Synchrony Degree of Dietary Energy and Nitrogen Release Influences Microbial Community, Fermentation, and Protein Synthesis in a Rumen Simulation System
Abstract
1. Introduction
2. Materials and Methods
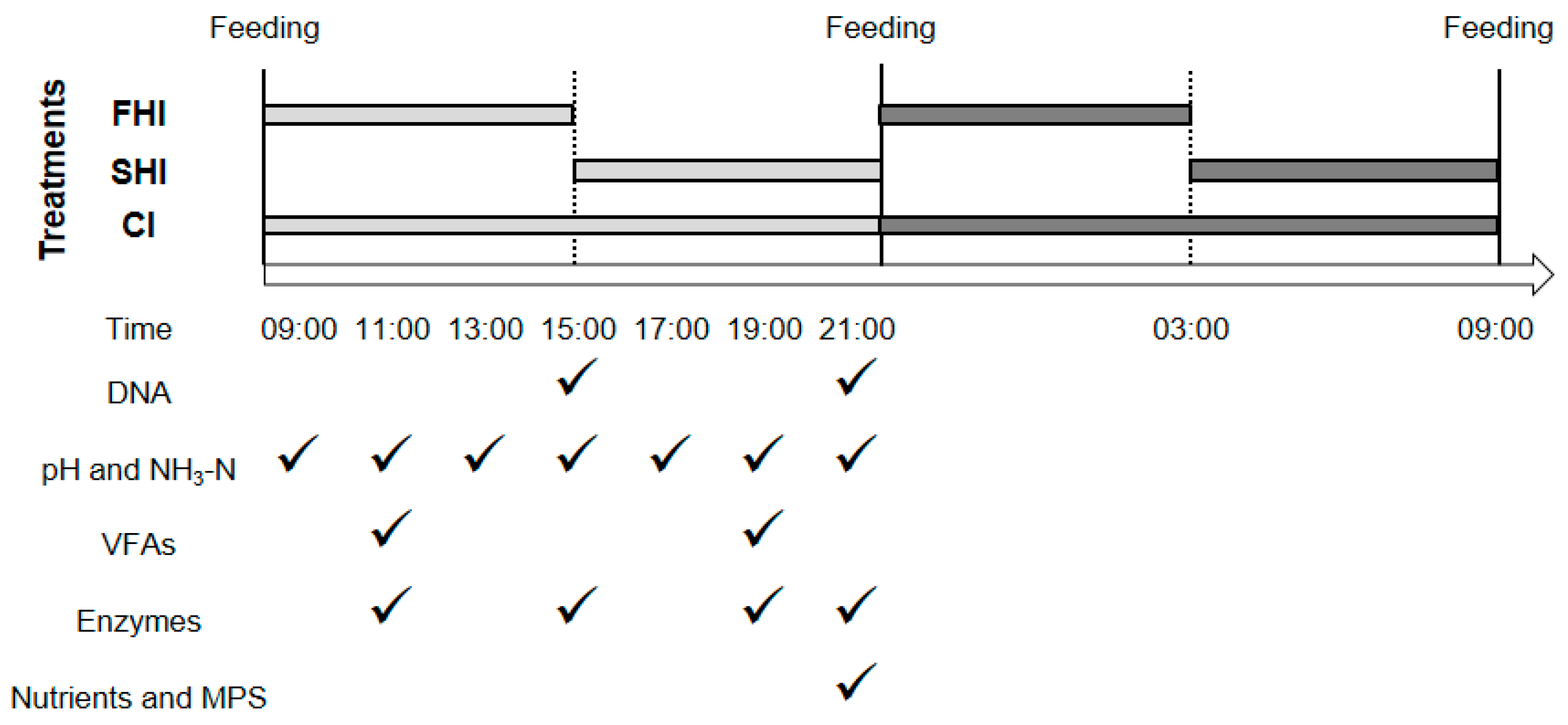
2.1. Experimental Design and Cultivation
2.2. Sampling
2.3. Microbial DNA Extraction and Quantitative PCR
2.4. Bacterial 16S rRNA Genes Amplification and Miseq Sequencing
2.5. Sequencing Data Analysis
2.6. Chemical Analysis
2.7. Statistical Analysis
2.8. Nucleotide Sequence Accession Number
3. Results
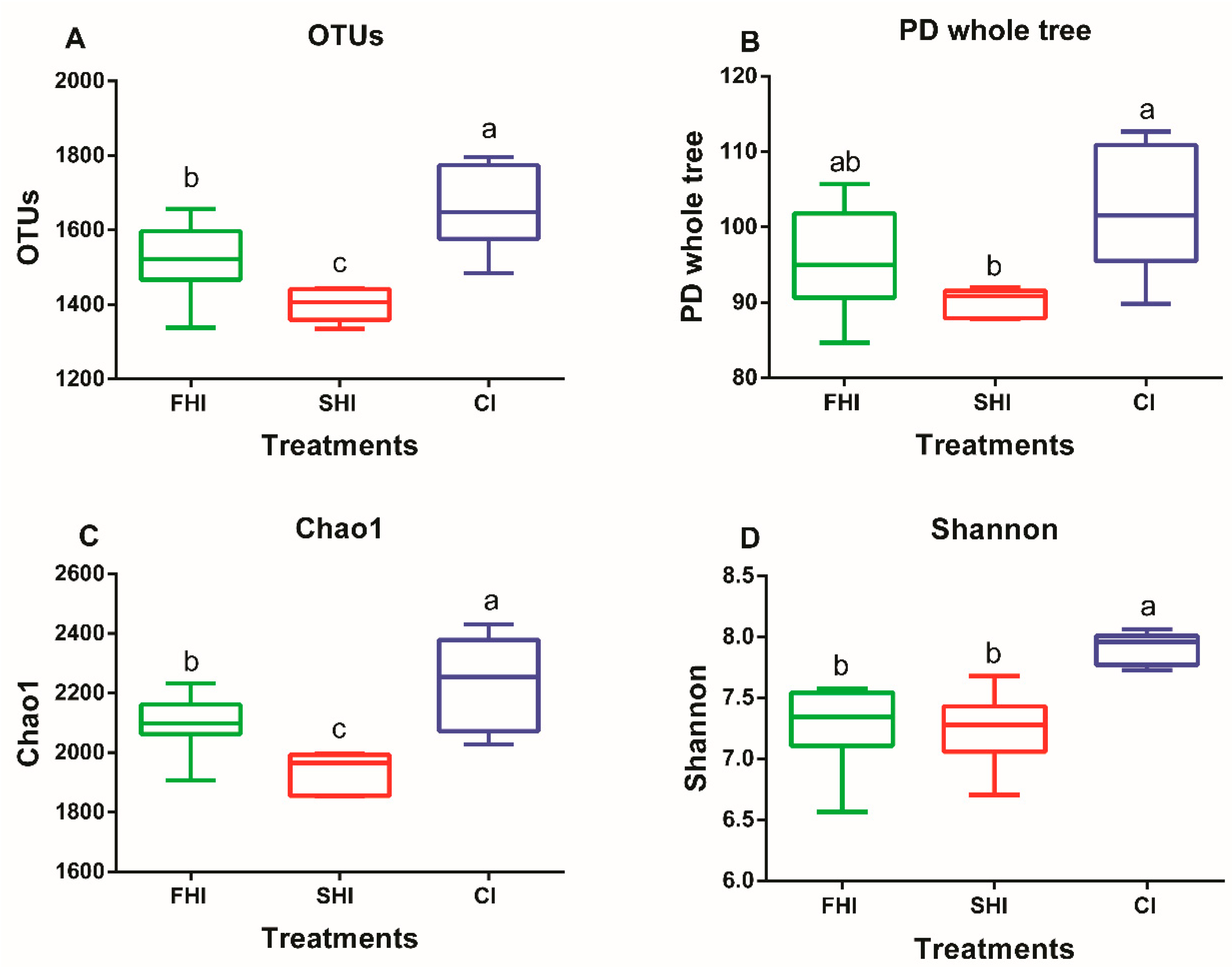
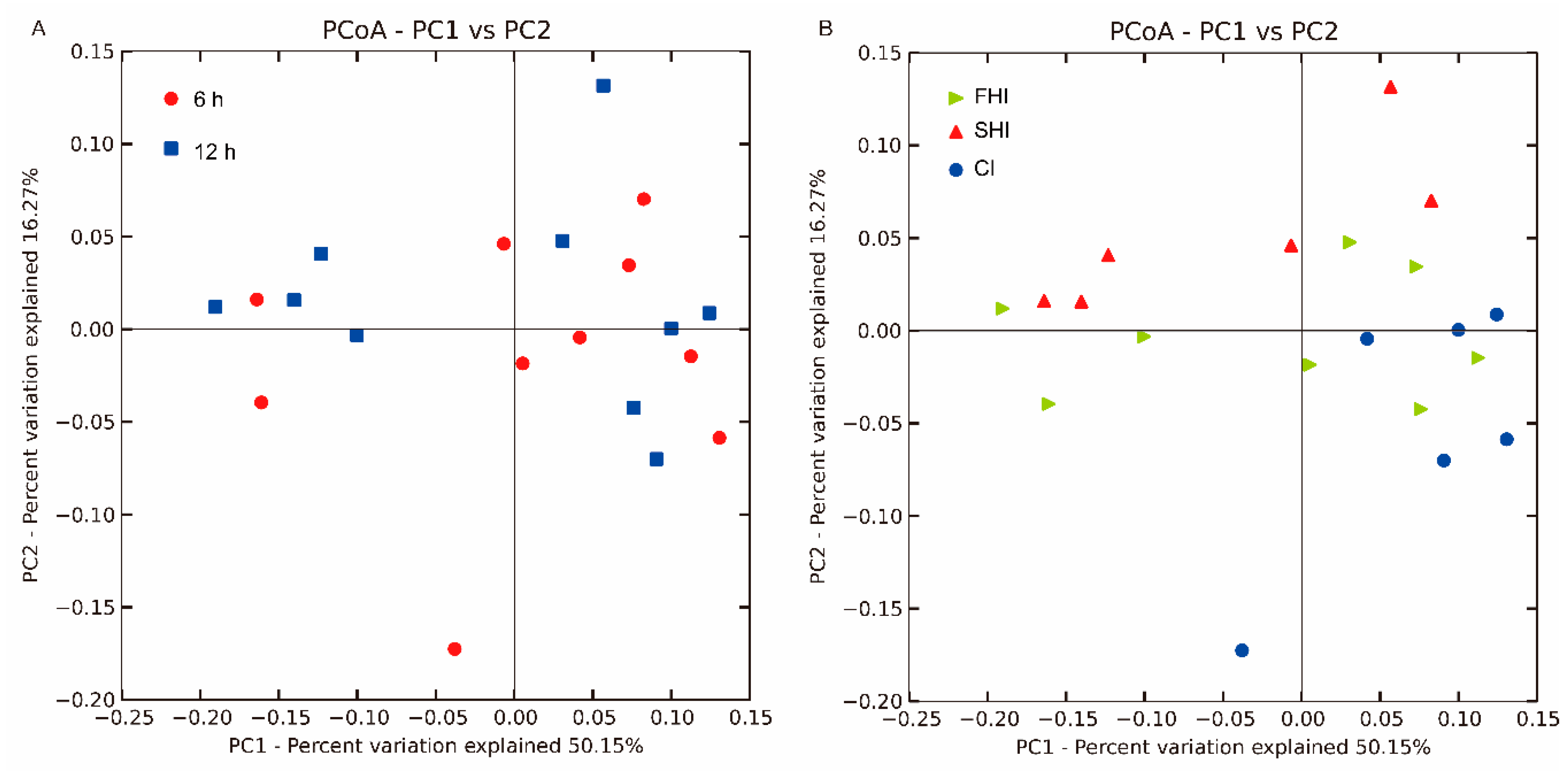
3.1. In Vitro Bacterial Diversity
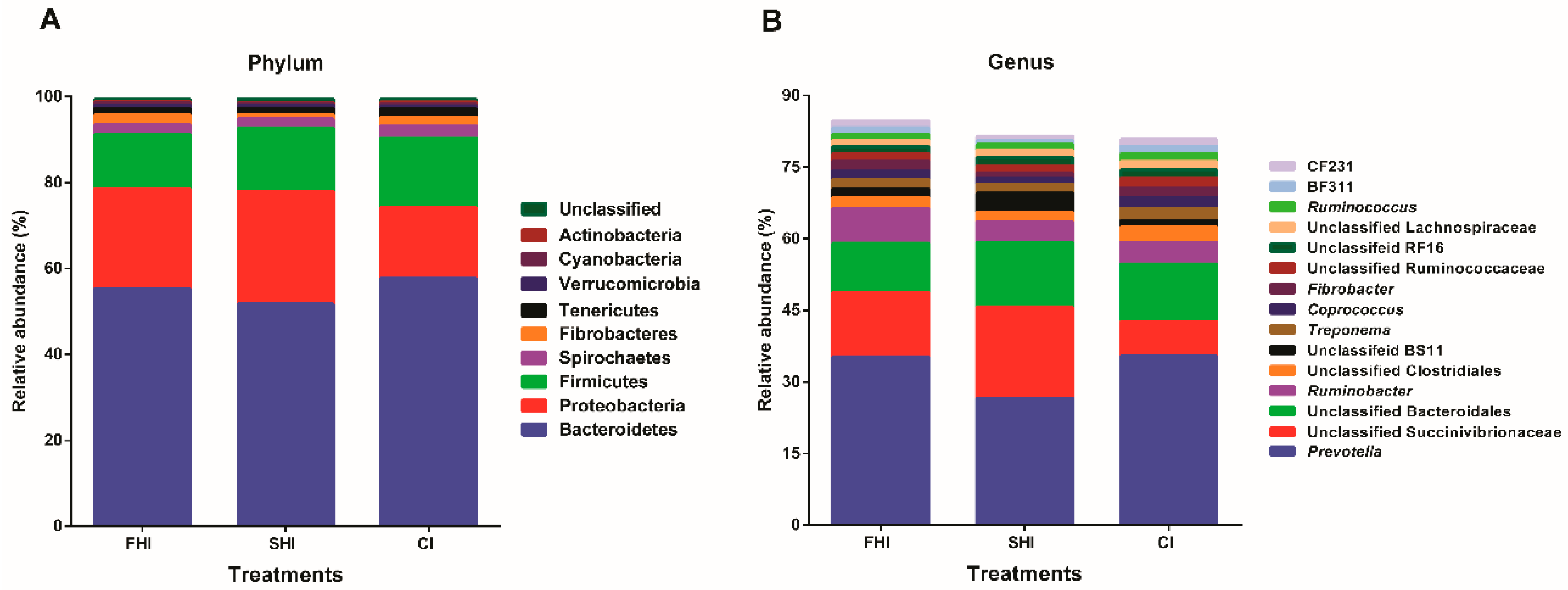
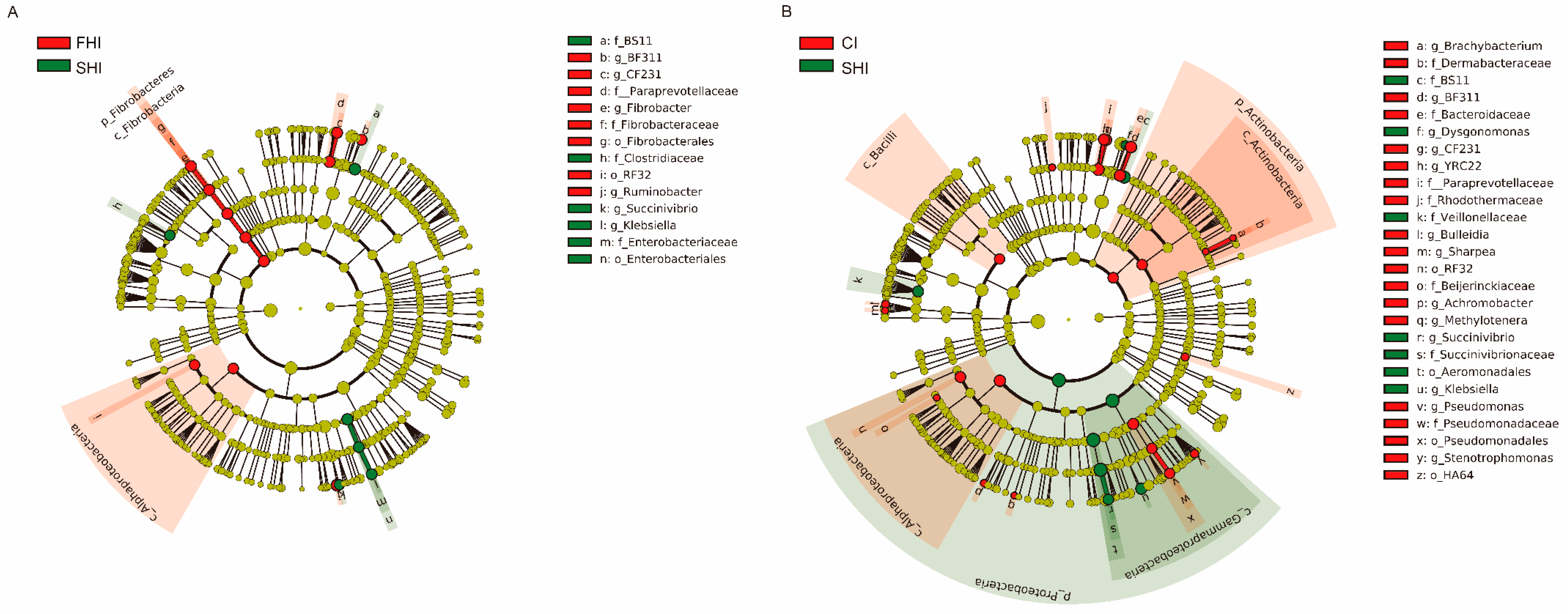
3.2. In Vitro Bacterial Composition
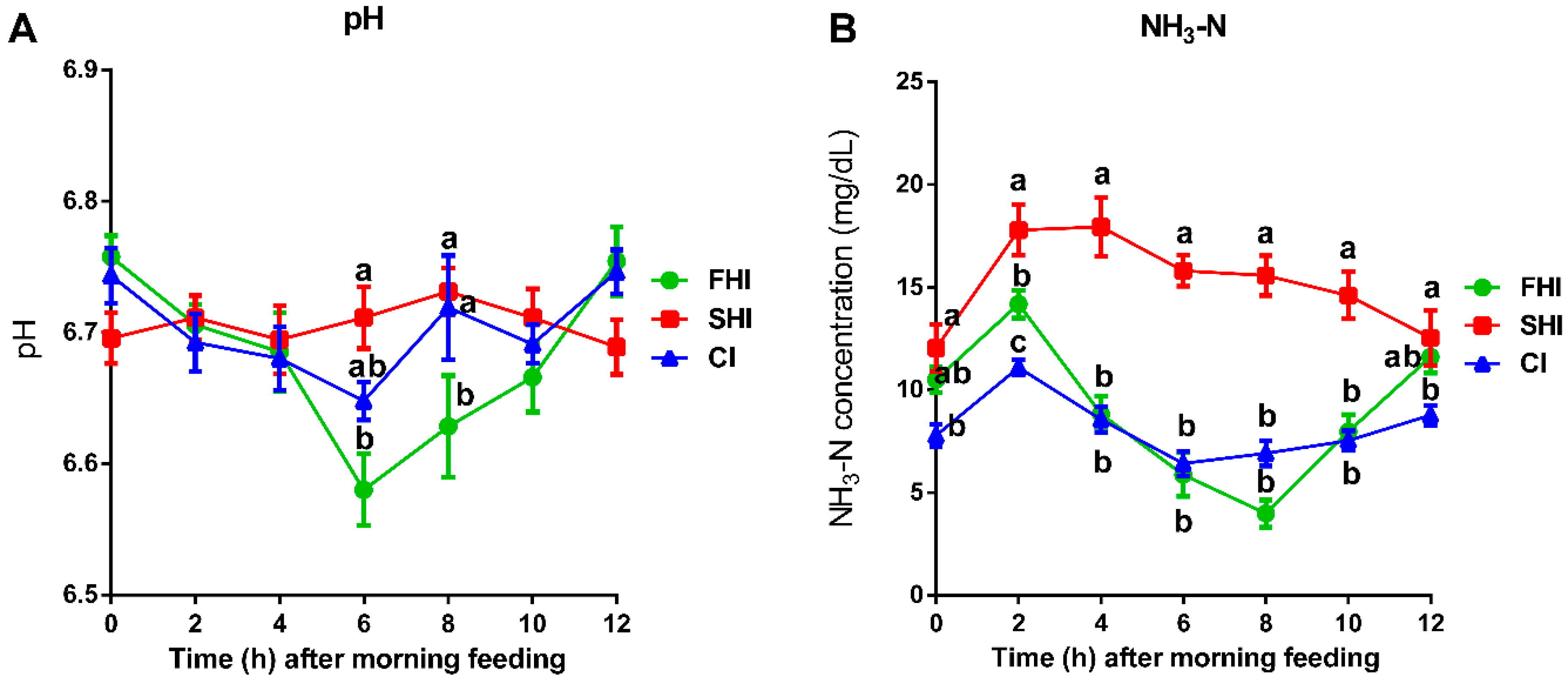
3.3. In Vitro Fermentation and Nutrient Digestion
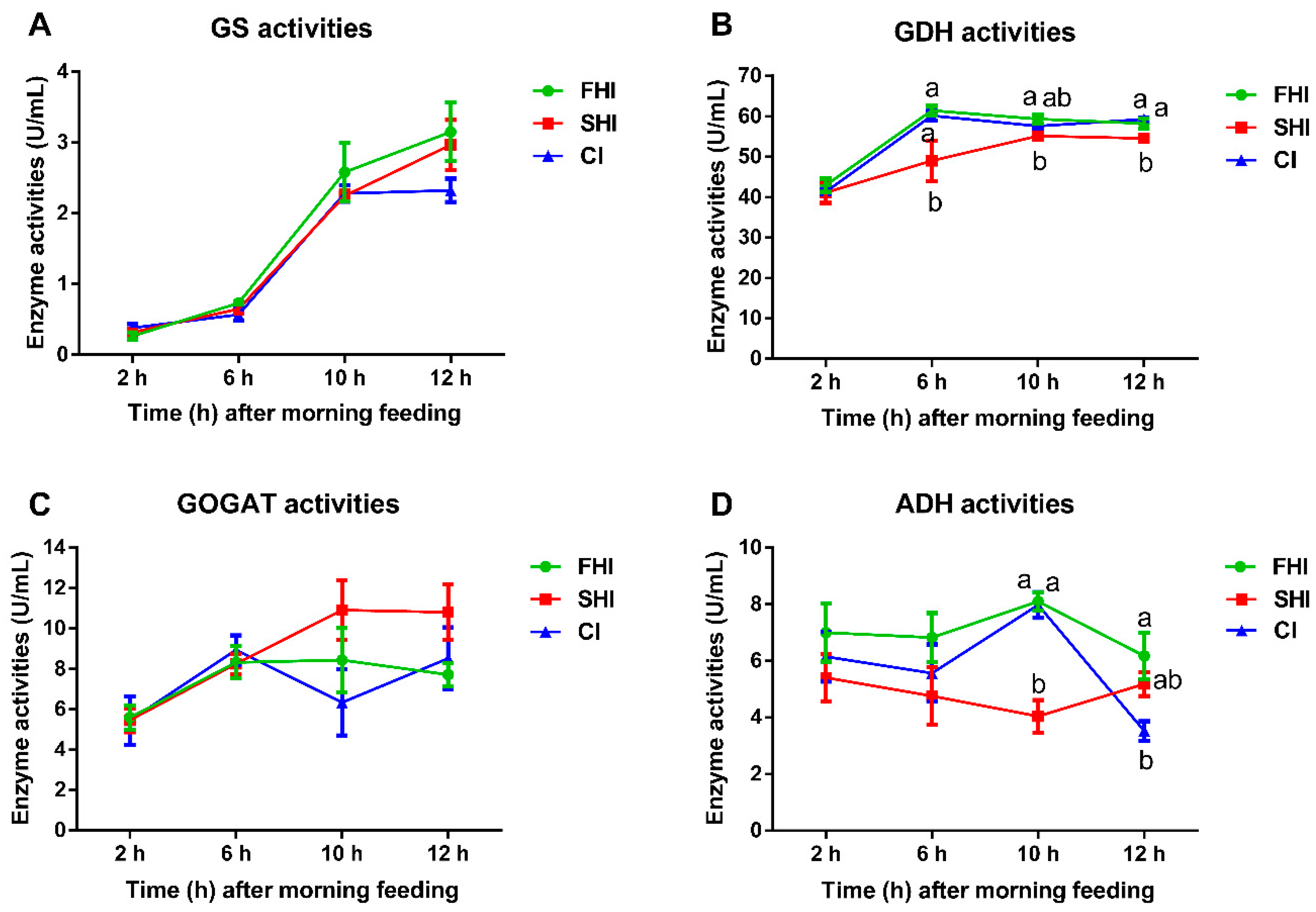
3.4. Enzyme Activity and Microbial Protein Synthesis
3.5. Correlations Between Bacterial Relative Abundance and Fermentation Parameters
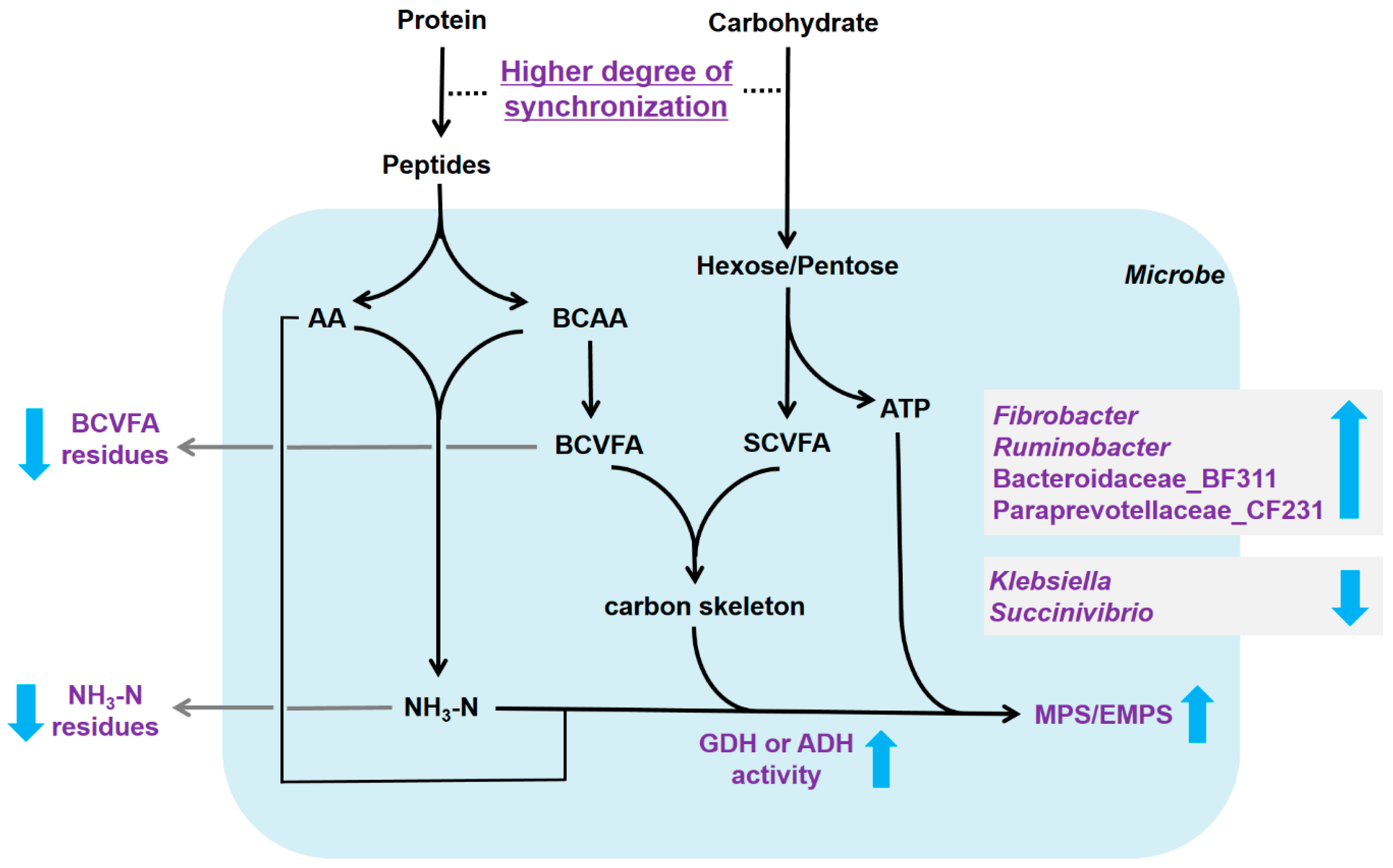
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haug, A.; Hostmark, A.T.; Harstad, O.M. Bovine milk in human nutrition—A review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef]
- Zhu, W.; Fu, Y.; Wang, B.; Wang, C.; Ye, J.A.; Wu, Y.M.; Liu, J.X. Effects of dietary forage sources on rumen microbial protein synthesis and milk performance in early lactating dairy cows. J. Dairy Sci. 2013, 96, 1727–1734. [Google Scholar] [CrossRef]
- Zhou, X.Q.; Zhang, Y.D.; Zhao, M.; Zhang, T.; Zhu, D.; Bu, D.P.; Wang, J.Q. Effect of dietary energy source and level on nutrient digestibility, rumen microbial protein synthesis, and milk performance in lactating dairy cows. J. Dairy Sci. 2015, 98, 7209–7217. [Google Scholar] [CrossRef]
- Schwab, C.G.; Satter, L.D.; Clay, B. Response to lactating dairy cows to abomasal infusion of amino acids. J. Dairy Sci. 1976, 59, 1254–1270. [Google Scholar] [CrossRef]
- Abbasi, I.H.R.; Abbasi, F.; Abd El-Hack, M.E.; Abdel-Latif, M.A.; Soomro, R.N.; Hayat, K.; Mohamed, M.A.E.; Bodinga, B.M.; Yao, J.; Cao, Y. Critical analysis of excessive utilization of crude protein in ruminants ration: Impact on environmental ecosystem and opportunities of supplementation of limiting amino acids-a review. Environ. Sci. Pollut. Res. Int. 2018, 25, 181–190. [Google Scholar] [CrossRef]
- White, R.R.; Roman-Garcia, Y.; Firkins, J.L. Meta-analysis of postruminal microbial nitrogen flows in dairy cattle. II. Approaches to and implications of more mechanistic prediction. J. Dairy Sci. 2016, 99, 7932–7944. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, S.; Wang, X.; Zhang, Y.; Zheng, N.; Wang, J. Ruminal Methanogen Community in Dairy Cows Fed Agricultural Residues of Corn Stover, Rapeseed, and Cottonseed Meals. J. Agric. Food Chem. 2016, 64, 5439–5445. [Google Scholar] [CrossRef]
- Hoover, W.H.; Stokes, S.R. Balancing carbohydrates and proteins for optimum rumen microbial yield. J. Dairy Sci. 1991, 74, 3630–3644. [Google Scholar] [CrossRef]
- Nocek, J.E.; Russell, J.B. Protein and Energy as an Integrated System. Relationship of Ruminal Protein and Carbohydrate Availability to Microbial Synthesis and Milk Production. J. Dairy Sci. 1988, 71, 2070–2107. [Google Scholar] [CrossRef]
- Chanjula, P.; Wanapat, M.; Wachirapakorn, C.; Rowlinson, P. Effect of Synchronizing Starch Sources and Protein (NPN) in the Rumen on Feed Intake, Rumen Microbial Fermentation, Nutrient Utilization and Performance of Lactating Dairy Cows. Asian-Australas. J. Anim. Sci. 2004, 17, 1400–1410. [Google Scholar] [CrossRef]
- Hall, M.B.; Huntington, G.B. Nutrient synchrony: Sound in theory, elusive in practice. J. Anim. Sci. 2008, 86, E287–E292. [Google Scholar] [CrossRef]
- Chumpawadee, S.; Sommart, K.; Vongpralub, T.; Pattarajinda, V. Effects of Synchronizing the Rate of Dietary Energy and Nitrogen Releasn on Ruminal Fermentation, Microbial Protein Synthesis, Blood Urea Nitrogen and Nutrient Digestibility in Beef Cattle. Asian-Australas. J. Anim. Sci. 2006, 19, 181–188. [Google Scholar] [CrossRef]
- Firkins, J.L. Maximizing microbial protein synthesis in the rumen. J. Nutr. 1996, 126, 1347S–1354S. [Google Scholar] [CrossRef]
- Reynolds, C.K.; Kristensen, N.B. Nitrogen recycling through the gut and the nitrogen economy of ruminants: An asynchronous symbiosis. J. Anim. Sci. 2008, 86, E293–E305. [Google Scholar] [CrossRef]
- Berthiaume, R.; Benchaar, C.; Chaves, A.V.; Tremblay, G.F.; Castonguay, Y.; Bertrand, A.; Belanger, G.; Michaud, R.; Lafreniere, C.; McAllister, T.A.; et al. Effects of nonstructural carbohydrate concentration in alfalfa on fermentation and microbial protein synthesis in continuous culture. J. Dairy Sci. 2010, 93, 693–700. [Google Scholar] [CrossRef]
- Henning, P.H.; Steyn, D.G.; Meissner, H.H. Effect of synchronization of energy and nitrogen supply on ruminal characteristics and microbial growth. J. Anim. Sci. 1993, 71, 2516–2528. [Google Scholar] [CrossRef]
- Kim, K.H.; Choung, J.J.; Chamberlain, D.G. Effects of varying the degree of synchrony of energy and nitrogen release in the rumen on the synthesis of microbial protein in lactating dairy cows consuming a diet of grass silage and a cereal-based concentrate. J. Sci. Food Agric. 1999, 79, 1441–1447. [Google Scholar] [CrossRef]
- Lu, Z.; Xu, Z.; Shen, Z.; Tian, Y.; Shen, H. Dietary Energy Level Promotes Rumen Microbial Protein Synthesis by Improving the Energy Productivity of the Ruminal Microbiome. Front. Microbiol. 2019, 10, 847. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Derakhshani, H.; Plaizier, J.C.; De Buck, J.; Barkema, H.W.; Khafipour, E. Association of bovine major histocompatibility complex (BoLA) gene polymorphism with colostrum and milk microbiota of dairy cows during the first week of lactation. Microbiome 2018, 6, 203. [Google Scholar] [CrossRef]
- Zhou, M.; Peng, Y.J.; Chen, Y.; Klinger, C.M.; Oba, M.; Liu, J.X.; Guan, L.L. Assessment of microbiome changes after rumen transfaunation: Implications on improving feed efficiency in beef cattle. Microbiome 2018, 6, 62. [Google Scholar] [CrossRef]
- Cheung, M.K.; Au, C.H.; Chu, K.H.; Kwan, H.S.; Wong, C.K. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 2010, 4, 1053–1059. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Wang, Y.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of Dietary Forage to Concentrate Ratios on Dynamic Profile Changes and Interactions of Ruminal Microbiota and Metabolites in Holstein Heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef]
- Shen, J.; Liu, Z.; Yu, Z.; Zhu, W. Monensin and Nisin Affect Rumen Fermentation and Microbiota Differently In Vitro. Front. Microbiol. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Jin, D.; Zhao, S.; Wang, P.; Zheng, N.; Bu, D.; Beckers, Y.; Wang, J. Insights into Abundant Rumen Ureolytic Bacterial Community Using Rumen Simulation System. Front. Microbiol. 2016, 7, 1006. [Google Scholar] [CrossRef]
- Sarnataro, C.; Petri, R.M.; Spanghero, M.; Zebeli, Q.; Klevenhusen, F. A nutritional and rumen ecological evaluation of the biorefinery by-product alfalfa silage cake supplemented with Scrophularia striata extract using the rumen simulation technique. J. Sci. Food Agric. 2019, 99, 4414–4422. [Google Scholar] [CrossRef]
- McDougall, E.I. Studies on ruminant saliva. 1. The composition and output of sheep’s saliva. Biochem. J. 1948, 43, 99–109. [Google Scholar] [CrossRef]
- Lapierre, H.; Lobley, G.E. Nitrogen Recycling in the Ruminant: A Review. J. Dairy Sci. 2001, 84, E223–E236. [Google Scholar] [CrossRef]
- Jin, E.; Bu, D.; Wang, J.; Jiang, Y.; Bao, W.; Shi, H.; Shen, W.; Li, F. Effects of plant essential oil on rumen fermentation and methanogenesis studied using a dual flow continuous culture system. Chin. J. Anim. Nutr. 2013, 25, 2303–2314. [Google Scholar]
- Shen, W.; Jiang, Y.; Wang, J.; Bu, D.; Sun, P.; Jin, E.; Bao, W.; Shi, H. Design and testing of rumen simulation system with discharging solid chime, liquid, and gas respectively. Trans. Chin. Soc. Agric. Eng. 2012, 28, 20–26. [Google Scholar]
- Dong, S.; Azarfar, A.; Zou, Y.; Li, S.; Wang, Y.; Cao, Z. Effects of sequence of nylon bags rumen incubation on kinetics of degradation in some commonly used feedstuffs in dairy rations. J. Integr. Agric. 2017, 16, 162–168. [Google Scholar] [CrossRef]
- Griffith, C.L.; Ribeiro, G.O., Jr.; Oba, M.; McAllister, T.A.; Beauchemin, K.A. Fermentation of Ammonia Fiber Expansion Treated and Untreated Barley Straw in a Rumen Simulation Technique Using Rumen Inoculum from Cattle with Slow versus Fast Rate of Fiber Disappearance. Front. Microbiol. 2016, 7, 1839. [Google Scholar] [CrossRef]
- Ramos-Morales, E.; Rossi, G.; Cattin, M.; Jones, E.; Braganca, R.; Newbold, C.J. The effect of an isoflavonid-rich liquorice extract on fermentation, methanogenesis and the microbiome in the rumen simulation technique. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, S.; Zhang, Y.; Sun, P.; Bu, D.; Wang, J. New Primers Targeting Full-Length Ciliate 18S rRNA Genes and Evaluation of Dietary Effect on Rumen Ciliate Diversity in Dairy Cows. Curr. Microbiol. 2015, 71, 650–657. [Google Scholar] [CrossRef]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef]
- Nelson, M.C.; Morrison, H.G.; Benjamino, J.; Grim, S.L.; Graf, J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE 2014, 9, e94249. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Mahnert, A.; Moissl-Eichinger, C.; Berg, G. Microbiome interplay: Plants alter microbial abundance and diversity within the built environment. Front. Microbiol. 2015, 6, 887. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, P.; Wu, Y.; Guo, P.; Liu, L.; Ma, N.; Levesque, C.; Chen, Y.; Zhao, J.; Zhang, J.; et al. Dietary Fiber Increases Butyrate-Producing Bacteria and Improves the Growth Performance of Weaned Piglets. J. Agric. Food Chem. 2018, 66, 7995–8004. [Google Scholar] [CrossRef]
- Wang, P.; Tan, Z.; Guan, L.; Tang, S.; Zhou, C.; Han, X.; Kang, J.; He, Z. Ammonia and amino acids modulates enzymes associated with ammonia assimilation pathway by ruminal microbiota in vitro. Livest. Sci. 2015, 178, 130–139. [Google Scholar] [CrossRef]
- AOAC. Association of Official Analytical Chemists, 17th ed.; AOAC International: Arlington, VA, USA, 2000. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Bach, A.; Yoon, I.K.; Stern, M.D.; Jung, H.G.; Chester-Jones, H. Effects of type of carbohydrate supplementation to lush pasture on microbial fermentation in continuous culture. J. Dairy Sci. 1999, 82, 153–160. [Google Scholar] [CrossRef]
- Littell, R.C.; Henry, P.R.; Ammerman, C.B. Statistical analysis of repeated measures data using SAS procedures. J. Anim. Sci. 1998, 76, 1216–1231. [Google Scholar] [CrossRef]
- Shen, J.S.; Song, L.J.; Sun, H.Z.; Wang, B.; Chai, Z.; Chacher, B.; Liu, J.X. Effects of corn and soybean meal types on rumen fermentation, nitrogen metabolism and productivity in dairy cows. Asian-Australas. J. Anim. Sci. 2015, 28, 351–359. [Google Scholar] [CrossRef]
- Gilbert, M.S.; van den Borne, J.J.; Berends, H.; Pantophlet, A.J.; Schols, H.A.; Gerrits, W.J. A titration approach to identify the capacity for starch digestion in milk-fed calves. Animal 2015, 9, 249–257. [Google Scholar] [CrossRef]
- Liu, C.; Li, D.; Chen, W.; Li, Y.; Wu, H.; Meng, Q.; Zhou, Z. Estimating ruminal crude protein degradation from beef cattle feedstuff. Sci. Rep. 2019, 9, 11368. [Google Scholar] [CrossRef]
- Maxin, G.; Ouellet, D.R.; Lapierre, H. Ruminal degradability of dry matter, crude protein, and amino acids in soybean meal, canola meal, corn, and wheat dried distillers grains. J. Dairy Sci. 2013, 96, 5151–5160. [Google Scholar] [CrossRef]
- Okeke, G.C.; Buchanan-Smith, J.G.; Grieve, D.G. Effect of sodium bicarbonate on rate of passage and degradation of soybean meal in postpartum dairy cows. J. Dairy Sci. 1983, 66, 1023–1031. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Li, S.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in Microbiota in Rumen Digesta and Feces Due to a Grain-Based Subacute Ruminal Acidosis (SARA) Challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef]
- Ye, H.; Liu, J.; Feng, P.; Zhu, W.; Mao, S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci. Rep. 2016, 6, 20329. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Santos, E.d.O.; Thompson, F. The Family Succinivibrionaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 639–648. [Google Scholar] [CrossRef]
- Suen, G.; Weimer, P.J.; Stevenson, D.M.; Aylward, F.O.; Boyum, J.; Deneke, J.; Drinkwater, C.; Ivanova, N.N.; Mikhailova, N.; Chertkov, O.; et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 2011, 6, e18814. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T., 2nd; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.K.; Zhu, W.; Pu, Y.Y.; Guan, L.L.; Liu, J.X. Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol. Ecol. 2014, 87, 576–585. [Google Scholar] [CrossRef]
- Sinclair, L.A.; Garnsworth, P.C.; Newbold, J.R.; Buttery, P.J. Effect of synchronizing the rate of dietary energy and nitrogen release on rumen fermentation and microbial protein synthesis in sheep. J. Agric. Sci. -Camb. 2009, 120, 251–263. [Google Scholar] [CrossRef]
- Witt, M.W.; Sinclair, L.A.; Wilkinson, R.G.; Buttery, P.J. The effects of synchronizing the rate of dietary energy and nitrogen supply to the rumen on the production and metabolism of sheep: Food characterization and growth and metabolism of ewe lambs given food ad libitum. Anim. Sci. 2016, 69, 223–235. [Google Scholar] [CrossRef]
- Piao, M.Y.; Kim, H.J.; Seo, J.K.; Park, T.S.; Yoon, J.S.; Kim, K.H.; Ha, J.K. Effects of synchronization of carbohydrate and protein supply in total mixed ration with korean rice wine residue on ruminal fermentation, nitrogen metabolism and microbial protein synthesis in holstein steers. Asian-Australas. J. Anim. Sci. 2012, 25, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.M. Response of forage fiber degradation by ruminal microorganisms to branched-chain volatile fatty acids, amino acids, and dipeptides. J. Dairy Sci. 2002, 85, 1183–1190. [Google Scholar] [CrossRef]
- Wang, P.; Tan, Z. Ammonia assimilation in rumen bacteria: A review. Anim. Biotechnol. 2013, 24, 107–128. [Google Scholar]
- Hespell, R.B. The Genera Succinivibrio and Succinimonas. In The Prokaryotes; Springer Science+Business Media: New York, NY, USA, 1992. [Google Scholar]
- Patterson, J.A.; Chalova, V.I.; Hespell, R.B.; Ricke, S.C. Dilution rates influence ammonia-assimilating enzyme activities and cell parameters of Selenomonas ruminantium strain D in continuous (glucose-limited) culture. J. Appl. Microbiol. 2010, 108, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Atasoglu, C.; Newbold, C.J.; Wallace, R.J. Incorporation of [15N] ammonia by the cellulolytic ruminal bacteria Fibrobacter succinogenes BL2, Ruminococcus albus SY3, and Ruminococcus flavefaciens 17. Appl. Environ. Microbiol. 2001, 67, 2819–2822. [Google Scholar] [CrossRef] [PubMed]
- Matheron, C.; Delort, A.-M.; Gaudet, G.; Liptaj, T.; Forano, E. Interactions between carbon and nitrogen metabolism in Fibrobacter succinogenes S85: A 1H and 13C nuclear magnetic resonance and enzymatic study. Appl. Environ. Microbiol. 1999, 65, 1941–1948. [Google Scholar] [CrossRef]
- Hobson, P.N.; Stewart, C.S. The Rumen Microbial Ecosystem; Blaclde Academic & Professional: London, UK, 1997. [Google Scholar]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef]
- Kumar, S.; Indugu, N.; Vecchiarelli, B.; Pitta, D.W. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front. Microbiol. 2015, 6, 781. [Google Scholar] [CrossRef]
- Kleiner, D. Energy expenditure for cyclic retention of NH3/NH4+ during N2 fixation by Klebsiella pneumoniae. FEBS Lett. 1985, 187, 237–239. [Google Scholar] [CrossRef]
- Brenchley, J.E.; Magasanik, B. Mutants of Klebsiella aerogenes lacking glutamate dehydrogenase. J. Bacteriol. 1974, 117, 544–550. [Google Scholar] [CrossRef]
- Deleo, A.B.; Magasanik, B. Identification of the structural gene for glutamine synthetase in Klebsiella aerogenes. J. Bacteriol. 1975, 121, 313–319. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Qiu, Q.; Shao, T.; Niu, W.; Xia, C.; Wang, H.; Li, Q.; Gao, Z.; Yu, Z.; Su, H.; et al. Dietary Alfalfa and Calcium Salts of Long-Chain Fatty Acids Alter Protein Utilization, Microbial Populations, and Plasma Fatty Acid Profile in Holstein Freemartin Heifers. J. Agric. Food Chem. 2017, 65, 10859–10867. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, N.M.; Koike, S.; Kobayashi, Y. Monitoring of gene expression in Fibrobacter succinogenes S85 under the co-culture with non-fibrolytic ruminal bacteria. Arch. Microbiol. 2015, 197, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Lu, Z.; Xu, Z.; Chen, Z.; Shen, Z. Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome 2017, 5, 123. [Google Scholar] [CrossRef]

| Items 1, % DM | Cow Diet 3 | RSS Diet |
|---|---|---|
| Corn silage | 22.4 | |
| Alfalfa hay | 14.5 | |
| Chinese wildrye | 8.8 | |
| Corn stover | 46.25 | |
| Ground corn | 22.9 | 25.00 |
| Soybean meal | 9.7 | 28.09 |
| Cottonseed meal | 4.6 | |
| DDGS | 4.6 | |
| Whole cottonseed | 8.9 | |
| Fat powder | 0.4 | |
| Salt | 0.5 | |
| Premix 2 | 2.5 | 0.66 |
| Chemical composition 1, % DM | ||
| CP | 15.5 | 19.2 |
| RDP | 8.4 | 12.3 |
| NDF | 30.2 | 43.7 |
| NFC | 39.0 | 29.5 |
| Starch | 26.1 | 20.8 |
| Comparison | Items | Taxa (Phylum; Family; Genus) | Relative Abundance in Treatments, % | LDA Score (log10) | ||
|---|---|---|---|---|---|---|
| FHI | SHI | CI | ||||
| FHI vs. SHI | FHI enriched | Bacteroidetes; Bacteroidaceae; BF311 | 1.39 | 0.78 | 1.56 | 3.50 |
| Bacteroidetes; Paraprevotellaceae; CF231 | 1.32 | 0.97 | 1.35 | 3.26 | ||
| Fibrobacteres; Fibrobacteraceae; Fibrobacter | 2.10 | 0.97 | 2.01 | 3.77 | ||
| Proteobacteria; Succinivibrionaceae; Ruminobacter | 7.19 | 4.25 | 4.61 | 4.18 | ||
| SHI enriched | Proteobacteria; Enterobacteriaceae; Klebsiella | 0.28 | 1.24 | 0.44 | 3.73 | |
| Proteobacteria; Succinivibrionaceae; Succinivibrio | 0.20 | 0.49 | 0.23 | 3.19 | ||
| CI vs. SHI | CI enriched | Bacteroidetes; Bacteroidaceae; BF311 | 1.39 | 0.78 | 1.56 | 3.58 |
| Bacteroidetes; Paraprevotellaceae; CF231 | 1.32 | 0.97 | 1.35 | 3.25 | ||
| Bacteroidetes; Paraprevotellaceae; YRC22 | 0.71 | 0.64 | 0.88 | 3.08 | ||
| Proteobacteria; Pseudomonadaceae; Pseudomonas | 0.17 | 0.09 | 0.32 | 3.09 | ||
| Proteobacteria; Xanthomonadaceae; Stenotrophomonas | 0.02 | 0.01 | 0.02 | 3.02 | ||
| SHI enriched | Bacteroidetes; Porphyromonadaceae; Dysgonomonas | < 0.01 | 0.03 | 0.01 | 3.01 | |
| Proteobacteria; Enterobacteriaceae; Klebsiella | 0.28 | 1.24 | 0.44 | 3.61 | ||
| Proteobacteria; Succinivibrionaceae; Succinivibrio | 0.20 | 0.49 | 0.23 | 3.13 | ||
| Time | Items | Treatments | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| FHI | SHI | CI | Treatment | Day | Treatment × Day | |||
| Daily | pH 1 | 6.68 | 6.71 | 6.70 | 0.027 | 0.62 | <0.01 | 0.53 |
| NH3-N, mg/dL 1 | 9.17 b | 15.28 a | 8.16 b | 2.126 | 0.01 | <0.01 | 0.01 | |
| 2 h | TVFAs, mmol/L | 77.58 | 72.31 | 79.51 | 2.489 | 0.05 | 0.98 | 0.44 |
| Acetate, % | 62.92 | 62.27 | 63.57 | 1.104 | 0.72 | 0.65 | 0.57 | |
| Propionate, % | 20.91 | 19.95 | 20.08 | 0.860 | 0.66 | 0.07 | 0.31 | |
| Butyrate, % | 11.25 | 11.85 | 11.82 | 0.432 | 0.52 | 0.85 | 0.78 | |
| Valerate, % | 2.15 | 2.15 | 2.15 | 0.118 | 0.87 | 0.03 | 0.26 | |
| Isobutyrate, % | 0.70 b | 0.84 a | 0.60 a | 0.035 | 0.01 | 0.04 | 0.34 | |
| Isovalerate, % | 2.06 b | 2.88 a | 1.81 b | 0.178 | 0.01 | 0.01 | 0.06 | |
| BCVFA, % | 2.76 b | 3.72 a | 2.41 b | 0.187 | <0.01 | <0.01 | 0.06 | |
| 10 h | TVFAs, mmol/L | 56.26 | 60.85 | 58.93 | 3.892 | 0.69 | 0.55 | 0.25 |
| Acetate, % | 62.87 | 63.33 | 64.19 | 1.529 | 0.81 | 0.08 | 0.30 | |
| Propionate, % | 20.83 a | 17.16 b | 18.22 b | 0.915 | 0.02 | 0.01 | 0.01 | |
| Butyrate, % | 11.89 | 12.41 | 12.68 | 0.757 | 0.73 | 0.08 | 0.75 | |
| Valerate, % | 2.00 | 2.03 | 2.21 | 0.127 | 0.44 | 0.97 | 0.65 | |
| Isobutyrate, % | 0.619 b | 0.76 a | 0.62 b | 0.033 | 0.01 | 0.82 | 0.28 | |
| Isovalerate, % | 1.78 b | 4.32 a | 2.08 b | 0.174 | <0.01 | 0.01 | 0.10 | |
| BCVFA, % | 2.40 b | 5.07 a | 2.71 b | 0.181 | <0.01 | 0.01 | 0.11 | |
| Items | Treatments | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|
| FHI | SHI | CI | Treatment | Day | Treatment × Day | ||
| MPS, g CP/d | 6.29 a | 4.45 b | 6.55 a | 0.219 | <0.01 | <0.01 | 0.19 |
| EMPS 1 | 40.47 a | 29.23 b | 40.29 a | 1.321 | 0.01 | 0.13 | 0.31 |
| Bacterial-N/available N 2, % | 49.02 a | 34.74 b | 49.19 a | 1.612 | <0.01 | 0.07 | 0.31 |
| Total bacteria, log10 16S rRNA gene copy numbers/mL fermenter fluid | |||||||
| 6 h | 10.92 | 10.90 | 11.26 | 0.297 | 0.59 | 0.64 | 0.99 |
| 12 h | 11.97 | 11.38 | 11.98 | 0.220 | 0.10 | 0.64 | 0.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zheng, N.; Shen, W.; Zhao, S.; Wang, J. Synchrony Degree of Dietary Energy and Nitrogen Release Influences Microbial Community, Fermentation, and Protein Synthesis in a Rumen Simulation System. Microorganisms 2020, 8, 231. https://doi.org/10.3390/microorganisms8020231
Zhang J, Zheng N, Shen W, Zhao S, Wang J. Synchrony Degree of Dietary Energy and Nitrogen Release Influences Microbial Community, Fermentation, and Protein Synthesis in a Rumen Simulation System. Microorganisms. 2020; 8(2):231. https://doi.org/10.3390/microorganisms8020231
Chicago/Turabian StyleZhang, Jun, Nan Zheng, Weijun Shen, Shengguo Zhao, and Jiaqi Wang. 2020. "Synchrony Degree of Dietary Energy and Nitrogen Release Influences Microbial Community, Fermentation, and Protein Synthesis in a Rumen Simulation System" Microorganisms 8, no. 2: 231. https://doi.org/10.3390/microorganisms8020231
APA StyleZhang, J., Zheng, N., Shen, W., Zhao, S., & Wang, J. (2020). Synchrony Degree of Dietary Energy and Nitrogen Release Influences Microbial Community, Fermentation, and Protein Synthesis in a Rumen Simulation System. Microorganisms, 8(2), 231. https://doi.org/10.3390/microorganisms8020231







