Isolation of an Obligate Mixotrophic Methanogen That Represents the Major Population in Thermophilic Fixed-Bed Anaerobic Digesters
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation Source and Sampling
2.2. Cultivation in Liquid Media
2.3. Cultivation on Solid Media
2.4. Gas Analyses
2.5. Phylogenetic Analyses
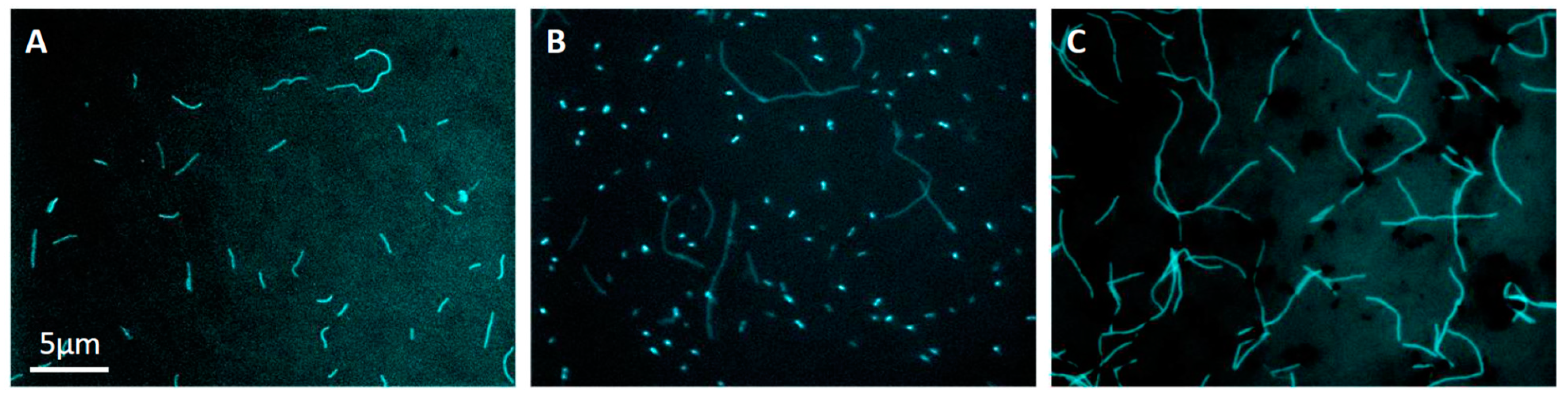
3. Results and Discussion
3.1. Colony Isolation
3.2. Limiting Dilution
3.3. Phylogenetic Characteristics
3.4. Growth Characteristics
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Weiland, P. Biogas production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Tatara, M.; Makiuchi, T.; Ueno, Y.; Goto, M.; Sode, K. Methanogenesis from acetate and propionate by thermophilic down-flow anaerobic packed-bed reactor. Bioresour. Technol. 2008, 99, 4786–4795. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Tang, Y.; Urakami, T.; Morimura, S.; Kida, K. Digestion performance and microbial community in full-scale methane fermentation of stillage from sweet potato-shochu production. J. Environ. Sci. 2014, 26, 423–431. [Google Scholar] [CrossRef]
- Kouzuma, A.; Tsutsumi, M.; Ishii, S.; Ueno, Y.; Abe, T.; Watanabe, K. Non-autotrophic methanogens dominate in anaerobic digesters. Sci. Rep. 2017, 7, 1510. [Google Scholar] [CrossRef] [PubMed]
- Zinder, S.H.; Sowers, K.R.; Ferry, J.G. Methanosarcina thermophila sp. nov., a thermophilic, acetotrophic, methane-producing bacterium. Int. J. Syst. Bacteriol. 1985, 35, 522–523. [Google Scholar] [CrossRef]
- Smith, D.R.; Doucette-Stamm, L.A.; Deloughery, C.; Lee, H.; Dubois, J.; Aldredge, T.; Bashirzadeh, R.; Blakely, D.; Cook, R.; Gilbert, K.; et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: Functional analysis and comparative genomics. J. Bacteriol. 1997, 179, 7135–7155. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 1999, 23, 13–38. [Google Scholar] [CrossRef] [PubMed]
- Ueoka, N.; Kouzuma, A.; Watanabe, K. Electrode plate-culture methods for colony isolation of exoelectrogens from anode microbiomes. Bioelectrochemistry 2018, 124, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Kosaka, T.; Hori, K.; Hotta, Y.; Watanabe, K. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl. Environ. Microbiol. 2005, 71, 7838–7845. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Kodama, Y.; Kaku, N. Diversity and abundance of bacteria in an underground oil-storage cavity. BMC Microbiol. 2002, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.A.; Teske, A.P. Diversity of methane-cycling archaea in hydrothermal sediment investigated by general and group-specific PCR primers. Appl. Environ. Microbiol. 2014, 81, 1426–1441. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Zinder, S.H.; Mah, R.A. Isolation and characterization of a thermophilic strain of Methanosarcina unable to use H2-CO2 for methanogenesis. Appl. Environ. Microbiol. 1979, 38, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Etchebehere, C.; Pavan, M.E.; Zorzópulos, J.; Soubes, M.; Muxí, L. Coprothermobacterplatensis sp. nov., a new anaerobic proteolytic thermophilic bacterium isolated from an anaerobic mesophilic sludge. Int. J. Syst. Bacteriol. 1998, 48, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Urushibata, W.; Morikawa, M. Studies on microbial symbiosis in a methanogenic high temperature subsurface petroleum reservoir. J. Environ. Biotechnol. 2016, 16, 31–38. [Google Scholar]
- Nakamura, K.; Takahashi, A.; Mori, C.; Tamaki, H.; Mochimaru, H.; Nakamura, K.; Takamizawa, K.; Kamagata, Y. Methanothermobacter tenebrarum sp. nov., a hydrogenotrophic, thermophilic methanogen isolated from gas-associated formation water of a natural gas field. Int. J. Syst. Evol. Microbiol. 2013, 63, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Dai, L.; Li, X.; Zhang, H.; Lu, Y. Isolation and characterization of Methanothermobacter crinale sp. nov., a novel hydrogenotrophic methanogen from the Shengli oil field. Appl. Environ. Microbiol. 2011, 77, 5212–5219. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagoya, M.; Kouzuma, A.; Ueno, Y.; Watanabe, K. Isolation of an Obligate Mixotrophic Methanogen That Represents the Major Population in Thermophilic Fixed-Bed Anaerobic Digesters. Microorganisms 2020, 8, 217. https://doi.org/10.3390/microorganisms8020217
Nagoya M, Kouzuma A, Ueno Y, Watanabe K. Isolation of an Obligate Mixotrophic Methanogen That Represents the Major Population in Thermophilic Fixed-Bed Anaerobic Digesters. Microorganisms. 2020; 8(2):217. https://doi.org/10.3390/microorganisms8020217
Chicago/Turabian StyleNagoya, Misa, Atsushi Kouzuma, Yoshiyuki Ueno, and Kazuya Watanabe. 2020. "Isolation of an Obligate Mixotrophic Methanogen That Represents the Major Population in Thermophilic Fixed-Bed Anaerobic Digesters" Microorganisms 8, no. 2: 217. https://doi.org/10.3390/microorganisms8020217
APA StyleNagoya, M., Kouzuma, A., Ueno, Y., & Watanabe, K. (2020). Isolation of an Obligate Mixotrophic Methanogen That Represents the Major Population in Thermophilic Fixed-Bed Anaerobic Digesters. Microorganisms, 8(2), 217. https://doi.org/10.3390/microorganisms8020217



