Planktonic Bacterial and Archaeal Communities in an Artificially Irrigated Estuarine Wetland: Diversity, Distribution, and Responses to Environmental Parameters
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection and Environmental Parameter Analysis
2.3. DNA Extraction and qPCR
2.4. Illumina 16S rRNA Gene Amplicon Sequencing
2.5. Data Analysis
3. Results
3.1. Characterization of the Environmental Parameters
3.2. Bacterial and Archaeal Community α-Diversity
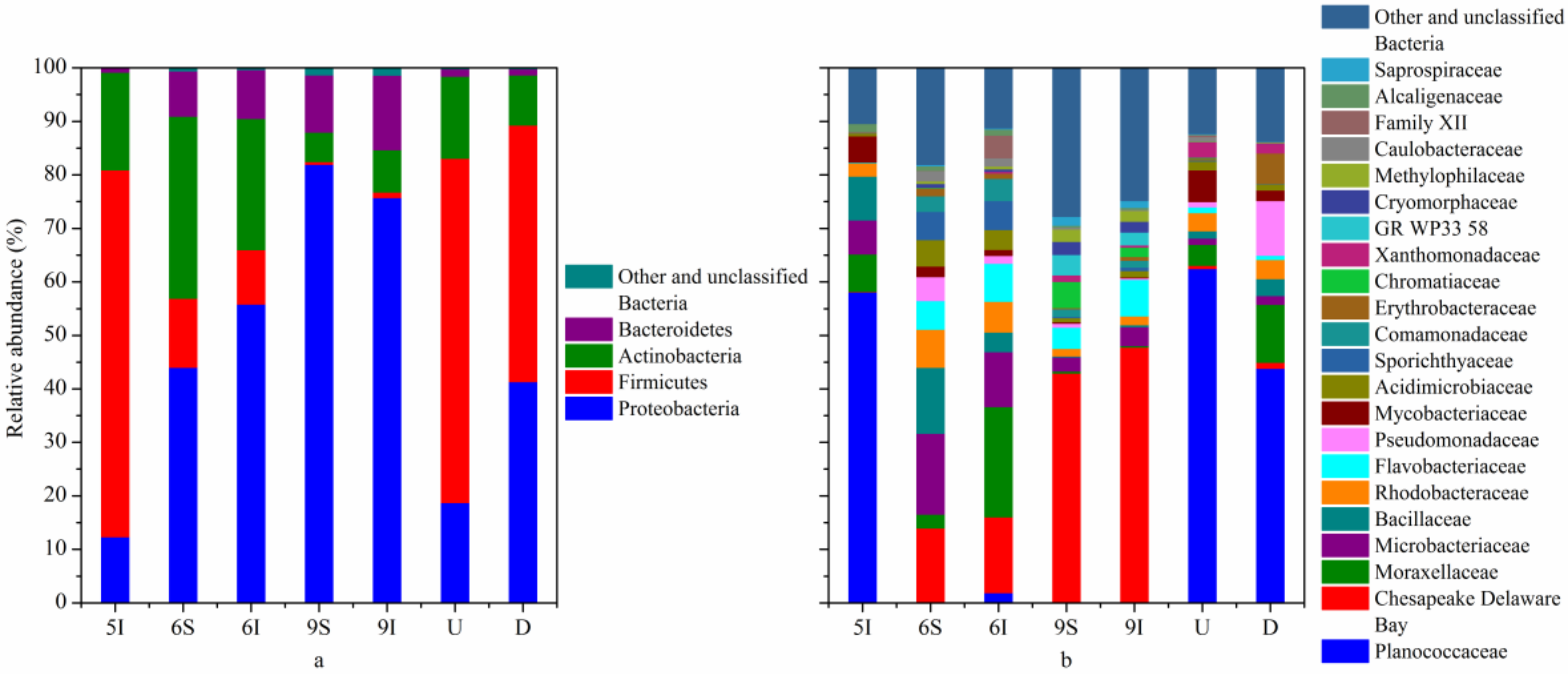
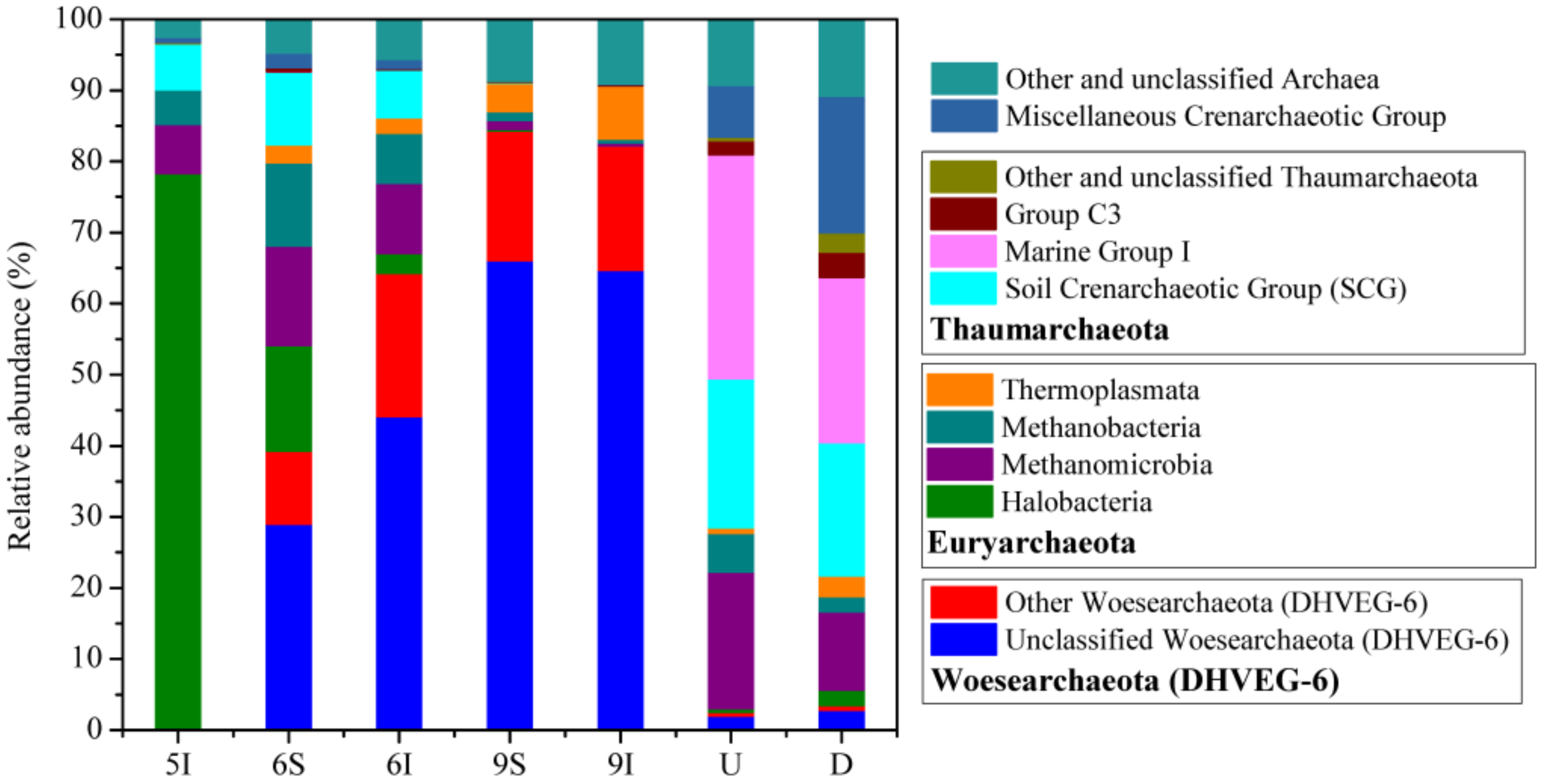
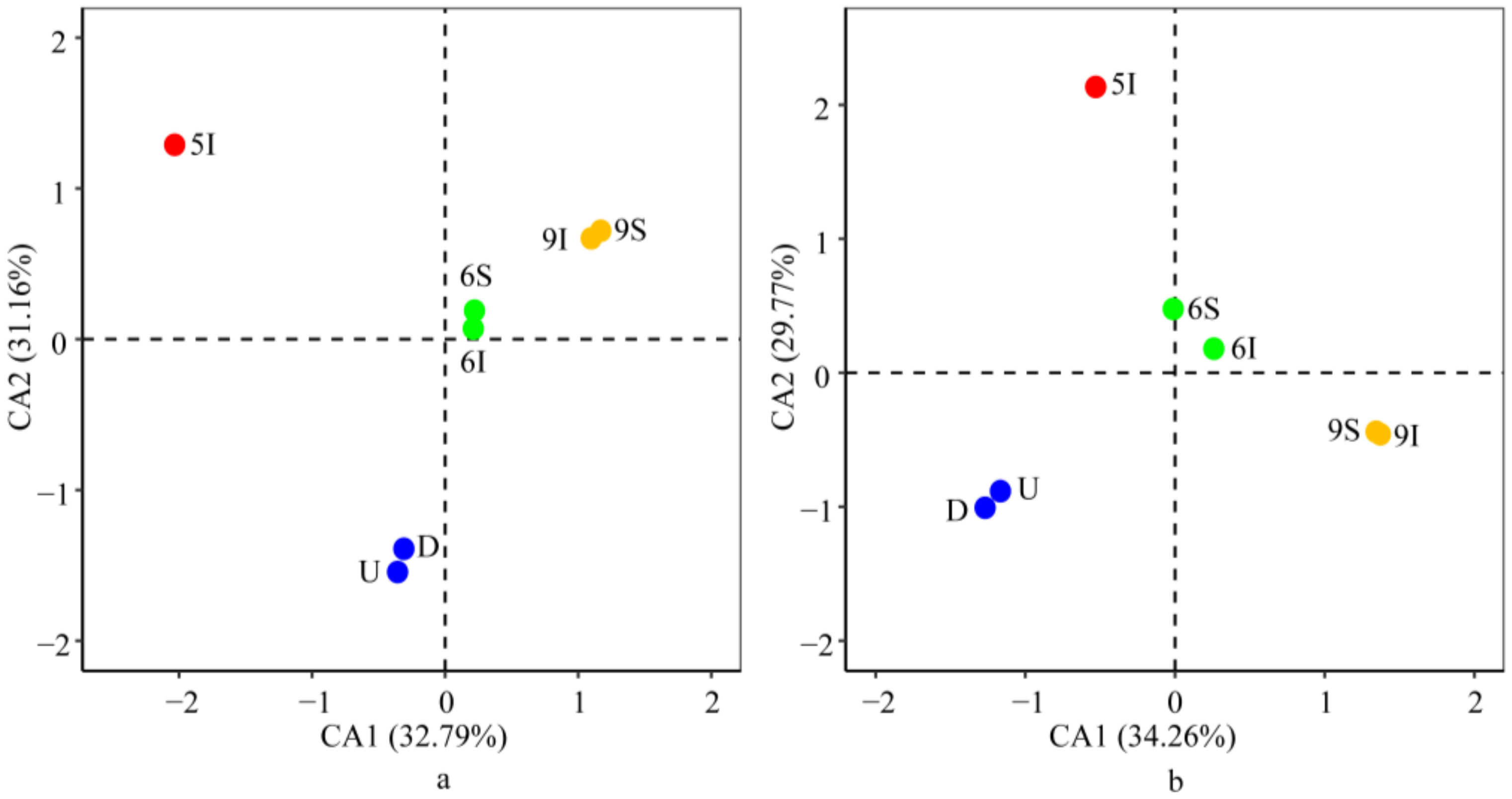
3.3. Bacterial and Archaeal Community Structures
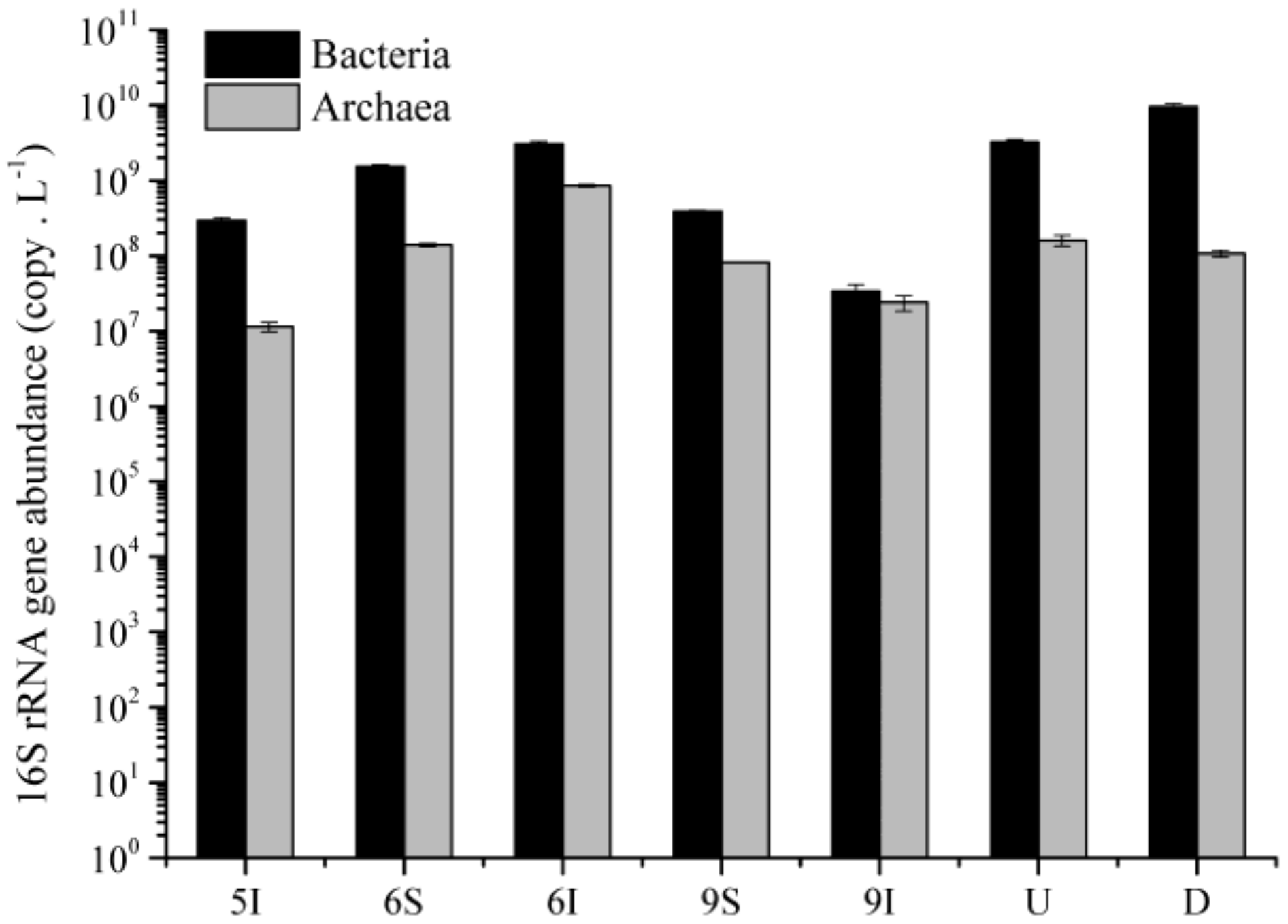
3.4. Abundances of the Bacterial and Archaeal 16S rRNA Genes
3.5. Effect of the Environmental Parameters on the Bacterial and Archaeal Communities
3.5.1. Effect of the Environmental Parameters on the Abundances and α-Diversity Indices of the Bacterial and Archaeal Communities
3.5.2. Correlations of the Detection Frequencies of the Dominant Bacterial and Archaeal Taxa with the Environmental Parameters
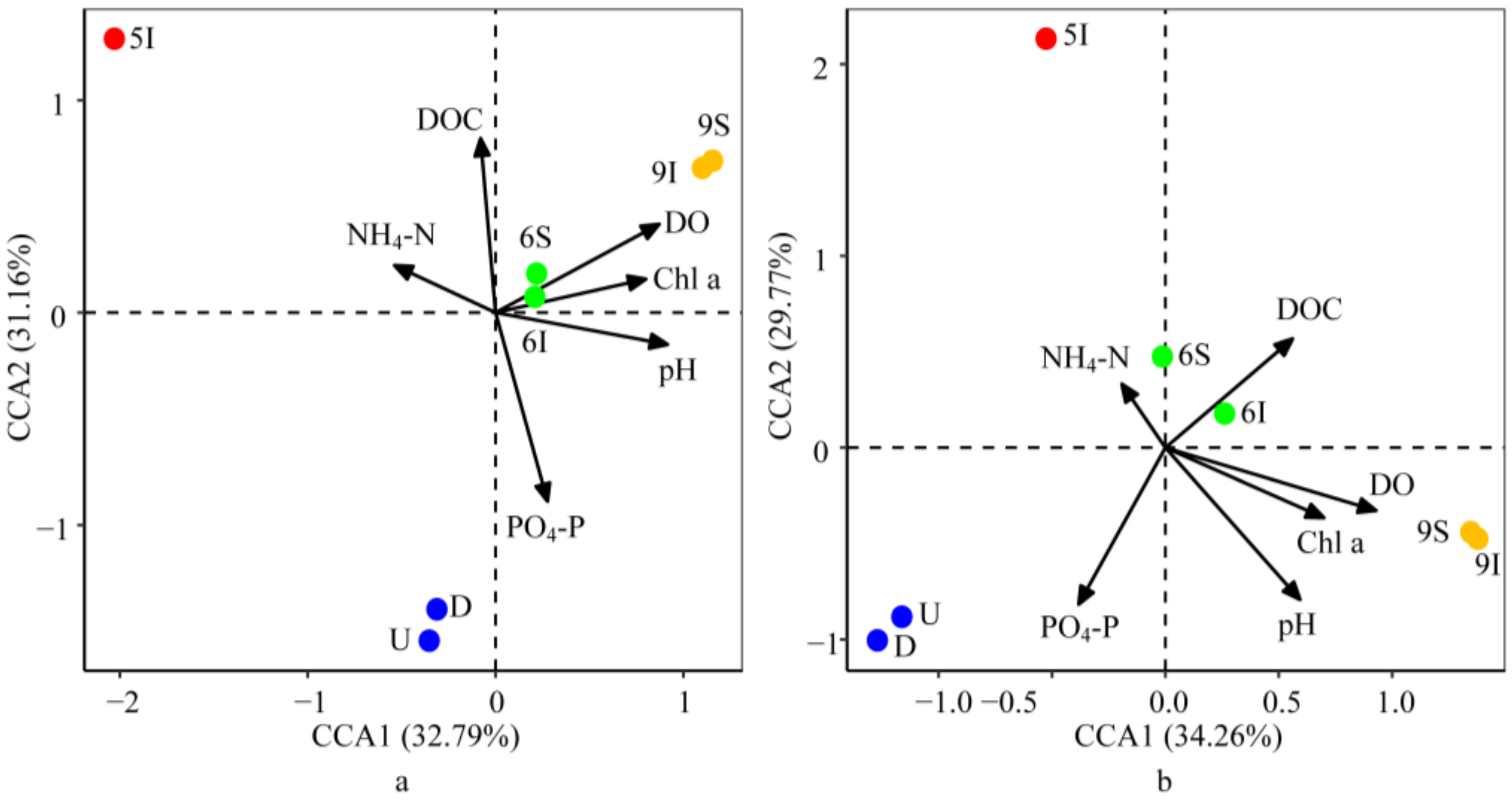
3.5.3. Effect of the Environmental Parameters on the Bacterial and Archaeal Community Structures
4. Discussion
4.1. Distinct Seasonal Dynamics of Planktonic Bacterial and Archaeal Communities in the Wetland
4.2. Negligible Effect of the Wetland Effluent on the Bacterial and Archaeal Communities in the Liaohe River
4.3. DO and pH as the Most Influential Factors in Shaping the Planktonic Bacterial and Archaeal Communities in the Liaohe River Estuarine Wetland (LEW)
4.4. Potential Roles of the Dominant Bacterial and Archaeal Taxa in the LEW
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chi, Y.; Zheng, W.; Shi, H.H.; Sun, J.K.; Fu, Z.Y. Spatial heterogeneity of estuarine wetland ecosystem health influenced by complex natural and anthropogenic factors. Sci. Total Environ. 2018, 634, 1445–1462. [Google Scholar] [CrossRef]
- Zhang, C.; Nie, S.; Liang, J.; Zeng, G.M.; Wu, H.P.; Hua, S.S.; Liu, J.Y.; Yuan, Y.J.; Xiao, H.B.; Deng, L.J.; et al. Effects of heavy metals and soil physicochemical properties on wetland soil microbial biomass and bacterial community structure. Sci. Total Environ. 2016, 557, 785–790. [Google Scholar] [CrossRef]
- Huang, X.; Bai, J.; Li, K.R.; Zhao, Y.G.; Tian, W.J.; Dang, J.J. Characteristics of two novel cold- and salt-tolerant ammonia-oxidizing bacteria from Liaohe Estuarine Wetland. Mar. Pollut. Bull. 2017, 114, 192–200. [Google Scholar] [CrossRef]
- Hua, G.F.; Cheng, Y.; Kong, J.; Li, M.; Zhao, Z.W. High-throughput sequencing analysis of bacterial community spatiotemporal distribution in response to clogging in vertical flow constructed wetlands. Bioresour. Technol. 2018, 248, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.Q.; Wang, H.; Chen, X.C.; Wang, R.Q.; Liu, J. Composition and distribution of microbial communities in natural river wetlands and corresponding constructed wetlands. Ecol. Eng. 2017, 98, 40–48. [Google Scholar] [CrossRef]
- Riva, V.; Mapelli, F.; Syranidou, E.; Crotti, E.; Choukrallah, R.; Kalogerakis, N.; Borin, S. Root bacteria recruited by Phragmites australis in constructed wetlands have the potential to enhance azo-dye phytodepuration. Microorganisms 2019, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- Mateu, M.G.; Park, C.E.; McAskill, C.P.; Baldwin, A.H.; Yarwood, S.A. Urbanization altered bacterial and archaeal composition in tidal freshwater wetlands near Washington DC, USA, and Buenos Aires, Argentina. Microorganisms 2019, 7, 72. [Google Scholar] [CrossRef]
- Franklin, R.B.; Morrissey, E.M.; Morina, J.C. Changes in abundance and community structure of nitrate-reducing bacteria along a salinity gradient in tidal wetlands. Pedobiologia 2017, 60, 21–26. [Google Scholar] [CrossRef]
- Zhang, H.X.; Zheng, S.L.; Ding, J.W.; Wang, O.M.; Liu, F.H. Spatial variation in bacterial community in natural wetland-river-sea ecosystems. J. Basic Microb. 2017, 57, 536–546. [Google Scholar] [CrossRef]
- Guan, X.Y.; Han, J.B.; Wang, B.; Dong, Y.; Jiang, B.; Chen, Z.; Yang, A.F.; Wang, Z.H.; Zhou, Z.C. Analysis of bacterial communities in Liaodong Bay Dalinghe estuarine wetland. Ecol. Environ. Sci. 2012, 21, 1063–1070. (In Chinese) [Google Scholar] [CrossRef]
- Yang, J.S.; Zhan, C.; Li, Y.Z.; Zhou, D.; Yu, Y.; Yu, J.B. Effect of salinity on soil respiration in relation to dissolved organic carbon and microbial characteristics of a wetland in the Liaohe River estuary, Northeast China. Sci. Total Environ. 2018, 642, 946–953. [Google Scholar] [CrossRef]
- Zhao, X.L.; Zhou, G.S.; Zhou, L.; Lv, G.H.; Jia, Q.Y.; Xie, Y.B. Characteristics of soil microbial community in bulrush wetlands of Panjin, Northeast China. Chin. J. Soil Sci. 2008, 39, 1376–1379. (In Chinese) [Google Scholar] [CrossRef]
- Li, T.T.; Hu, H.; Li, Z.Y.; Zhang, J.Y.; Li, D. The impact of irrigation on bacterial community composition and diversity in Liaohe estuary wetland. J. Ocean Univ. China 2018, 17, 855–863. [Google Scholar] [CrossRef]
- He, T.; Guan, W.; Luan, Z.Y.; Xie, S.G. Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment. Appl. Microbiol. Biotechnol. 2016, 100, 1479–1488. [Google Scholar] [CrossRef]
- Yang, F.; Zhao, D.Z.; Suo, A.N. The study of Shuangtaizihekou wetland landscape temporal and spatial changes. Remote Sens. Technol. Appl. 2008, 23, 51–56. (In Chinese) [Google Scholar]
- Li, L.; Zou, L.; Yang, Y.; Q., R.Q.; Dai, Q.Y.; Jian, H.M. Diurnal variations of nutrient composition and structure in reed wetland of Liaohe river estuary. Trans. Oceanol. Limnol. 2018, 2, 80–87. (In Chinese) [Google Scholar]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Porat, I.; Vishnivetskaya, T.A.; Mosher, J.J.; Brandt, C.C.; Yang, Z.M.K.; Brooks, S.C.; Liang, L.Y.; Drake, M.M.; Podar, M.; Brown, S.D.; et al. Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb. Ecol. 2010, 60, 784–795. [Google Scholar] [CrossRef]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.Y.; Aas, J.A.; Paster, B.J.; DeSantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z.H. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef]
- Ohene-Adjei, S.; Teather, R.M.; Ivanj, M.; Forster, R.J. Postinoculation protozoan establishment and association patterns of methanogenic archaea in the ovine rumen. Appl. Environ. Microb. 2007, 73, 4609–4618. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Reyon, D.; Tsai, S.Q.; Khayter, C.; Foden, J.A.; Sander, J.D.; Joung, J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012, 30, 460. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Wu, H.P.; Zeng, G.M.; Liang, J.; Guo, S.L.; Dai, J.; Lu, L.H.; Wei, Z.; Xu, P.A.; Li, F.; Yuan, Y.J.; et al. Effect of early dry season induced by the Three Gorges Dam on the soil microbial biomass and bacterial community structure in the Dongting Lake wetland. Ecol. Indic. 2015, 53, 129–136. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Al-Kharusi, S.; Gkorezis, P.; Prigent, S.; Headley, T. Bacterial communities in the rhizosphere of Phragmites australis from an oil-polluted wetland. Arch. Agron. Soil Sci. 2018, 64, 360–370. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zhang, X.L.; Liu, Y.; Xie, S.G.; Liu, Y.G. Bacterioplankton communities in a high-altitude freshwater wetland. Ann. Microbiol. 2014, 64, 1405–1411. [Google Scholar] [CrossRef]
- Faulwetter, J.L.; Burr, M.D.; Parker, A.E.; Stein, O.R.; Camper, A.K. Influence of season and plant species on the abundance and diversity of sulfate reducing bacteria and ammonia oxidizing bacteria in constructed wetland microcosms. Microb. Ecol. 2013, 65, 111–127. [Google Scholar] [CrossRef]
- Wang, X.L.; Xu, L.G.; Wan, R.G.; Chen, Y.W. Seasonal variations of soil microbial biomass within two typical wetland areas along the vegetation gradient of Poyang Lake, China. Catena 2016, 137, 483–493. [Google Scholar] [CrossRef]
- Li, B.X.; Chen, H.L.; Li, N.N.; Wu, Z.; Wen, Z.G.; Xie, S.G.; Liu, Y. Spatio-temporal shifts in the archaeal community of a constructed wetland treating river water. Sci. Total Environ. 2017, 605, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.J.; Lai, G.L.; Chi, C.Q.; Zhao, J.Y.; Yan, Y.C.; Nie, Y.; Wu, X.L. Purification of eutrophic water containing chlorpyrifos by aquatic plants and its effects on planktonic bacteria. Chemosphere 2018, 193, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Lyon, S.R.; Leddy, M.; Jacobson-Meyers, M. Impact of plant density and microbial composition on water quality from a free water surface constructed wetland. J. Appl. Microbiol. 2007, 102, 921–936. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Meng, H.; Liu, Y.; Gu, J.D.; Li, M. Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S rRNA gene sequencing. Front. Microbiol. 2017, 8, 2148. [Google Scholar] [CrossRef]
- Long, Y.; Yi, H.; Chen, S.L.; Zhang, Z.K.; Cui, K.; Bing, Y.X.; Zhuo, Q.F.; Li, B.X.; Xie, S.G.; Guo, Q.W. Influences of plant type on bacterial and archaeal communities in constructed wetland treating polluted river water. Environ. Sci. Pollut. Res. 2016, 23, 19570–19579. [Google Scholar] [CrossRef]
- Long, Y.; Zhang, Z.K.; Pan, X.K.; Li, B.X.; Xie, S.G.; Guo, Q.W. Substrate influences on archaeal and bacterial assemblages in constructed wetland microcosms. Ecol. Eng. 2016, 94, 437–442. [Google Scholar] [CrossRef]
- Baik, K.; Park, S.; Kim, E.; Bae, K.; Ann, J.H.; Ka, J.O.; Chun, J.; Seong, C. Diversity of bacterial community in freshwater of Woopo wetland. J. Microbiol. 2008, 46, 647–655. [Google Scholar] [CrossRef]
- Buesing, N.; Filippini, M.; Burgmann, H.; Gessner, M.O. Microbial communities in contrasting freshwater marsh microhabitats. FEMS Microbiol. Ecol. 2009, 69, 84–97. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, G.; Tang, X.M.; Shao, K.Q.; Bayartu, S.; Dai, J.Y. Bacterial community changes along a salinity gradient in a Chinese wetland. Can. J. Microbiol. 2013, 59, 611–619. [Google Scholar] [CrossRef]
- Elsayed, O.F.; Maillard, E.; Vuilleumier, S.; Imfeld, G. Bacterial communities in batch and continuous-flow wetlands treating the herbicide S-metolachlor. Sci. Total Environ. 2014, 499, 327–335. [Google Scholar] [CrossRef] [PubMed]
- De Figueiredo, D.R.; Pereira, M.J.; Moura, A.; Silva, L.; Barrios, S.; Fonseca, F.; Henriques, I.; Correia, A. Bacterial community composition over a dry winter in meso- and eutrophic Portuguese water bodies. FEMS Microbiol. Ecol. 2007, 59, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.C.; Dong, H.L.; Deng, S.C.; Yu, B.S.; Huang, Q.Y.; Wu, Q.L. Response of archaeal community structure to environmental changes in lakes on the Tibetan Plateau, Northwestern China. Geomicrobiol. J. 2009, 26, 289–297. [Google Scholar] [CrossRef]
- Fan, L.M.; Song, C.; Meng, S.L.; Qiu, L.P.; Zheng, Y.; Wu, W.; Qu, J.H.; Li, D.D.; Zhang, C.; Hu, G.D.; et al. Spatial distribution of planktonic bacterial and archaeal communities in the upper section of the tidal reach in Yangtze River. Sci. Rep. UK 2016, 6, 39147. [Google Scholar] [CrossRef]
- Hu, A.Y.; Yang, X.Y.; Chen, N.W.; Hou, L.Y.; Ma, Y.; Yu, C.P. Response of bacterial communities to environmental changes in a mesoscale subtropical watershed, Southeast China. Sci. Total Environ. 2014, 472, 746–756. [Google Scholar] [CrossRef]
- Liu, J.W.; Yu, S.L.; Zhao, M.X.; He, B.Y.; Zhang, X.H. Shifts in archaeaplankton community structure along ecological gradients of Pearl Estuary. FEMS Microbiol. Ecol. 2014, 90, 424–435. [Google Scholar] [CrossRef]
- Hu, A.Y.; Hou, L.Y.; Yu, C.P. Biogeography of planktonic and benthic archaeal communities in a subtropical eutrophic estuary of China. Microb. Ecol. 2015, 70, 322–335. [Google Scholar] [CrossRef]
- Gorrasi, S.; Pesciaroli, C.; Barghini, P.; Pasqualetti, M.; Fenice, M. Structure and diversity of the bacterial community of an Arctic estuarine system (Kandalaksha Bay) subject to intense tidal currents. J. Mar. Syst. 2019, 196, 77–85. [Google Scholar] [CrossRef]
- Farnell-Jackson, E.A.; Ward, A.K. Seasonal patterns of viruses, bacteria and dissolved organic carbon in a riverine wetland. Freshwater Biol. 2003, 48, 841–851. [Google Scholar] [CrossRef]
- Liu, X.B.; Yao, T.D.; Kang, S.C.; Jiao, N.A.Z.; Zeng, Y.H.; Liu, Y.Q. Bacterial community of the largest oligosaline lake, Namco on the Tibetan Plateau. Geomicrobiol. J. 2010, 27, 669–682. [Google Scholar] [CrossRef]
- Logue, J.B.; Langenheder, S.; Andersson, A.F.; Bertilsson, S.; Drakare, S.; Lanzen, A.; Lindstrom, E.S. Freshwater bacterioplankton richness in oligotrophic lakes depends on nutrient availability rather than on species-area relationships. ISME J. 2012, 6, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Van Horn, D.J.; Sinsabaugh, R.L.; Takacs-Vesbach, C.D.; Mitchell, K.R.; Dahm, C.N. Response of heterotrophic stream biofilm communities to a gradient of resources. Aquat. Microb. Ecol. 2011, 64, 149–161. [Google Scholar] [CrossRef]
- Heino, J.; Tolkkinen, M.; Pirttila, A.M.; Aisala, H.; Mykra, H. Microbial diversity and community-environment relationships in boreal streams. J. Biogeogr. 2014, 41, 2234–2244. [Google Scholar] [CrossRef]
- Wei, G.S.; Li, M.C.; Li, F.G.; Li, H.; Gao, Z. Distinct distribution patterns of prokaryotes between sediment and water in the Yellow River estuary. Appl. Microbiol. Biot. 2016, 100, 9683–9697. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Woodhouse, J.N.; Te, S.H.; Gin, K.Y.H.; He, Y.L.; Xu, C.; Chen, L. Seasonal variation in the bacterial community composition of a large estuarine reservoir and response to cyanobacterial proliferation. Chemosphere 2018, 202, 576–585. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Kim, M.; Lai-Hoe, A.; Shukor, N.A.A.; Rahim, R.A.; Go, R.; Adams, J.M. pH dominates variation in tropical soil archaeal diversity and community structure. FEMS Microbiol. Ecol. 2013, 86, 303–311. [Google Scholar] [CrossRef]
- Guan, W.; Yin, M.; He, T.; Xie, S.G. Influence of substrate type on microbial community structure in vertical-flow constructed wetland treating polluted river water. Environ. Sci. Pollut. Res. 2015, 22, 16202–16209. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Yan, C.; Zhang, H.; Li, B.; Wang, D.; Zhao, Y.J.; Zheng, Z. Effects of influent C/N ratios on CO2 and CH4 emissions from vertical subsurface flow constructed wetlands treating synthetic municipal wastewater. J. Hazard. Mater. 2012, 203, 188–194. [Google Scholar] [CrossRef]
- Liu, D.Y.; Ding, W.X.; Yuan, J.J.; Xiang, J.; Lin, Y.X. Substrate and/or substrate-driven changes in the abundance of methanogenic archaea cause seasonal variation of methane production potential in species-specific freshwater wetlands. Appl. Microbiol. Biotechnol. 2014, 98, 4711–4721. [Google Scholar] [CrossRef]
- Gabarro, J.; Hernandez-del Amo, E.; Gich, F.; Ruscalleda, M.; Balaguer, M.D.; Colprim, J. Nitrous oxide reduction genetic potential from the microbial community of an intermittently aerated partial nitritation SBR treating mature landfill leachate. Water Res. 2013, 47, 7066–7077. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Chen, J.F.; Wu, Z.; Wu, S.F.; Xie, S.G.; Liu, Y. Seasonal and spatial dynamics of denitrification rate and denitrifier community in constructed wetland treating polluted river water. Int. Biodeterior. Biodegrad. 2018, 126, 143–151. [Google Scholar] [CrossRef]
- Kim, B.K.; Jung, M.Y.; Yu, D.S.; Park, S.J.; Oh, T.K.; Rhee, S.K.; Kim, J.F. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J. Bacteriol. 2011, 193, 5539–5540. [Google Scholar] [CrossRef] [PubMed]
- Mosier, A.C.; Allen, E.E.; Kim, M.; Ferriera, S.; Francis, C.A. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J. Bacteriol. 2012, 194, 2121–2122. [Google Scholar] [CrossRef]
- Ling, N.; Deng, K.Y.; Song, Y.; Wu, Y.C.; Zhao, J.; Raza, W.; Huang, Q.W.; Shen, Q.R. Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol. Res. 2014, 169, 570–578. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, H.J.; Ngo, H.H.; Guo, W.S.; Zhang, J.; Liu, C.; Liang, S.; Hu, Z.; Yang, Z.C.; Zhao, C.C. Microbial abundance and community in subsurface flow constructed wetland microcosms: Role of plant presence. Environ. Sci. Pollut. Res. 2016, 23, 4036–4045. [Google Scholar] [CrossRef] [PubMed]

| Sample * | OTUs † | Shannon † | Chao 1 † | Pielou † | Good’s Coverage (%) † | |
|---|---|---|---|---|---|---|
| Bacteria | 5I | 192 | 3.53 | 259.00 | 0.46 | 99.55 |
| 6S | 367 | 5.66 | 406.47 | 0.66 | 99.43 | |
| 6I | 330 | 5.47 | 493.38 | 0.65 | 99.26 | |
| 9S | 375 | 4.55 | 473.06 | 0.53 | 99.26 | |
| 9I | 379 | 4.46 | 437.13 | 0.52 | 99.44 | |
| U | 271 | 3.17 | 350.00 | 0.39 | 99.41 | |
| D | 265 | 4.11 | 331.16 | 0.51 | 99.48 | |
| Archaea | 5I | 69 | 3.94 | 100.00 | 0.64 | 99.97 |
| 6S | 528 | 7.02 | 544.88 | 0.78 | 99.89 | |
| 6I | 754 | 7.79 | 778.43 | 0.81 | 99.79 | |
| 9S | 532 | 7.81 | 546.53 | 0.86 | 99.91 | |
| 9I | 805 | 8.14 | 828.66 | 0.84 | 99.84 | |
| U | 281 | 5.02 | 290.56 | 0.62 | 99.94 | |
| D | 270 | 5.90 | 300.17 | 0.73 | 99.87 |
| Genera | 5I | 6S | 6I | 9S | 9I | U | D |
|---|---|---|---|---|---|---|---|
| Planococcus | - | - | 1.48 | 0.15 | 0.27 | 61.32 | 42.24 |
| Psychrobacter | 7.00 | 1.01 | 17.03 | - | - | 3.47 | 10.48 |
| Psychrobacillus | 34.76 | - | 0.12 | - | - | - | 0.98 |
| Bacillus | 8.13 | 12.36 | 3.64 | 0.18 | 0.30 | 0.90 | 1.20 |
| Sporosarcina | 23.08 | - | - | - | - | - | - |
| Pseudomonas | - | 4.49 | 1.46 | 0.74 | 0.28 | 0.98 | 10.23 |
| Mycobacterium | 4.95 | 1.95 | 1.14 | 0.28 | 0.17 | 5.93 | 1.98 |
| CL500-29 marine group | 0.41 | 4.05 | 3.09 | 0.75 | 1.08 | 1.48 | 0.55 |
| Flavobacterium | - | 2.25 | 4.31 | 1.97 | 1.91 | 0.53 | - |
| Altererythrobacter | - | 1.17 | 0.84 | 0.24 | 0.72 | 0.32 | 5.74 |
| hgcI clade | - | 3.48 | 3.76 | 0.22 | 0.64 | 0.10 | - |
| Rheinheimera | - | 0.14 | - | 4.88 | 1.71 | 0.17 | - |
| Acinetobacter | - | 1.56 | 3.57 | - | - | 0.36 | 0.37 |
| Limnohabitans | - | 2.17 | 2.95 | 0.10 | 0.15 | - | - |
| Paracoccus | - | 0.36 | 1.10 | - | - | 2.67 | 1.01 |
| Stenotrophomonas | - | - | 0.29 | - | - | 2.87 | 1.80 |
| Exiguobacterium | - | - | 4.26 | - | - | 0.35 | 0.13 |
| Brevundimonas | - | 1.78 | 1.10 | 0.32 | 0.18 | 0.39 | - |
| Lutibacter | - | - | - | - | 3.64 | - | - |
| NS3a marine group | - | 1.43 | 1.77 | 0.12 | 0.26 | - | - |
| Owenweeksia | - | 0.46 | 0.36 | 1.88 | 0.73 | - | - |
| Candidatus Pelagibacter | - | - | - | - | - | 0.79 | 1.87 |
| Rhodobacter | 0.22 | 0.96 | 0.57 | 0.30 | 0.19 | 0.18 | 0.13 |
| BAL58 marine group | - | 0.42 | 0.76 | 0.68 | 0.63 | - | - |
| MWH-UniP1 aquatic group | - | 0.61 | 0.96 | 0.38 | 0.37 | - | - |
| GKS98 freshwater group | 1.56 | 0.24 | 0.16 | - | - | - | - |
| OM43 clade | - | 0.44 | 0.51 | 0.30 | 0.55 | - | - |
| Methylophilus | - | - | - | 0.98 | 0.88 | - | - |
| Methylophaga | - | - | 0.24 | 0.68 | 0.22 | - | 0.70 |
| Citricoccus | 1.26 | - | - | - | - | 0.17 | 0.11 |
| Community | α-Diversity Index | R2 | p | n | Explanatory Variables (β-Weights) |
|---|---|---|---|---|---|
| Bacteria | Shannon | 0.995 | 0.002 | 7 | DIN/PO4-P (−0.941) ***, DOC (−0.454) * |
| Chao 1 | 0.901 | 0.01 | 7 | DO (0.641) *, NH4-N (−0.547) * | |
| Pielou | 1.000 | <0.001 | 7 | T (0.478) ***, DOC (−0.636) ***, DIN (−0.530) ***, SAL (−0.118) *** | |
| Archaea | Shannon | 0.938 | 0.004 | 7 | TP (−1.029) ***, DOC (−0.412) * |
| Chao 1 | 0.749 | 0.012 | 7 | Chla (0.865) * | |
| Pielou | 0.860 | 0.003 | 7 | TP (−0.927) ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Mi, T.; Yu, Z.; Ma, M.; Zhen, Y. Planktonic Bacterial and Archaeal Communities in an Artificially Irrigated Estuarine Wetland: Diversity, Distribution, and Responses to Environmental Parameters. Microorganisms 2020, 8, 198. https://doi.org/10.3390/microorganisms8020198
Li M, Mi T, Yu Z, Ma M, Zhen Y. Planktonic Bacterial and Archaeal Communities in an Artificially Irrigated Estuarine Wetland: Diversity, Distribution, and Responses to Environmental Parameters. Microorganisms. 2020; 8(2):198. https://doi.org/10.3390/microorganisms8020198
Chicago/Turabian StyleLi, Mingyue, Tiezhu Mi, Zhigang Yu, Manman Ma, and Yu Zhen. 2020. "Planktonic Bacterial and Archaeal Communities in an Artificially Irrigated Estuarine Wetland: Diversity, Distribution, and Responses to Environmental Parameters" Microorganisms 8, no. 2: 198. https://doi.org/10.3390/microorganisms8020198
APA StyleLi, M., Mi, T., Yu, Z., Ma, M., & Zhen, Y. (2020). Planktonic Bacterial and Archaeal Communities in an Artificially Irrigated Estuarine Wetland: Diversity, Distribution, and Responses to Environmental Parameters. Microorganisms, 8(2), 198. https://doi.org/10.3390/microorganisms8020198






