Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Feedstock Materials
2.2. Starting Up of the Digesters (Test 1)
2.3. Verification Experiments (Test 2)
2.4. Digestate and Biogas Analysis
2.5. Microbial Community Analysis
2.6. Data Preparation and Statistical Analysis
3. Results
3.1. Test 1―Starting up of the Digesters
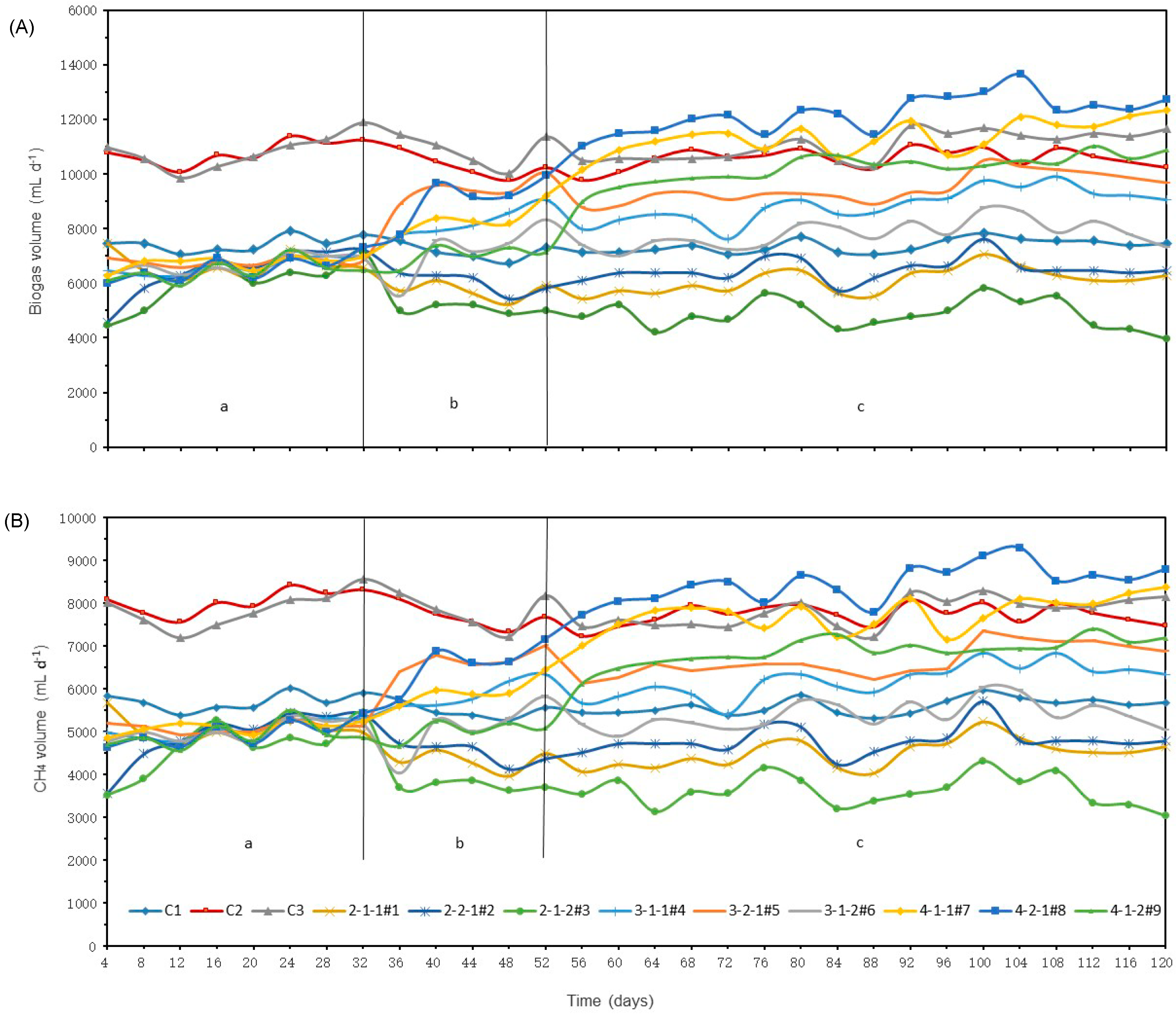
3.1.1. Biogas and Methane Production
3.1.2. Variation of pH, total alkalinity and ammonia
3.2. Test 2―Verification experiments
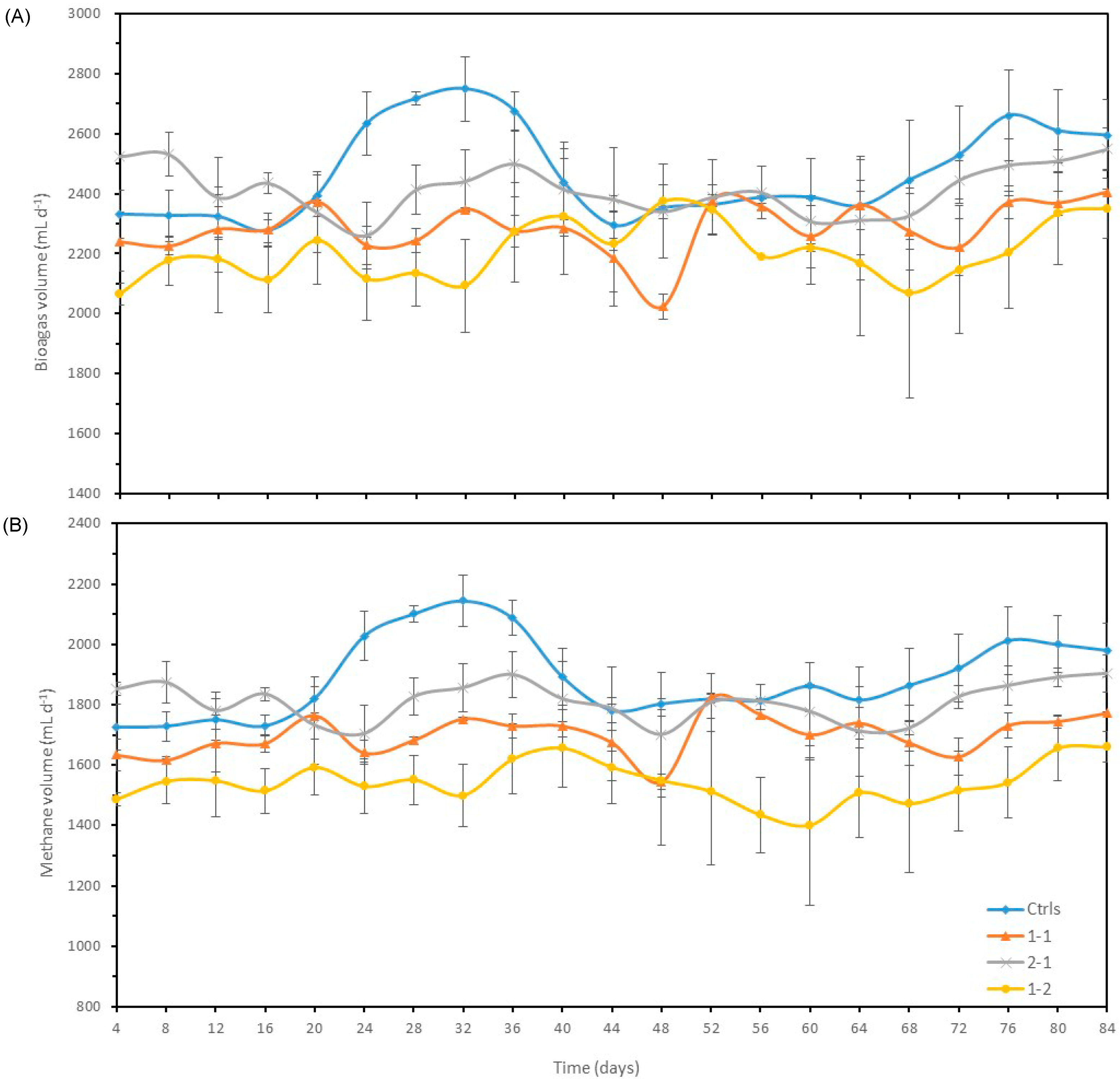
3.2.1. Biogas and methane production
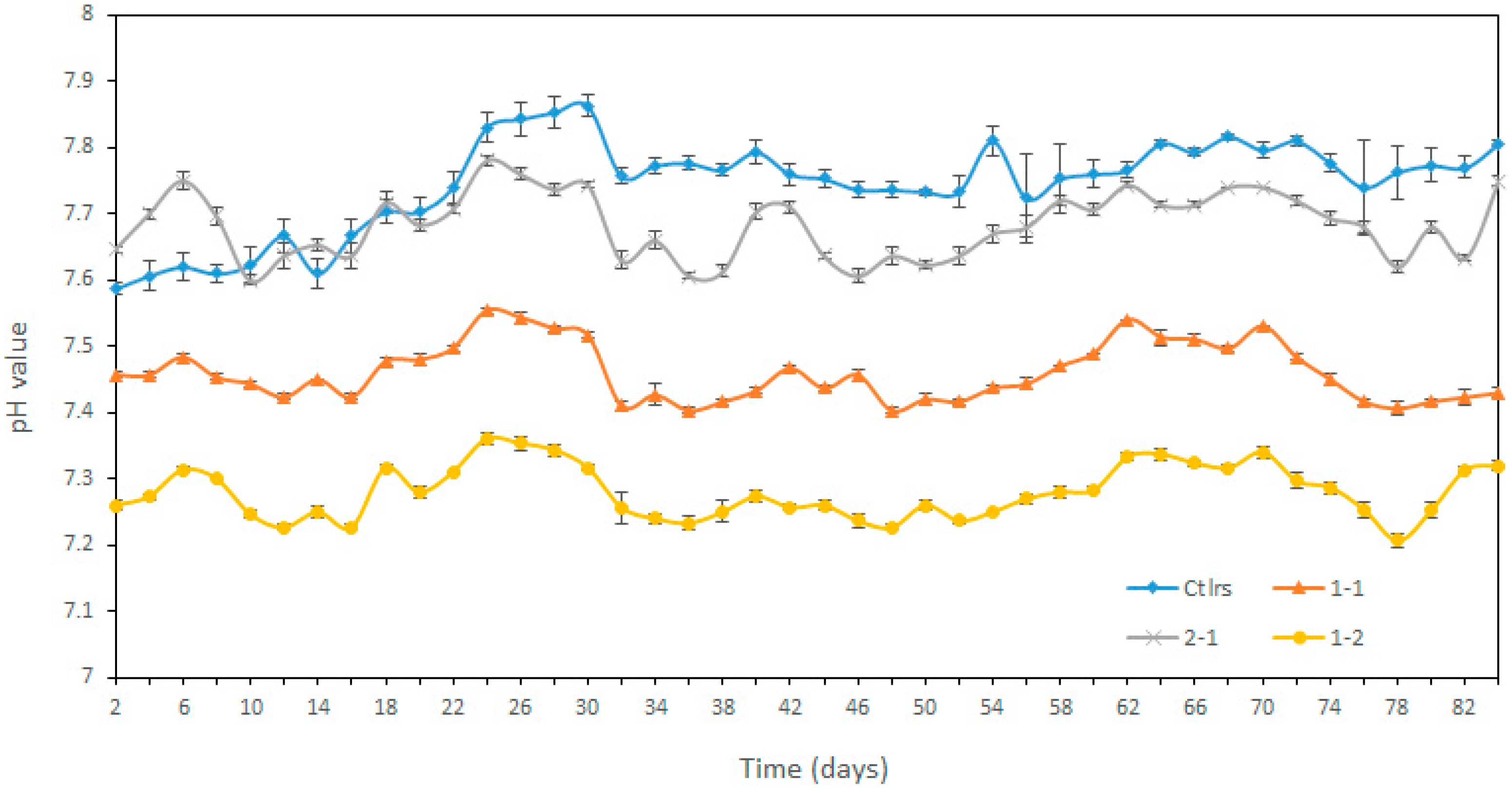
3.2.2. Variation of pH, Total Alkalinity and Ammonia
3.2.3. Variation of Carbon, Nitrogen, C/N Ratio, Cellulose and Lignin Degradation Rates
3.2.4. Estimation of Carbon Conversion Efficiency
3.3. Microbial Community Analysis
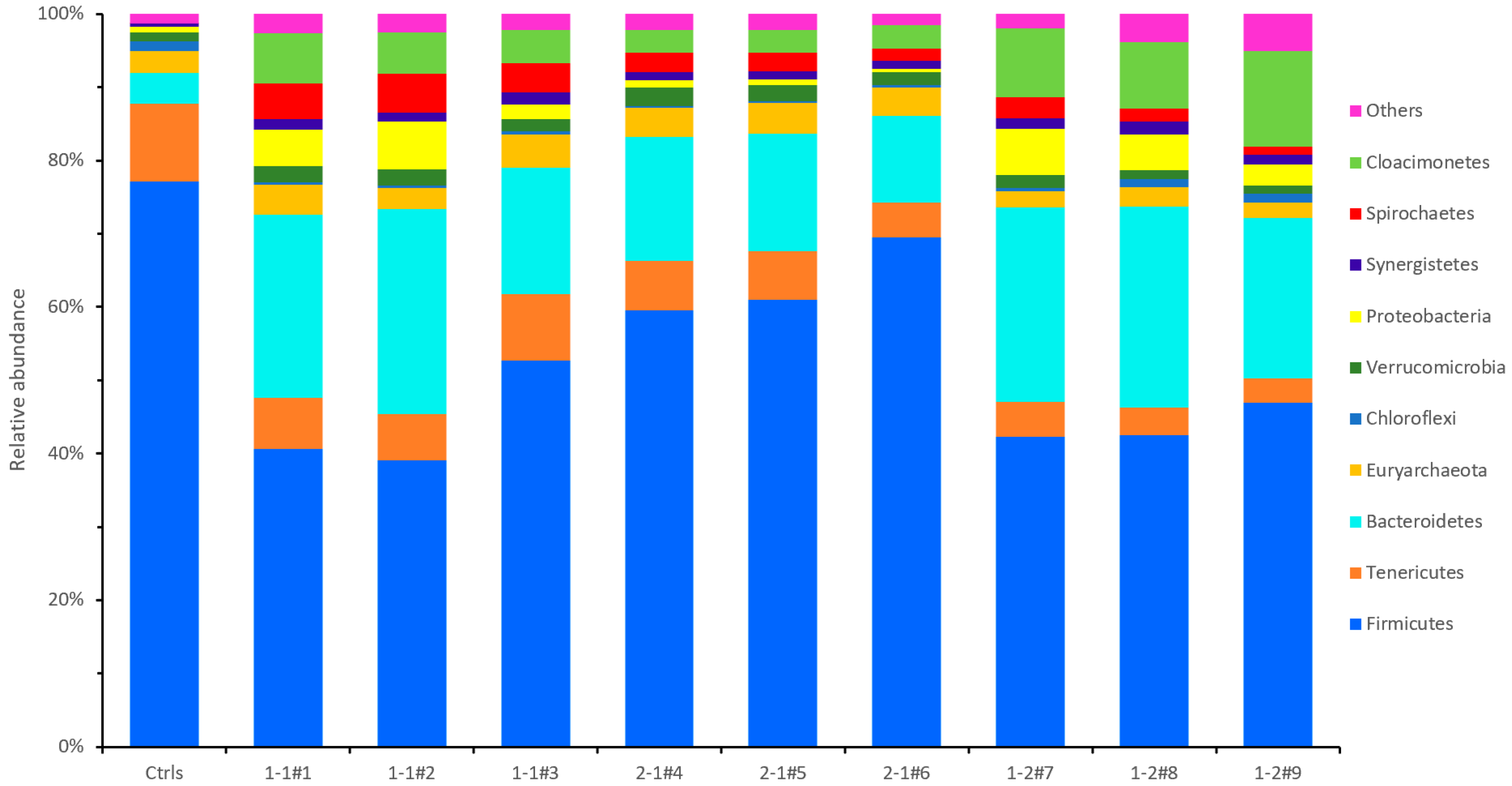
3.3.1. Microbial Community at Phylum Level
3.3.2. Microbial Community at Genus Level
4. Discussion
4.1. Biogas and Methane Production
4.2. Total Alkalinity and Ammonia Concentration
4.3. C/N Ratio, Cellulose and Lignin Degradation Rate
4.4. Microbial Community at Phylum Level
4.5. Microbial Community at Genus Level
4.6. Outlook of Full-Scale Anaerobic Co-Digestion SM with CS
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion processes: A review. Biores. Tech. 2008, 99, 4044–4064. [Google Scholar] [CrossRef]
- Riaño, B.; Garcíagonzález, M.C. On-farm treatment of swine manure based on solid-liquid separation and biological nitrification-denitrification of the liquid fraction. J. Environ. Manag. 2014, 132, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Lyberatos, G.; Skiadas, I.V. Modelling of anaerobic digestion-a review. Glob. Nest. Int. J. 1999, 1, 63–76. [Google Scholar]
- Wang, M.; Zhang, X.Y.; Zhou, J.; Yuan, Y.X.; Dai, Y.M.; Li, D.; Liu, X.F.; Yan, Z.Y. The dynamic changes and interactional networks of prokaryotic community between co-digestion and mono-digestions of corn stalk and pig manure. Biores. Tech. 2017, 225, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Mata-Alvarez, J.; Dosta, J.; Romero-Güiza, M.S.; Fonoll, X.; Peces, M.; Astals, S. A critical review on anaerobic co-digestion achievements between 2010 and 2013. Renew. Sustain. Energy Rev. 2014, 36, 412–427. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: http://faostat3.fao.org/ (accessed on 1 January 2014).
- Sekhon, R.S.; Breitzman, M.W.; Silva, R.R.; Santoro, N.; Rooney, W.L.; De-Leon, N.; Kaeppler, S.M. Stover composition in maize and sorghum reveals remarkable genetic variation and plasticity for carbohydrate accumulation. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Spyridon, M.; Keri, B.C.; Francisco, J.A.; Kipling, S.B.; Jeff, M.N.; James, R.F.; Douglas, L.K. Carbohydrate and nutrient composition of corn stover from three southeastern USA locations. Biomass Bioenergy 2016, 85, 153–158. [Google Scholar]
- Fujita, M.; Scharer, J.M.; Mooyoung, M. Effect of corn stover addition on the anaerobic digestion of swine manure. Agric. Wastes 1980, 2, 177–184. [Google Scholar] [CrossRef]
- Wu, X.; Yao, W.Y.; Zhu, J.; Miller, C. Biogas and CH4 productivity by co-digesting swine manure with three crop residues as an external carbon source. Biores. Tech. 2010, 101, 4042–4047. [Google Scholar] [CrossRef]
- Cuetos, M.J.; Fernández, C.; Gómez, X.; Morán, A. Anaerobic co-digestion of swine manure with energy crop residues. Biotech. Bioprocess Eng. 2011, 16, 1044–1052. [Google Scholar] [CrossRef]
- Zhang, T.; Mao, C.L.; Zhai, N.; Wang, X.; Yang, G. Influence of initial pH on thermophilic anaerobic co-digestion of swine manure and maize stalk. Waste Manag. 2015, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.M.; Yang, A.Y.; Bao, J.; Ma, R.X.; Yan, L.; Wang, Y.J.; Wang, W.D. Bioreactor performance and microbial community dynamics in a production-scale biogas plant in Northeast of China. Int. J. Agri. Biol. Eng. 2017, 10, 191–201. [Google Scholar]
- EPA. Total, fixed, and Volatile Solids in Water, Solids, and Biosolids; United States Environmental Protection Agency: Washington, DC, USA, 2001; Method 1684; p. 16.
- AOAC. AOAC Official Method 973.18, Fiber (Acid Detergent) and Lignin in Animal Feed, 16th ed.; Official Methods of Analysis of AOAC International; ASA-SSA Inc.: Arlington TX, USA, 1997; pp. 28–29. [Google Scholar]
- Nogueira, R.G.S.; Teng, T.L.; Wang, H.Q.; Rodrigues, P.H.M. Performance, microbial community analysis and fertilizer value of anaerobic co-digestion of cattle manure with waste kitchen oil. Appl. Eng. Agric. 2019, (in press). [CrossRef]
- Ericsson, A.C.; Davis, D.J.; Franklin, C.W.; Hagan, C.E. Exoelectrogenic capacity of host microbiota predicts lymphocyte recruitment to the gut. Physiol. Genom. 2015, 47, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Agler, M.T.; Wrenn, B.A.; Zinder, S.H.; Angenent, L.T. Waste to bioproduct conversion with undefined mixed cultures: The carboxylate platform. Trends Biotech. 2011, 29, 70–78. [Google Scholar] [CrossRef]
- Nkemka, V.N.; Murto, M. Evaluation of biogas production from seaweed in batch tests and in UASB reactors combined with the removal of heavy metals. J. Environ. Manag. 2010, 91, 1573–1579. [Google Scholar] [CrossRef]
- Koster, I.W.; Lettinga, G. Anaerobic digestion at extreme ammonia concentrations. Biol. Wastes 1988, 25, 51–59. [Google Scholar] [CrossRef]
- Albertson, O.E. Ammonia nitrogen and the anaerobic environment. J. Water Pollut. Control. Fed. 1961, 33, 978–995. [Google Scholar]
- Hailin, T.; Loannis, A.F.; Konstantinos, K.; Irini, A. Effect of different ammonia sources on aceticlastic and hydrogenotrophic methanogens. Biores. Tech. 2017, 250, 390–397. [Google Scholar]
- Melbinger, N.R.; Donnellon, J.; Zablatzky, H.R. Toxic effects of ammonia nitrogen in high-rate digestion [with Discussion]. J. Water Pollut. Control. Fed. 1971, 43, 1658–1670. [Google Scholar]
- Velsen, A.F.M.V. Adaptation of methanogenic sludge to high ammonia-nitrogen concentrations. Water Res. 1979, 13, 995–999. [Google Scholar] [CrossRef]
- Sievers, D.M.; Brune, D.E. Carbon/nitrogen ratio and anaerobic digestion of swine waste. Trans. ASAE 1978, 21, 537–541. [Google Scholar] [CrossRef]
- Bondesson, P.M.; Galbe, M.; Zacchi, G. Ethanol and biogas production after steam pretreatment of corn stover with or without the addition of sulphuric acid. Biotech. Biofuels 2013, 6, 11–22. [Google Scholar] [CrossRef]
- Cheng, J.; Ding, L.; Lin, R.; Yue, L.; Liu, J.Z.; Zou, J.H.; Cen, K. Fermentative biohydrogen and biomethane co-production from mixture of food waste and sewage sludge: Effects of physiochemical properties and mix ratios on fermentation performance. Appl. Energy 2016, 184, 1–8. [Google Scholar] [CrossRef]
- Kröber, M.; Bekel, T.; Diaz, N.N.; Goesmann, A.; Jaenicke, S.; Krause, L. Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J. Biotechnol. 2009, 142, 38–49. [Google Scholar] [CrossRef]
- Schlüter, A.; Bekel, T.; Diaz, N.N.; Dondrup, M.; Eichenlaub, R.; Gartemann, K.H.; Goesmann, A. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotech. 2008, 136, 77–90. [Google Scholar] [CrossRef]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. The division “Synergistes”. Anaerobe 2007, 13, 99–106. [Google Scholar] [CrossRef]
- Rivière, D. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef]
- Krause, L.; Diaz, N.N.; Edwards, R.A.; Gartemann, K.H.; Krömeke, H.; Neuweger, H. Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotech. 2008, 136, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. The revised road map to the manual. In Bergey’s Manual® of Systematic Bacteriology; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: Boston, MA, USA, 2005; pp. 159–187. [Google Scholar]
- Skennerton, C.T.; Haroon, M.F.; Briegel, A.; Shi, J.; Jensen, G.J.; Tyson, G.W.; Orphan, V.J. Phylogenomic analysis of Candidatus ‘Izimaplasma’ species: Free-living representatives from a Tenericutes clade found in methane seeps. ISME J. 2016, 10, 2679–2692. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kang, H.J.; Lee, Y.H.; Lee, T.J.; Han, K.; Choi, Y. Monitoring bacterial community structure and variability in time scale in full-scale anaerobic digesters. J. Environ. Monit. 2012, 14, 1893–1905. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, J.H.; Kang, H.J.; Lee, Y.H.; Lee, T.J.; Park, H.D. Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Biores. Tech. 2013, 145, 25–32. [Google Scholar] [CrossRef]
- Rogosa, M. Peptococcaceae, a new family to include the gram-positive, anaerobic cocci of the genera Peptococcus, Peptostreptococcus, and Ruminococcus. Int. J. Syst. Bacteriol. 1971, 21, 234–237. [Google Scholar] [CrossRef][Green Version]
- He, Q.; Li, L.; Zhao, X.F.; Li, Q.; Wu, D.; Peng, X.Y. Investigation of foaming causes in three mesophilic food waste digesters: Reactor performance and microbial analysis. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ravachol, J.; Borne, R.; Meynialsalles, I.; Soucaille, P.; Pagès, S.; Tardif, C. Combining free and aggregated cellulolytic systems in the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Biotech. Biofuels 2015, 8, 114–118. [Google Scholar] [CrossRef]
- Braun, F.; Hamelin, J.; Bonnafous, A.; Delgenès, N.; Steyer, J.P.; Patureau, D. Similar PAH fate in anaerobic digesters inoculated with three microbial communities accumulating either volatile fatty acids or methane. PLoS ONE 2015, 10, e0125552. [Google Scholar] [CrossRef]
- Bouanane-Darenfed, A.; Fardeau, M.L.; Grégoire, P.; Joseph, M.; Kebbouchegana, S.; Benayad, T. Caldicoprobacter algeriensis sp. nov. a new thermophilic anaerobic, xylanolytic bacterium isolated from an algerian hot spring. Curr. Microbiol. 2011, 62, 826–832. [Google Scholar] [CrossRef]
- Ziganshina, E.E.; Ibragimov, E.M.; Vankov, P.Y.; Miluykov, V.A.; Ziganshin, A.M. Comparison of anaerobic digestion strategies of nitrogen-rich substrates: Performance of anaerobic reactors and microbial community diversity. Waste Manag. 2016, 59, 160–171. [Google Scholar] [CrossRef]
- Nyonyo, T.; Shinkai, T.; Mitsumori, M. Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen. FEMS Microbiol. Ecol. 2014, 88, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Gregor, D.Z.; Natasa, U.Z.; Milenko, R. Full-scale anaerobic digestion of organic waste and municipal sludge. Biomass Bioenergy 2008, 32, 162–167. [Google Scholar]
- Alessandro, C.; Francesco, D.B.; Sonia, L. Dry anaerobic digestion of cow manure and agricultural products in a full-scale plant: Efficiency and comparison with wet fermentation. Waste Manag. 2018, 71, 704–710. [Google Scholar]
- Cavinato, C.; Fatone, F.; Bolzonella, D.; Pavan, P. Thermophilic anaerobic co-digestion of cattle manure with agro-wastes and energy crops: Comparison of pilot and full scale experiences. Biores. Tech. 2010, 101, 545–550. [Google Scholar] [CrossRef]

| pH | Moisture (%) | TS (%) c | VS (%) d | TC (%) e | TN (%) f | Cellulose (w/w%) | lignin (w/w%) | |
|---|---|---|---|---|---|---|---|---|
| SM a | 5.33 ± 0.23 | 69.87 ± 3.27 | 25.89 ± 0.09 | 82.0 2 ± 0.18 | 13.30 ± 1.40 | 1.25 ± 0.11 | 8.10 | 3.31 |
| CS b | - | 8.80 ± 0.10 | 90.95 ± 0.05 | 95.13 ± 0.25 | 41.95 ± 0.25 | 0.49 ± 0.03 | 46.10 | 10.97 |
| Units | C 1 a | C 2 | C 3 | 2-1-1#1 | 2-2-1#2 | 2-1-2#3 | 3-1-1#4 | 3-2-1#5 | 3-1-2#6 | 4-1-1#7 | 4-2-1#8 | 4-1-2#9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VS digestate | g L−1 | 14.5 | 28.6 | 39.9 | 22.3 | 20.1 | 20.5 | 26.3 | 27.3 | 31.2 | 35.9 | 39.5 | 42.4 |
| VS reduction | % | 65.5 | 54.6 | 52.5 | 46.9 | 52.1 | 51.2 | 58.3 | 56.7 | 50.5 | 57.3 | 53.0 | 49.5 |
| Biogas | mL d−1 | 1856 | 2658 | 2804 | 1559 | 1640 | 1223 | 2258 | 2400 | 1983 | 2878 | 3112 | 2612 |
| Biogas | mL g−1 VS des b | 97.8 | 112.0 | 92.2 | 114.7 | 108.5 | 82.4 | 89.2 | 97.4 | 90.4 | 86.7 | 101.3 | 91.0 |
| CH4 content | % | 75.6 | 73.1 | 70.5 | 73.6 | 73.6 | 74.3 | 70.3 | 70.3 | 69.3 | 67.9 | 69.1 | 67.3 |
| CH4 | mL d−1 | 1405 | 1942 | 1971 | 1148 | 1209 | 909 | 1576 | 1689 | 1368 | 1952 | 2147 | 1749 |
| CH4 | mL g−1 VS des | 74.0 | 81.8 | 64.8 | 84.4 | 80.0 | 61.3 | 62.2 | 68.6 | 62.3 | 58.8 | 69.9 | 61.0 |
| pH | - | 7.61 | 7.87 | 7.86 | 7.27 | 7.38 | 7.08 | 7.38 | 7.57 | 7.17 | 7.52 | 7.72 | 7.27 |
| TA c | mg L−1 | 10,500 | 16,000 | 18,000 | 6500 | 7000 | 5000 | 7500 | 9000 | 6500 | 9000 | 11,500 | 7500 |
| Ammonia | mg L−1 | 1600 | 2400 | 4000 | 800 | 800 | 800 | 800 | 800 | 800 | 800 | 1600 | 800 |
| a control groups. b VS-destroyed. c total alkalinity. | |||||||||||||
| Units | Ctrls a | 1-1 | 2-1 | 1-2 | |
|---|---|---|---|---|---|
| VS digestate | g/L | 28.47 ± 0.82 | 26.23 ± 0.12 | 26.80 ± 0.36 | 31.43 ± 0.47 |
| VS reduction | % | 54.81 ± 1.30 | 58.36 ± 0.20 | 57.46 ± 0.56 | 50.11 ± 0.75 |
| Biogas | mL d−1 | 2415.2 ± 37.42 | 2284.4 ± 23.62 | 2414.8 ± 83.38 | 2166.0 ± 181.62 |
| Biogas | mL g−1 VS des | 101.41 ± 2.71 | 90.04 ± 0.70 | 96.72 ± 4.32 | 99.51 ± 8.85 |
| CH4 content | % | 76.56 ± 1.35 | 74.41 ± 1.40 | 75.02 ± 1.17 | 71.25 ± 1.25 |
| CH4 | mL d−1 | 1849.0 ± 31.08 | 1699.8 ± 11.21 | 1811.6 ± 66.99 | 1542.3 ± 118.53 |
| CH4 | mL g−1 VS des | 77.63 ± 1.85 | 67.00 ± 0.33 | 72.56 ± 3.40 | 70.84 ± 5.73 |
| pH | - | 7.75 ± 0.07 | 7.46 ± 0.04 | 7.68 ± 0.05 | 7.28 ± 0.04 |
| TAb | mg L−1 | 9500 ± 354 | 7333 ± 118 | 7667 ± 236 | 6083 ± 118 |
| Ammonia | mg L−1 | 4700 ± 141 | 1467 ± 340 | 2667 ± 499 | 867 ± 94 |
| Raw materials | Cellulose (w/w%) | Lignin (w/w%) | |||||
|---|---|---|---|---|---|---|---|
| TC (%) | TN (%) | C/N | Substrate | Digestate | Substrate | Digestate | |
| Ctrls | 4.09 | 0.38 | 10.64 | 34.26 | - | 13.11 | - |
| 1-1 | 3.59 | 0.21 | 17.10 | 40.39 | 25.01 | 12.54 | 12.22 |
| 2-1 | 3.76 | 0.27 | 13.93 | 38.35 | 19.78 | 13.03 | 8.73 |
| 1-2 | 3.41 | 0.15 | 22.73 | 42.36 | 27.86 | 12.73 | 12.37 |
| Raw materials | TC of SM (g) | TC of CS (g) | TC of Digestate (%) | TC of Digestate (g) | TC of CH4 (g) | ECCE (%) b | |||
|---|---|---|---|---|---|---|---|---|---|
| SM (g) | CS (g) | Distilled water (g) | |||||||
| Ctrlsc | 19.42 | - | 46.08 | 2.58 | - | 0.63 | 8.25 | 1.02 | 36.78 |
| 1-1 | 9.70 | 2.40 | 53.40 | 1.29 | 1.01 | 0.49 | 6.42 | 0.91 | 39.57 |
| 2-1 | 12.95 | 1.60 | 50.95 | 1.72 | 0.67 | 1.29 | 16.89 | 0.97 | 40.59 |
| 1-2 | 6.48 | 3.20 | 55.82 | 0.86 | 1.34 | 0.54 | 7.07 | 0.83 | 37.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Lim, T.T.; Duong, C.; Zhang, W.; Xu, C.; Yan, L.; Mei, Z.; Wang, W. Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis. Microorganisms 2020, 8, 188. https://doi.org/10.3390/microorganisms8020188
Wang H, Lim TT, Duong C, Zhang W, Xu C, Yan L, Mei Z, Wang W. Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis. Microorganisms. 2020; 8(2):188. https://doi.org/10.3390/microorganisms8020188
Chicago/Turabian StyleWang, Haipeng, Teng Teeh Lim, Cuong Duong, Wei Zhang, Congfeng Xu, Lei Yan, Zili Mei, and Weidong Wang. 2020. "Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis" Microorganisms 8, no. 2: 188. https://doi.org/10.3390/microorganisms8020188
APA StyleWang, H., Lim, T. T., Duong, C., Zhang, W., Xu, C., Yan, L., Mei, Z., & Wang, W. (2020). Long-Term Mesophilic Anaerobic Co-Digestion of Swine Manure with Corn Stover and Microbial Community Analysis. Microorganisms, 8(2), 188. https://doi.org/10.3390/microorganisms8020188





