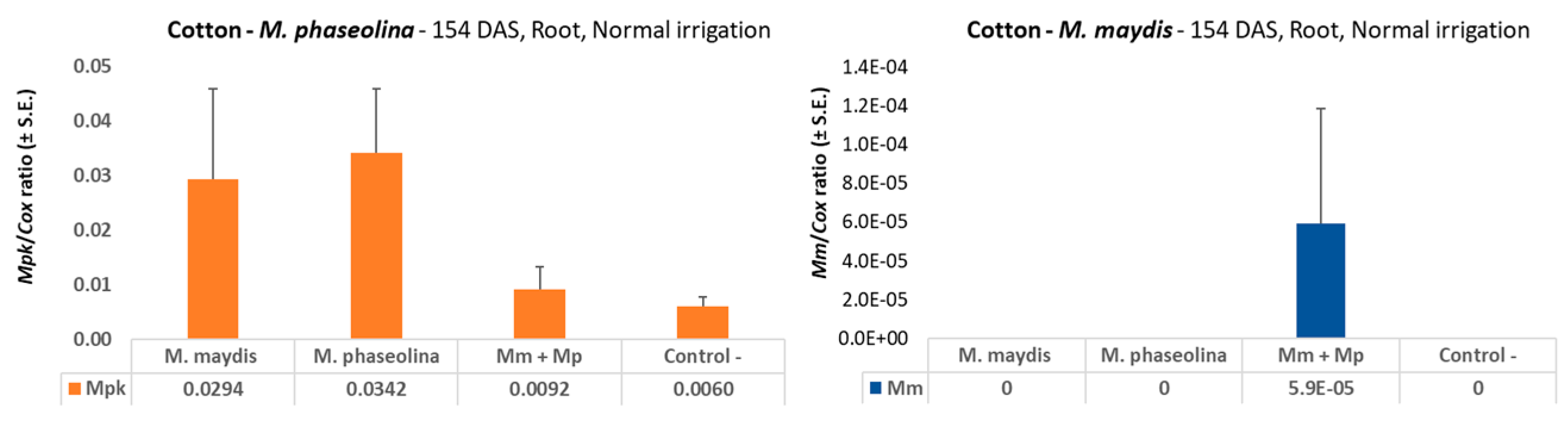
According to the data presented here, this may well be the case. Tracking the effect of M. maydis inoculation on cotton plants throughout the whole season in this two-year study supplies a large and consistent dataset. The data presented show that M. maydis inoculation at a regular irrigation regime did not affect the cotton growth parameters measured or the yield. On the contrary, in the 2018 experiment, in most measures, this inoculation resulted in higher growth and yield values at the end of the season. In the 2019 experiment, under drought stress, M. maydis infection led to a decreased weight and height of the above-ground parts of the cotton plants (without any measurable effect on yield production). This result may suggest that some opportunistic behaviors of this pathogen could exist. This assumption should be tested and established in future studies.
Formerly, results from random amplified polymorphic DNA (RAPD) analysis showed that
M.
phaseolina isolates from maize, cotton, and other crop hosts were differentiated from each other based on the host from which they initially isolated [
41,
43]. Moreover, isolates within each host that have different sensitivities to chlorate (a powerful oxidizer) also distinctly separated [
41,
43]. The researchers’ studies of a field with a long history of constant crop rotation led them to suggest that host specialization developed in this fungus. However, host specialization in
M.
phaseolina appears to occur with maize, but not with sorghum, cotton, or soybean, as indicated by the results of cross-inoculation experiments [
41]. The colonization of maize roots was significantly higher with maize isolates than with isolates from other hosts. Since
M. maydis distribution is limited to about only eight countries and is considered to be an exotic pathogen in most of the world, the above-mentioned knowledge about its host specialization and chlorate sensitivity is absent. It was only recently reported that the
M. maydis host range is wider than we had thought [
10].
It would be very interesting and economically significant to study the differentiation between
M. maydis populations originating from different host species. Such studies, which were already carried out in maize in Egypt, had shown that isolates of
M. maydis differ in pathogenicity, morphology, and route of infection [
44]. Most importantly, it was reported in Egypt and Spain that this pathogen could undergo pathogenic changes that can result in highly virulent strains [
34,
45]. The Egyptian isolates of
M. maydis were classified into four clonal lineages, which revealed not only diversity in virulence and colonization ability on maize but also fascinating competitiveness relationships [
34]. The most virulent lineage, when tested alone, was the least competitive on susceptible maize accessions when incorporated into the soil as a component of the mixed inocula of all four lineages. In contrast, one of the less virulent lineages dominated (70% of infections) and appeared to be the most competitive.
Thus, it was suggested [
34] that the most virulent strain may have an advantage in this interspecies competition when the field is seeded with resistance maize cultivar. However, in fields in which susceptible maize hybrids are cultivated, the other less virulent
M. maydis strains may become more prevalent, and the most virulent lineage may be relatively rare. It should be taken into consideration that environmental aspects (biological and physical) are involved.
M. maydis strains may differentiate due to their ability to grow either in the soil or in the host plant and due to their sensitivity to other soil microbes or their metabolic products.
The reciprocal suppression that was documented between
M. maydis and
M. phaseolina species in this study raises the importance of inspecting the effectiveness of preventing treatments while considering the pathogen specialization and the circumstances under which several pathogens are involved. To emphasize this, in spring 2018, a commercial field (Neot Mordechai field, Hula Valley, Upper Galilee, northern Israel) was planted with the sensitive maize cultivar Prelude cv. This field had a long history of late wilt incidences [
5] and was therefore protected by a recently developed and highly effective chemical method [
6]. Despite the use of the potent commercial mixture of Azoxystrobin and Difenoconazole (Syngenta, Basel, Switzerland), at the end of the season, only meager yields were achieved. The yield reduction was most probably the consequence of another fungal disease caused by
F. verticillioides and
F. oxysporum, which led to dehydration and yield loss (Ofir Degani et al., unpublished results). These Fusarium species are indeed universal and important vascular wilt disease-causing fungal pathogens [
46]. To support this conclusion, in other plants in that field that were not affected by the
Fusarium spp. disease, the chemical treatment almost wholly abolished any sign of late wilt disease.
It has now become evident that optimal water supply can reduce late wilt disease in maize. Indeed, our past experience [
6,
30,
33] and a review of the literature support the conclusion that high water potential is one of the most important agrotechnical aims for restricting late wilt disease progression [
17,
47,
48,
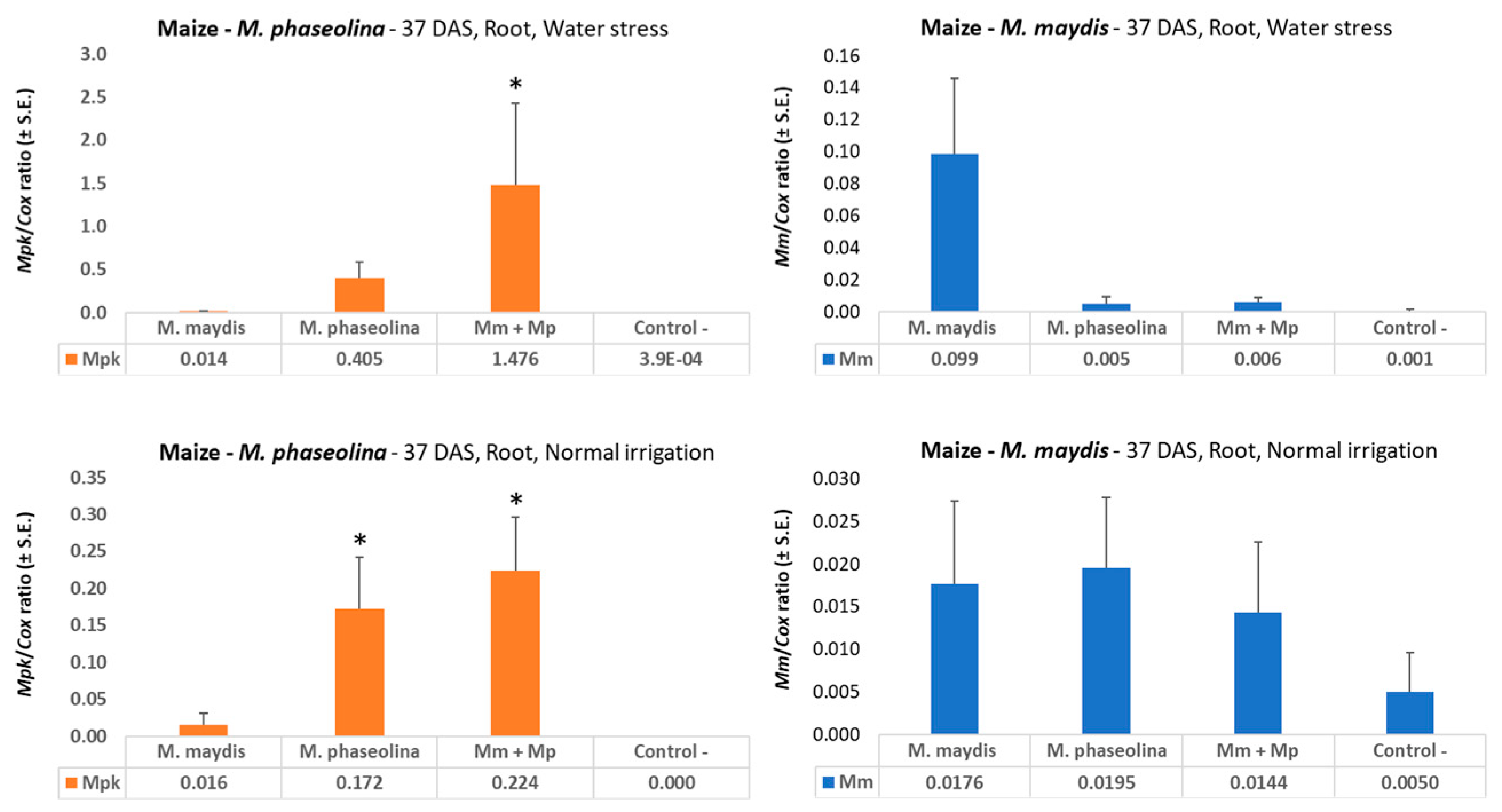
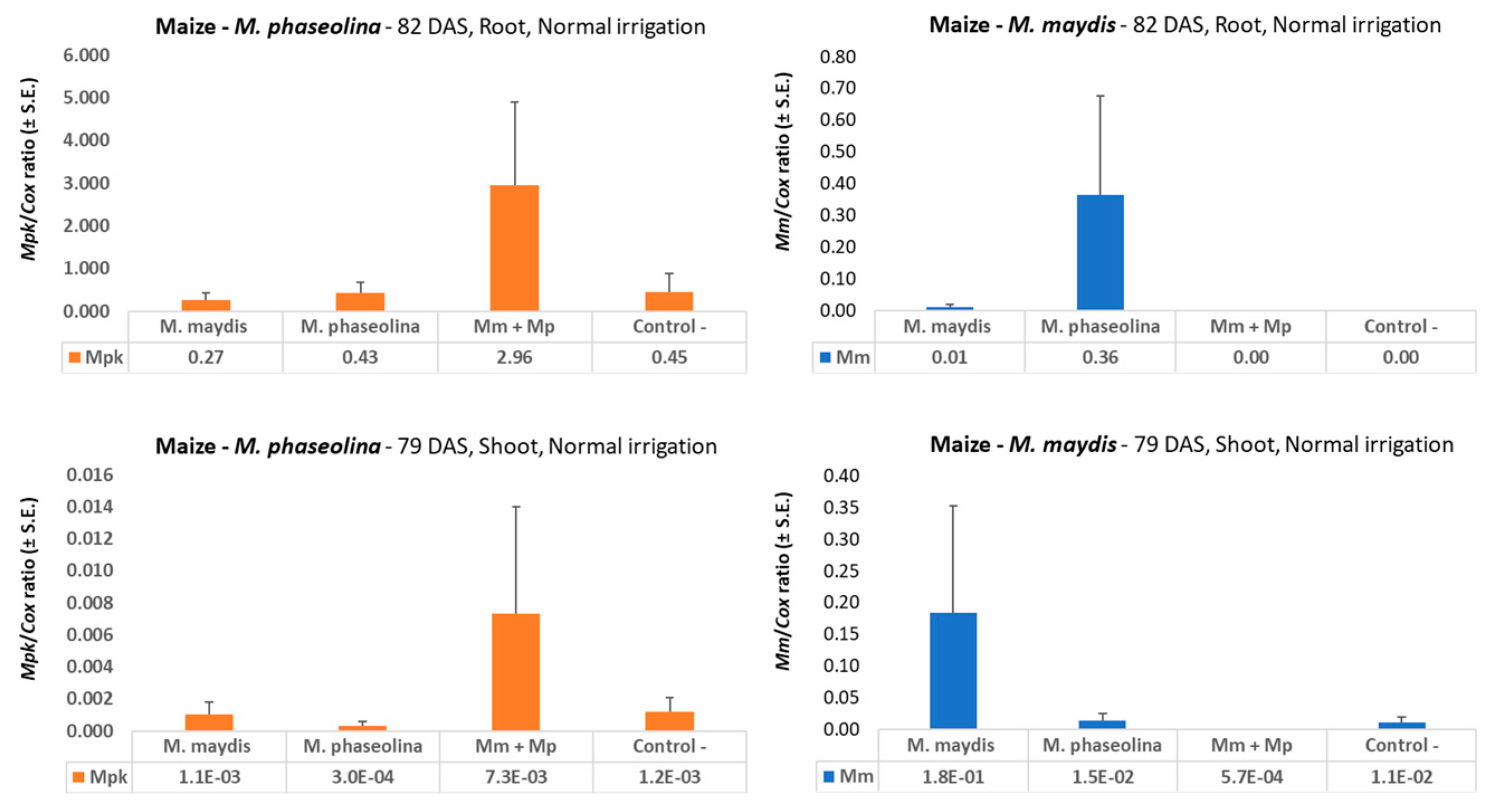
49]. The results presented in the current study are in line with this conclusion, with nearly 10 times higher
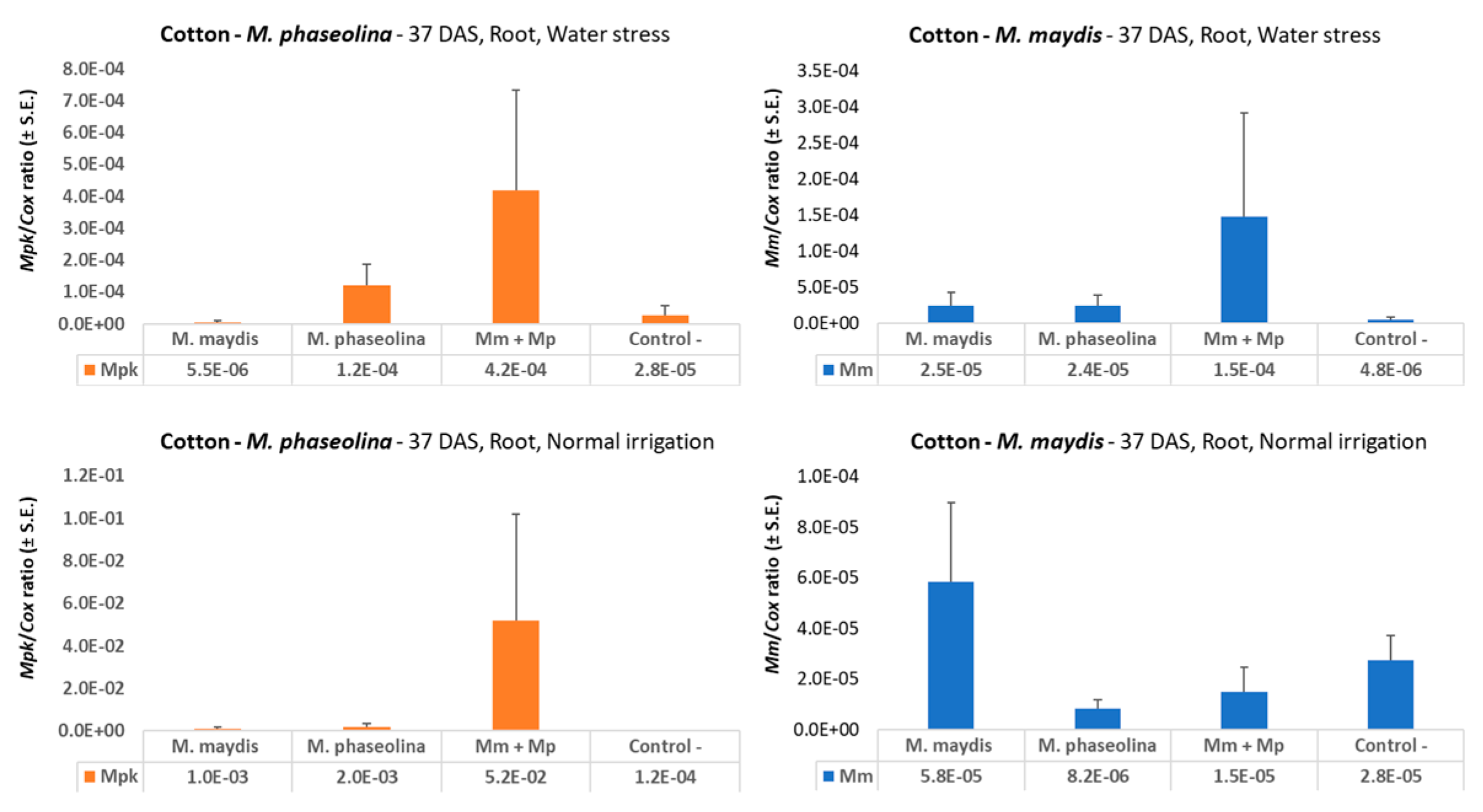
M. maydis DNA in drought-stressed maize sprouts (37 DAS,
Figure 2). Interestingly, drought pressure also caused about three times higher
M.
phaseolina DNA in young maize plants, but both pathogens did not flourish under a restricted water regime in cotton (
Figure 6) compared to regular irrigation conditions. A possible explanation is that the applied restricted water regime did not produce drought pressure as expected. Indeed the cotton plants in the decrease water irrigation pots were healthy and had an apparent healthy development.