SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs
Abstract
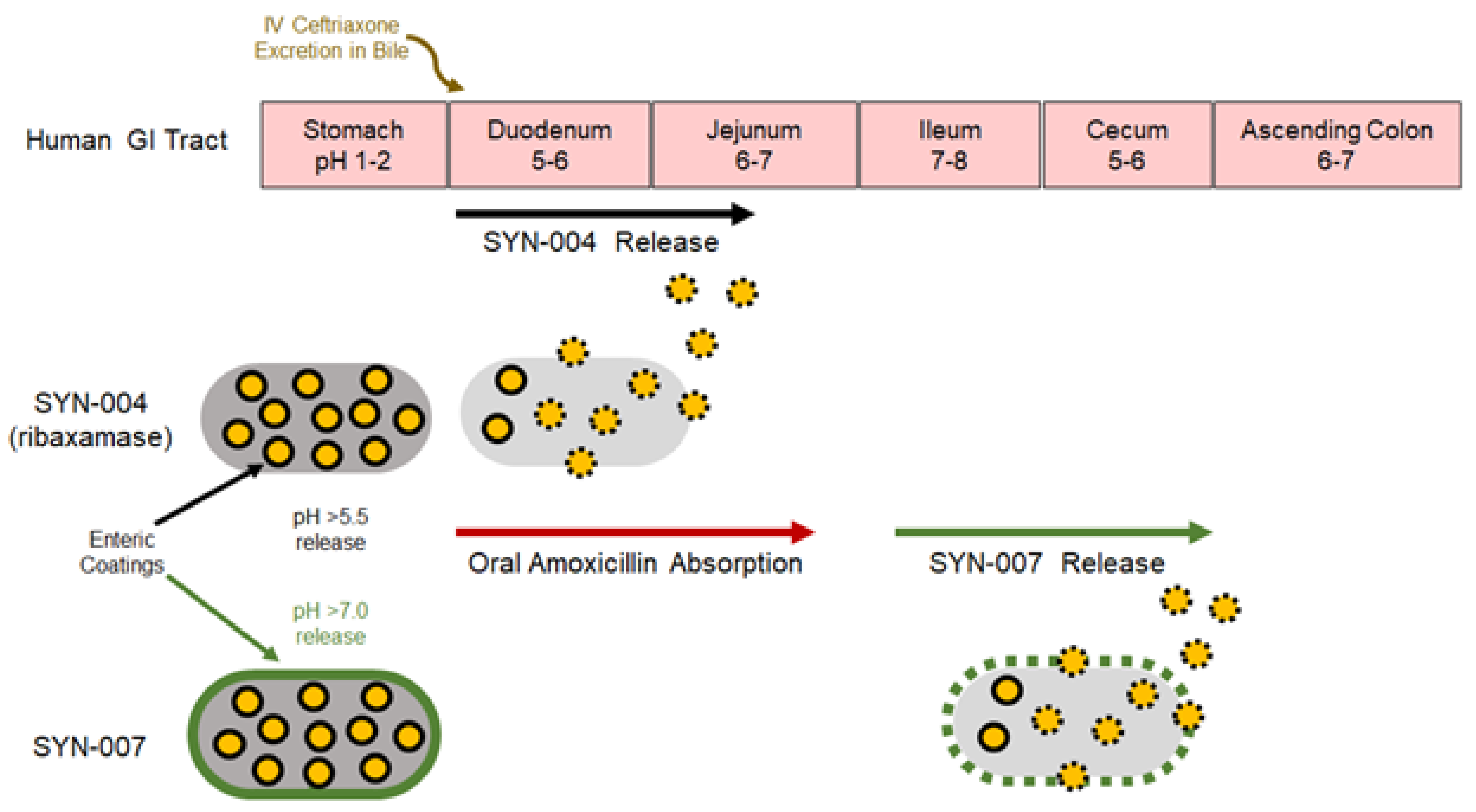
1. Introduction
2. Materials and Methods
2.1. Test Article
2.2. Animals and Test Article Administration
2.3. Amoxicillin Serum Measurement
2.4. Fecal DNA Extraction, Whole Genome Shotgun Sequencing, and Metagenomic Analyses
2.5. Data Availability
3. Results
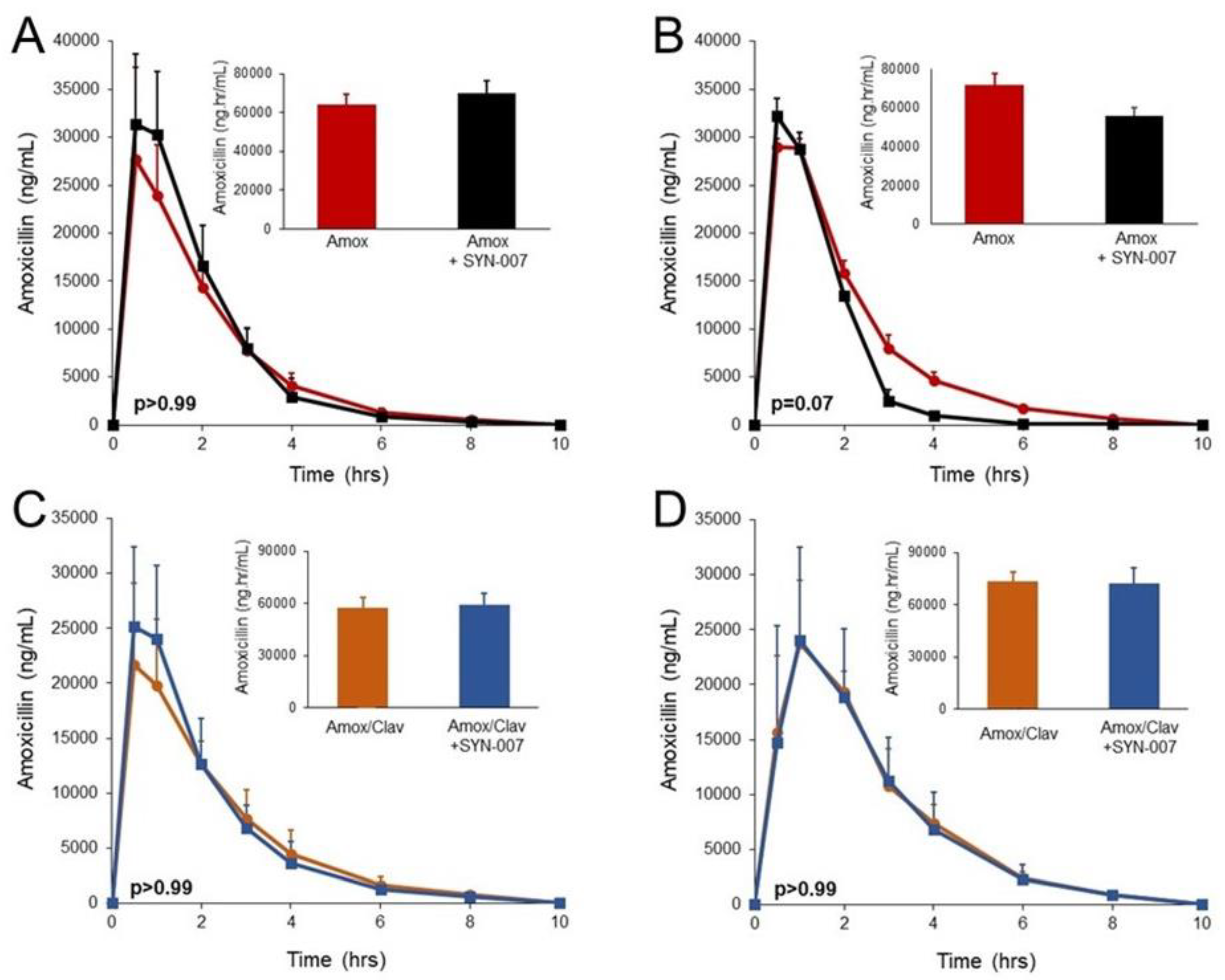
3.1. SYN-007 did not Significantly Affect Oral Amoxicillin Systemic Absorption
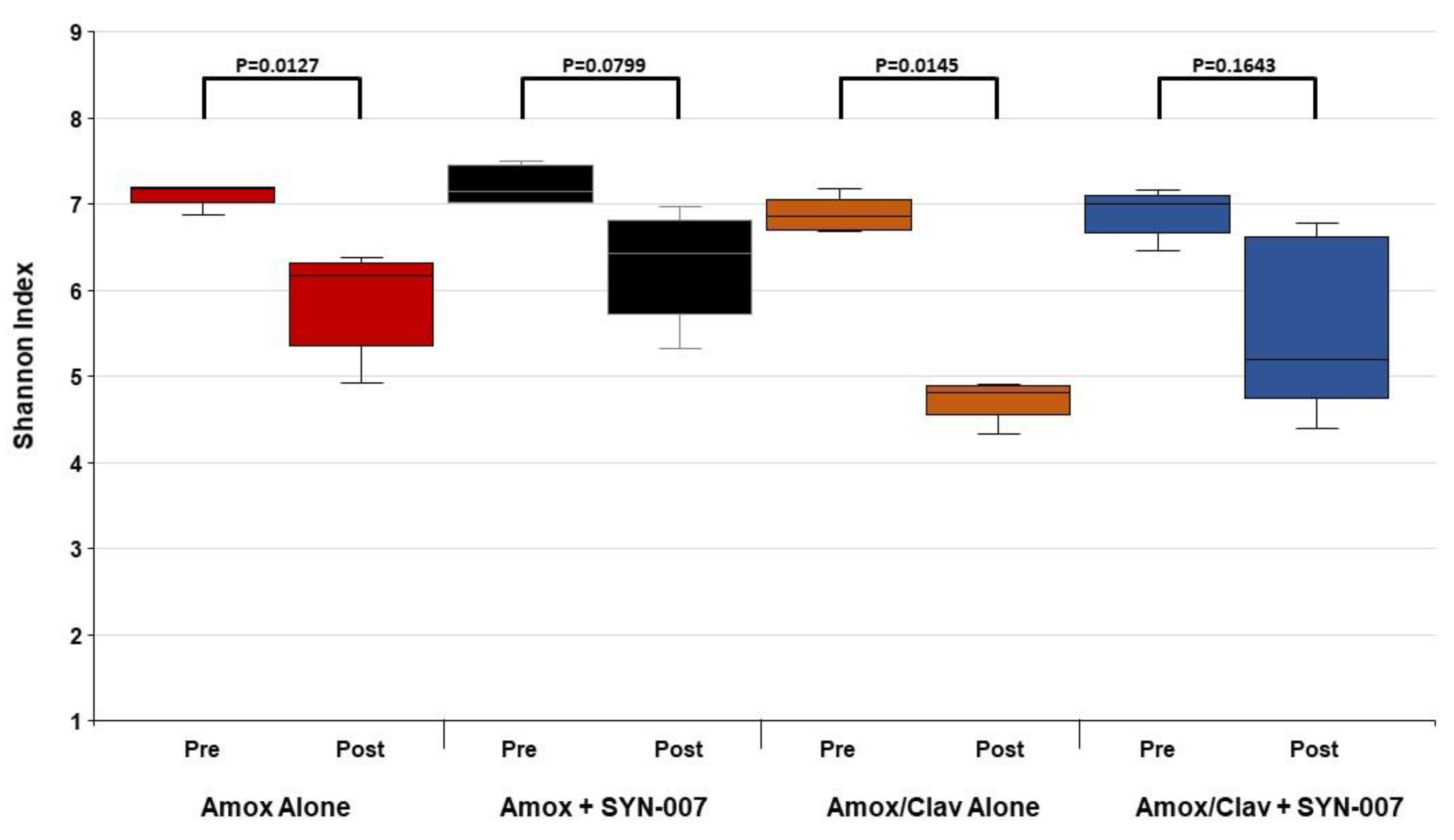
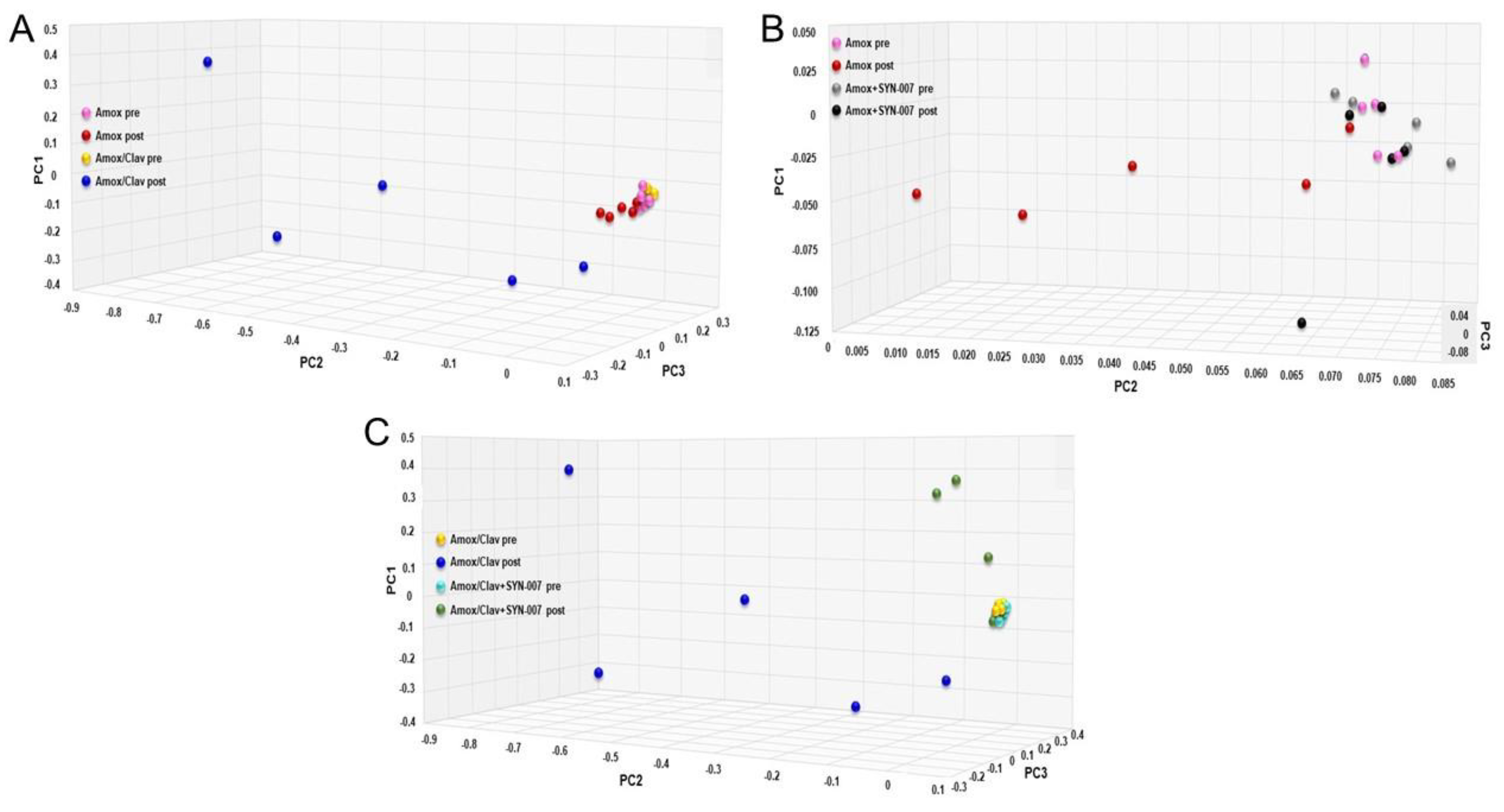
3.2. SYN-007 Reduced Oral Amoxicillin-Mediated Microbiome Damage
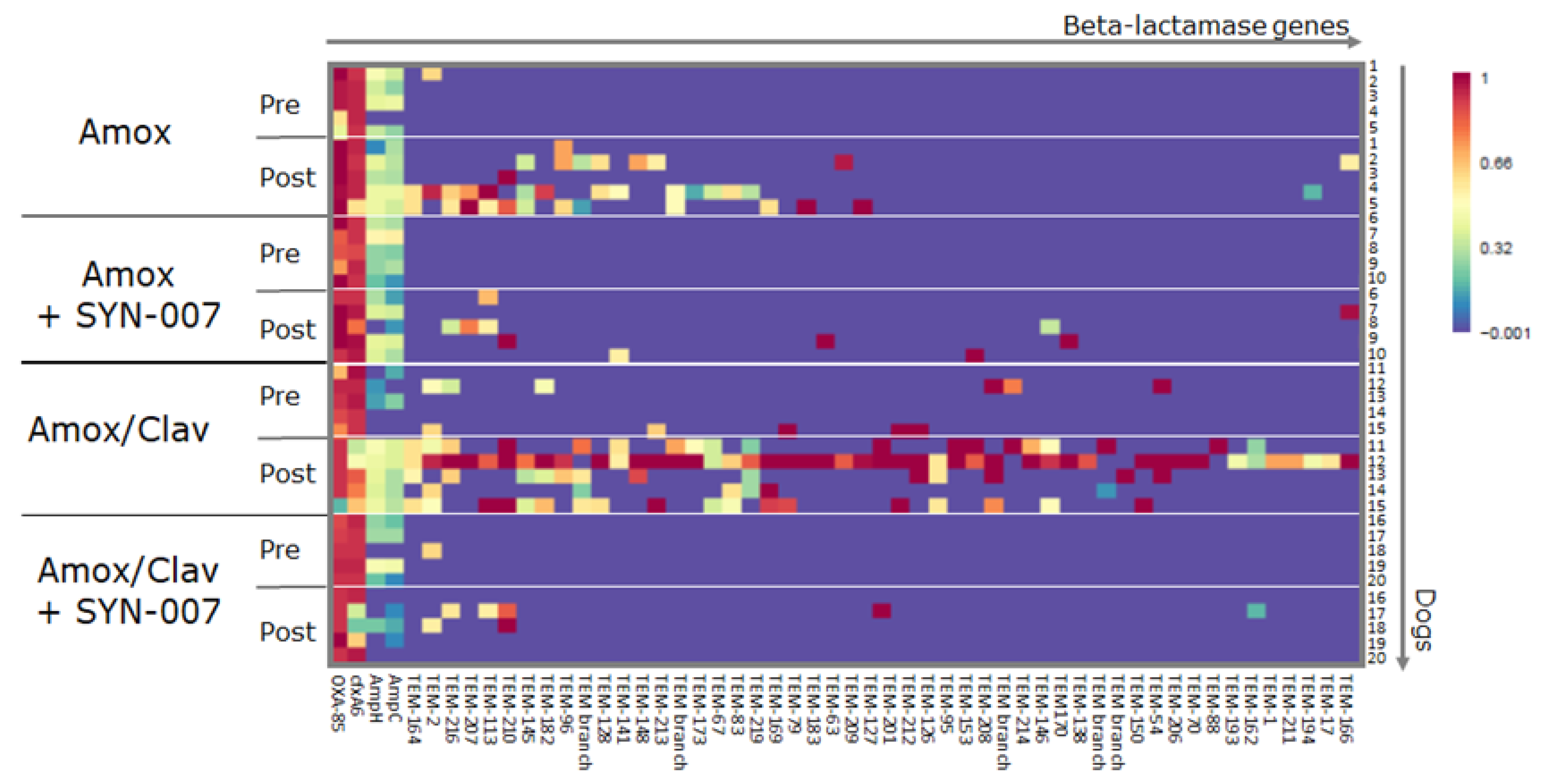
3.3. SYN-007 Attenuated Antibiotic Resistance Gene Propagation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knecht, H.; Neulinger, S.C.; Heinsen, F.A.; Knecht, C.; Schilhabel, A.; Schmitz, R.A.; Zimmermann, A.; dos Santos, V.M.; Ferrer, M.; Rosenstiel, P.C.; et al. Effects of beta-lactam antibiotics and fluoroquinolones on human gut microbiota in relation to Clostridium difficile associated diarrhea. PLoS ONE 2014, 9, e89417. [Google Scholar] [CrossRef] [PubMed]
- Crandon, J.L.; Nicolau, D.P. Pharmacodynamic Approaches to Optimizing Beta-Lactam Therapy. Crit. Care Clin. 2011, 27, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.; Zahar, J.R.; Lesprit, P.; Ruppe, E.; Leone, M.; Chastre, J.; Lucet, J.C.; Paugam-Burtz, C.; Brun-Buisson, C.; Timsit, J.F.; et al. Elaboration of a consensual definition of de-escalation allowing a ranking of beta-lactams. Clin. Microbiol. Infect. 2015, 21, 649. [Google Scholar] [PubMed]
- Vardakas, K.Z.; Trigkidis, K.K.; Boukouvala, E.; Falagas, M.E. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2016, 48, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Watson, T.; Hickok, J.; Fraker, S.; Korwek, K.; Poland, R.E.; Septimus, E. Evaluating the Risk Factors for Hospital-Onset Clostridium difficile Infections in a Large Healthcare System. Clin. Infect. Dis. 2018, 66, 1957–1959. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Stobberingh, E.E.; Savelkoul, P.H.M.; Wolffs, P.F.G. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013, 4, 87. [Google Scholar] [CrossRef]
- Stecher, B.; Maier, L.; Hardt, W.D. ’Blooming’ in the gut: How dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013, 11, 277–284. [Google Scholar] [CrossRef]
- Bristol, A.; Hubert, S.; Hofmann, F.; Baer, H. Formulation development of SYN-004 (ribaxamase) oral solid dosage form, a beta-lactamase to prevent intravenous antibiotic-associated dysbiosis of the colon. Int. J. Pharm. 2017, 534, 25–34. [Google Scholar] [CrossRef]
- Connelly, S.; Hubert, S.; Subramanian, P.; Kaleko, M.; Bristol, J.; Hasan, N.A.; Colwell, R.R. SYN-004 (ribaxamase), an Oral Beta-Lactamase, Mitigates Antibiotic-Mediated Dysbiosis in a Porcine Gut Microbiome Model. J. Appl. Microbiol. 2017, 123, 66–79. [Google Scholar] [CrossRef]
- Kokai-Kun, J.; Connelly, S. Ribaxamase, an orally administered β-lactamase, protects the gut microbiome in patients treated with ceftriaxone. J. Transl. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Kokai-Kun, J.F.; Roberts, T.; Coughlin, O.; Sicard, E.; Rufiange, M.; Fedorak, R.; Carter, C.; Adams, M.H.; Longstreth, J.; Whalen, H.; et al. The oral beta-lactamase SYN-004 (Ribaxamase) Degrades Ceftriaxone Excreted into the Intestine in Phase 2a Clinical Studies. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Kokai-Kun, J.F.; Roberts, T.; Coughlin, O.; Le, C.; Whalen, H.; Stevenson, R.; Wacher, V.J.; Sliman, J. Use of ribaxamase (SYN-004), a β-lactamase, to prevent Clostridium difficile infection in β-lactam-treated patients: A double-blind, phase 2b, randomised placebo-controlled trial. Lancet Infect. Dis. 2019, 19, 487–496. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Outpatient Antibiotic Prescriptions United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. [Google Scholar]
- Connelly, S.; Fanelli, B.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Low dose oral beta-lactamase protects the gut microbiome from oral beta-lactam-mediated damage in dogs. AIMS Public Health 2019, 6, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Connelly, S.; Fanelli, B.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage. Microorganisms 2019, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Barr, W.H.; Zola, E.M.; Candler, E.L.; Hwang, S.M.; Tendolkar, A.V.; Shamburek, R.; Parker, B.; Hilty, M.D. Differential absorption of amoxicillin from the human small and large intestine. Clin. Pharmacol. Ther. 1994, 56, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Hubert, S.; Chadwick, A.; Wacher, V.; Coughlin, O.; Kokai-Kun, J.; Bristol, A. Development of a Modified-Release Formulation of Lovastatin Targeted to Intestinal Methanogens Implicated in Irritable Bowel Syndrome With Constipation. J. Pharm. Sci. 2018, 107, 662–671. [Google Scholar] [CrossRef]
- Kuehn, J.; Ismael, Z.; Long, P.F.; Barker, C.I.; Sharland, M. Reported Rates of Diarrhea Following Oral Penicillin Therapy in Pediatric Clinical Trials. J. Pediatr. Pharmacol. Ther. 2015, 20, 90–104. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010, 11, 367. [Google Scholar] [CrossRef]
- Schroeder, P.J.; Jenkins, D.G. How robust are popular beta diversity indices to sampling error? Ecosphere 2018, 9, e02100. [Google Scholar] [CrossRef]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Kaleko, M.; Bristol, J.A.; Hubert, S.; Parsley, T.; Widmer, G.; Tzipori, S.; Subramanian, P.; Hasan, N.; Koski, P.; Kokai-Kun, J.; et al. Development of SYN-004, an oral beta-lactamase treatment to protect the gut microbiome from antibiotic-mediated damage and prevent Clostridium difficile infection. Anaerobe 2016, 41, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D. IPSAT P1A, a class A beta-lactamase therapy for the prevention of penicillin-induced disruption to the intestinal microflora. Curr. Opin. Investig. Drugs 2009, 10, 838–844. [Google Scholar] [PubMed]
- Hoberman, A.; Paradise, J.L.; Rockette, H.E.; Jeong, J.H.; Kearney, D.H.; Bhatnagar, S.; Shope, T.R.; Muñiz, G.; Martin, J.M.; Kurs-Lasky, M.; et al. Reduced-Concentration Clavulanate for Young Children with Acute Otitis Media. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Bulow, C.; Langdon, A.; Hink, T.; Wallace, M.; Reske, K.A.; Patel, S.; Sun, X.; Seiler, S.; Jones, S.; Kwon, J.H.; et al. Impact of Amoxicillin-Clavulanate followed by Autologous Fecal Microbiota Transplantation on Fecal Microbiome Structure and Metabolic Potential. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, T.A.; Pallav, K.; Dowd, S.E.; Villafuerte-Galvez, J.; Vanga, R.R.; Castillo, N.E.; Hansen, J.; Dennis, M.; Leffler, D.A.; Kelly, C.P. Prospective randomized controlled study on the effects of Saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes 2017, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Portrait, V.; Cottenceau, G.; Pons, A. A Fusobacterium mortiferum strain produces a bacteriocin-like substance(s) inhibiting Salmonella enteritidis. Lett. Appl. Microbiol. 2000, 31, 115–117. [Google Scholar] [CrossRef]
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef]
- Grønvold, A.M.R.; L’Abée-Lund, T.M.; Sørum, H.; Skancke, E.; Yannarell, A.C.; Mackie, R.I. Changes in fecal microbiota of healthy dogs administered amoxicillin. FEMS Microbiol. Ecol. 2010, 71, 313–326. [Google Scholar] [CrossRef]
- Schmidt, V.M.; Pinchbeck, G.; McIntyre, K.M.; Nuttall, T.; McEwan, N.; Dawson, S.; Williams, N.J. Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J. Antimicrob. Chemother. 2018, 73, 3305–3316. [Google Scholar] [CrossRef]
- Connelly, S.; Subramanian, P.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Distinct consequences of amoxicillin and ertapenem exposure in the porcine gut microbiome. Anaerobe 2018, 53, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Connelly, S.; Fanelli, B.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs. Microorganisms 2020, 8, 152. https://doi.org/10.3390/microorganisms8020152
Connelly S, Fanelli B, Hasan NA, Colwell RR, Kaleko M. SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs. Microorganisms. 2020; 8(2):152. https://doi.org/10.3390/microorganisms8020152
Chicago/Turabian StyleConnelly, Sheila, Brian Fanelli, Nur A. Hasan, Rita R. Colwell, and Michael Kaleko. 2020. "SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs" Microorganisms 8, no. 2: 152. https://doi.org/10.3390/microorganisms8020152
APA StyleConnelly, S., Fanelli, B., Hasan, N. A., Colwell, R. R., & Kaleko, M. (2020). SYN-007, an Orally Administered Beta-Lactamase Enzyme, Protects the Gut Microbiome from Oral Amoxicillin/Clavulanate without Adversely Affecting Antibiotic Systemic Absorption in Dogs. Microorganisms, 8(2), 152. https://doi.org/10.3390/microorganisms8020152




