Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Sites and Samples
2.2. Physicochemical Analysis
2.3. Photopigment Analysis
2.4. Quinone Profiling
2.5. Phase-Contrast and Epifluorescence Microscopy
2.6. Enumeration of Viable Phototrophs
2.7. Isolation and Phylogenetic Identification of PNSB
2.8. DNA Extraction from Bloom Samples
2.9. Real-Time Quantitative PCR (RT-qPCR)
2.10. pufM Gene Clone Library Analysis
2.11. Genome Analysis
2.12. Phylogenomic Analysis
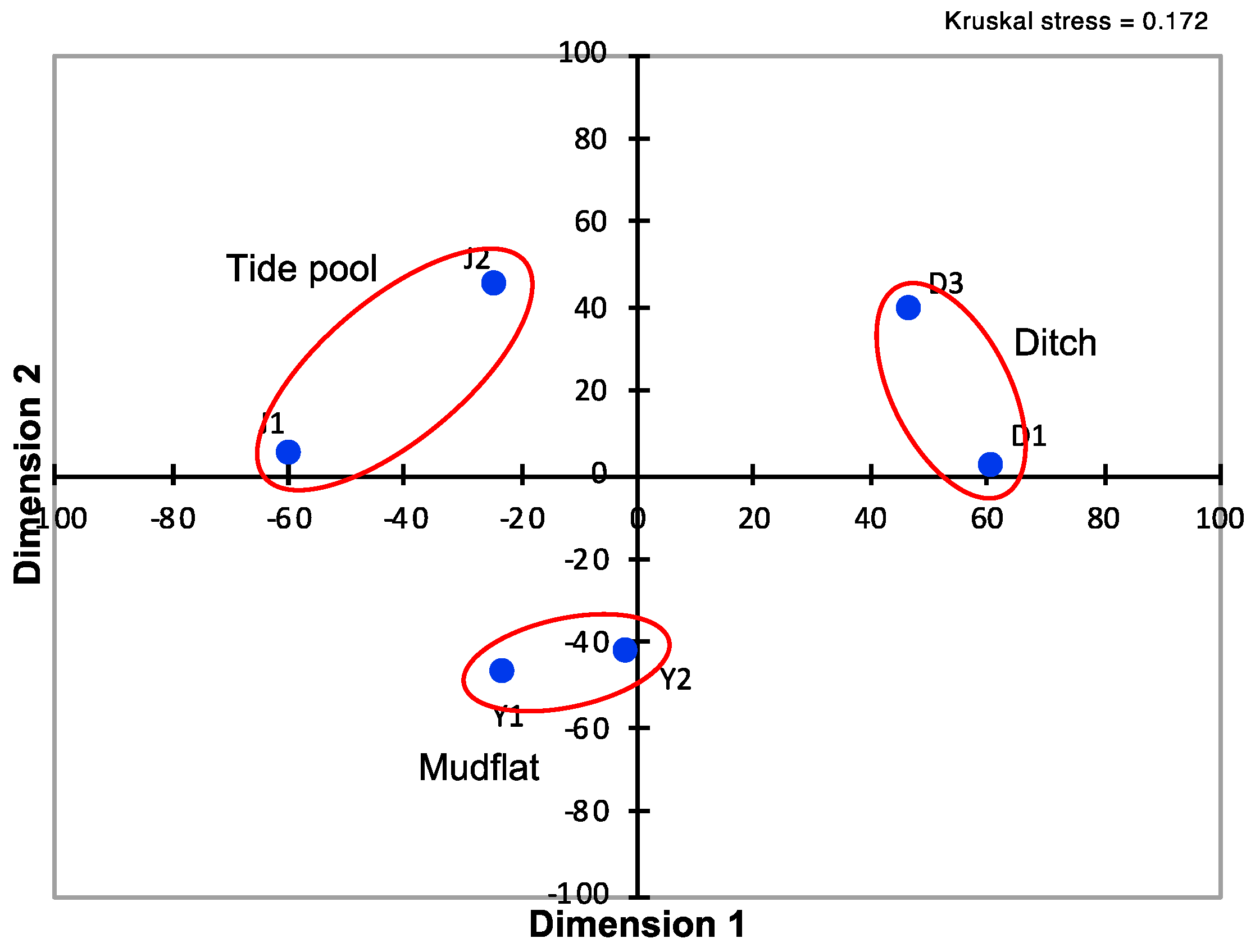
2.13. Statistical and Numerical Analysis
2.14. Accession Numbers
3. Results
3.1. Appearance and General Characteristics of Colored Blooms
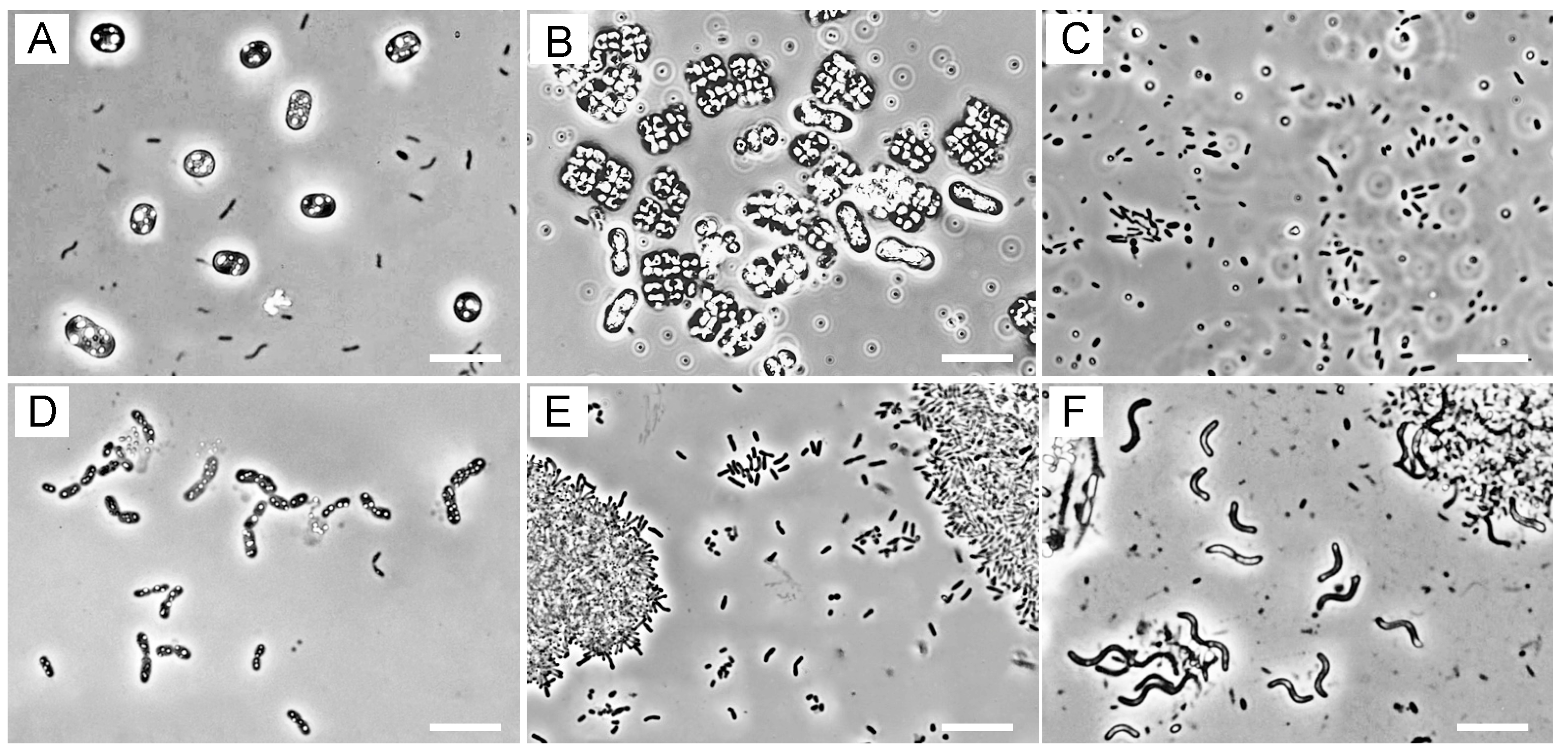
3.2. Microscopic Observations
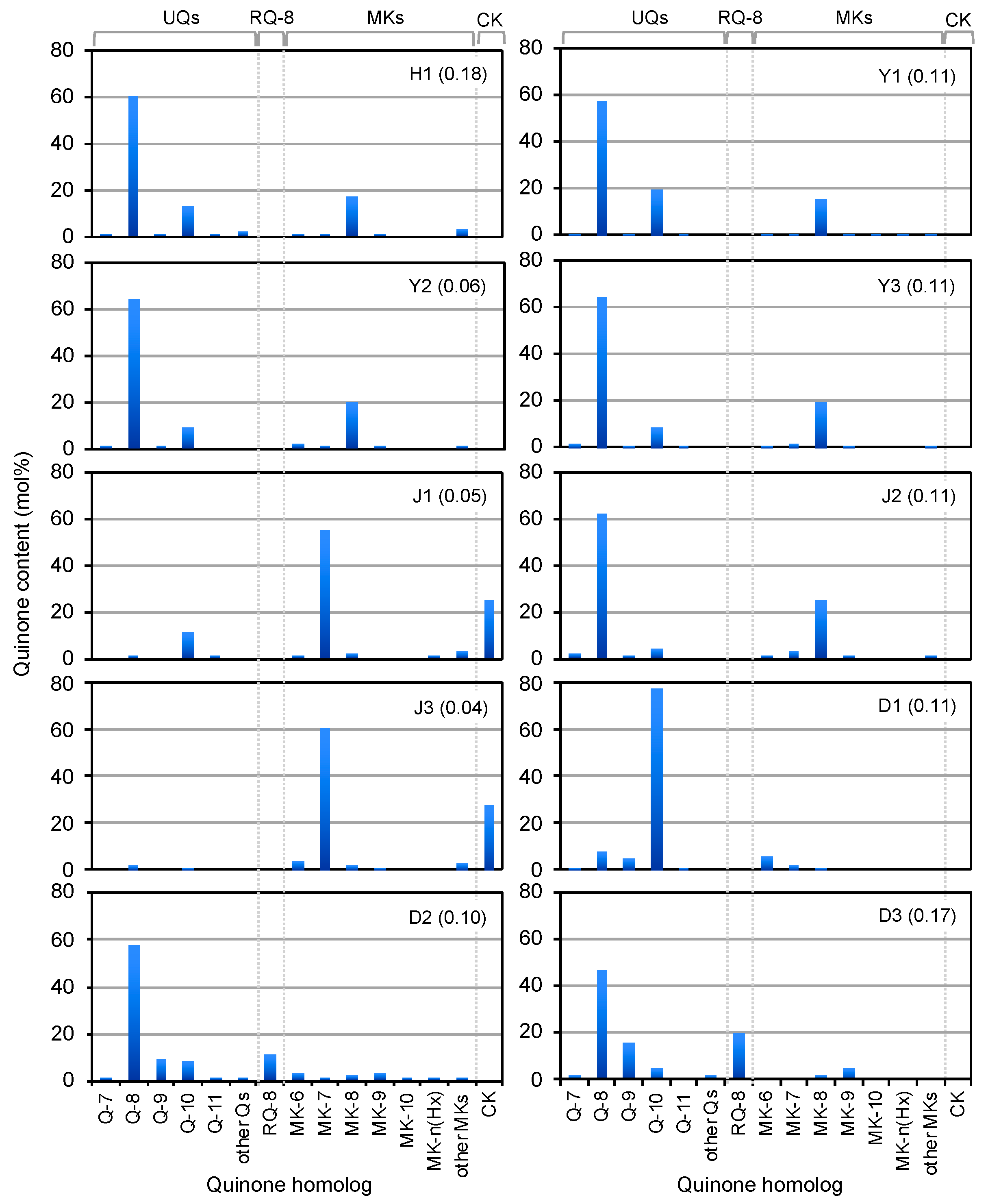
3.3. Photopigment and Quinone Profiles
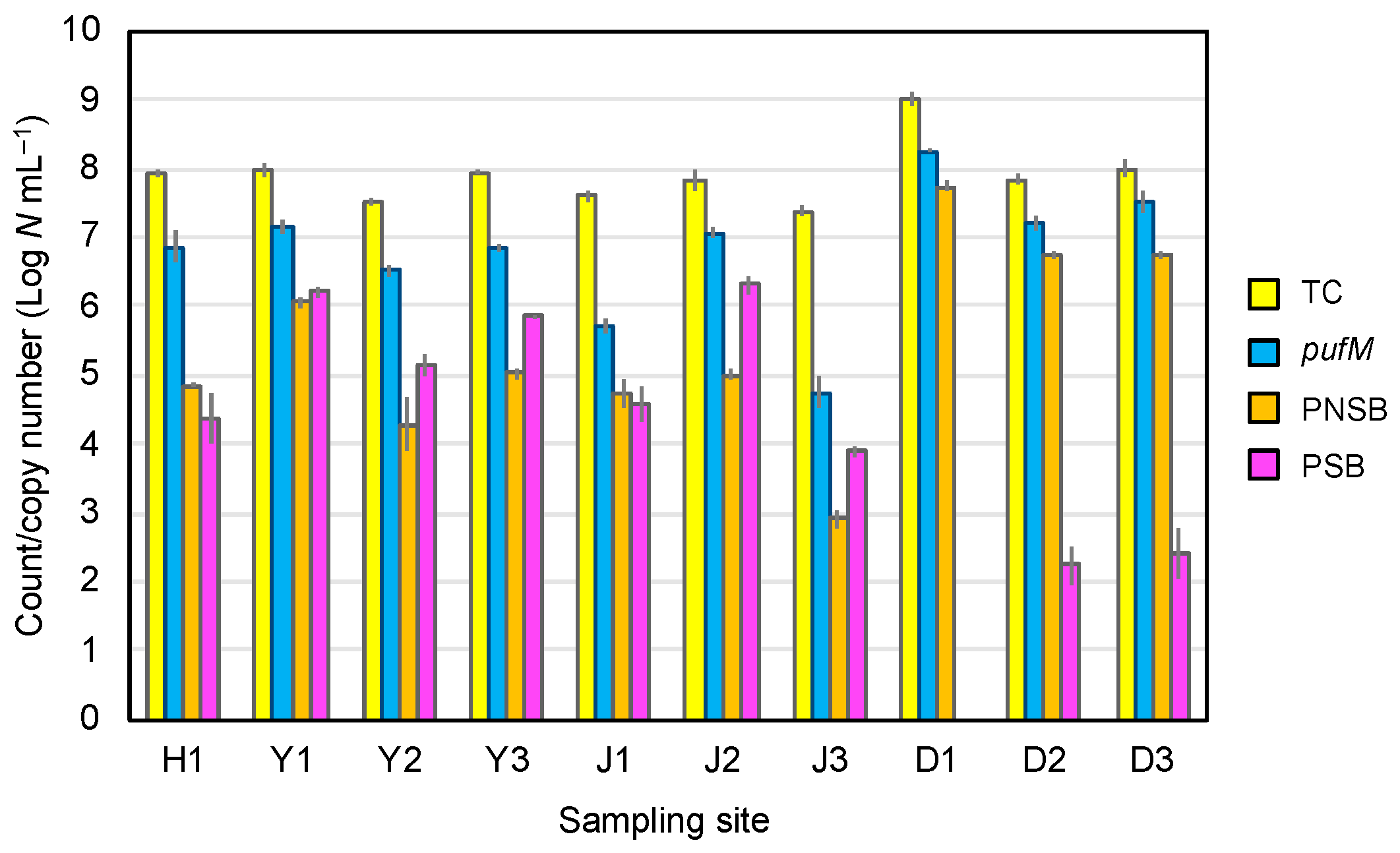
3.4. PNSB and PSB Populations
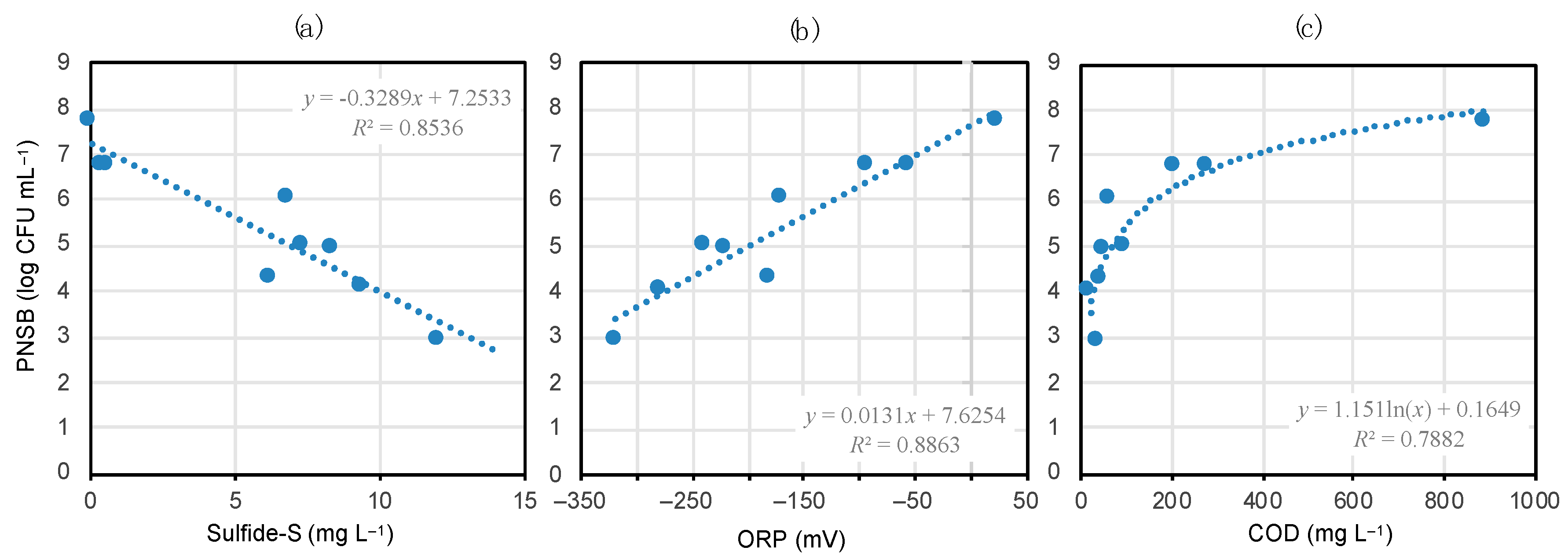
3.5. Environmental Factors Affecting PNSB
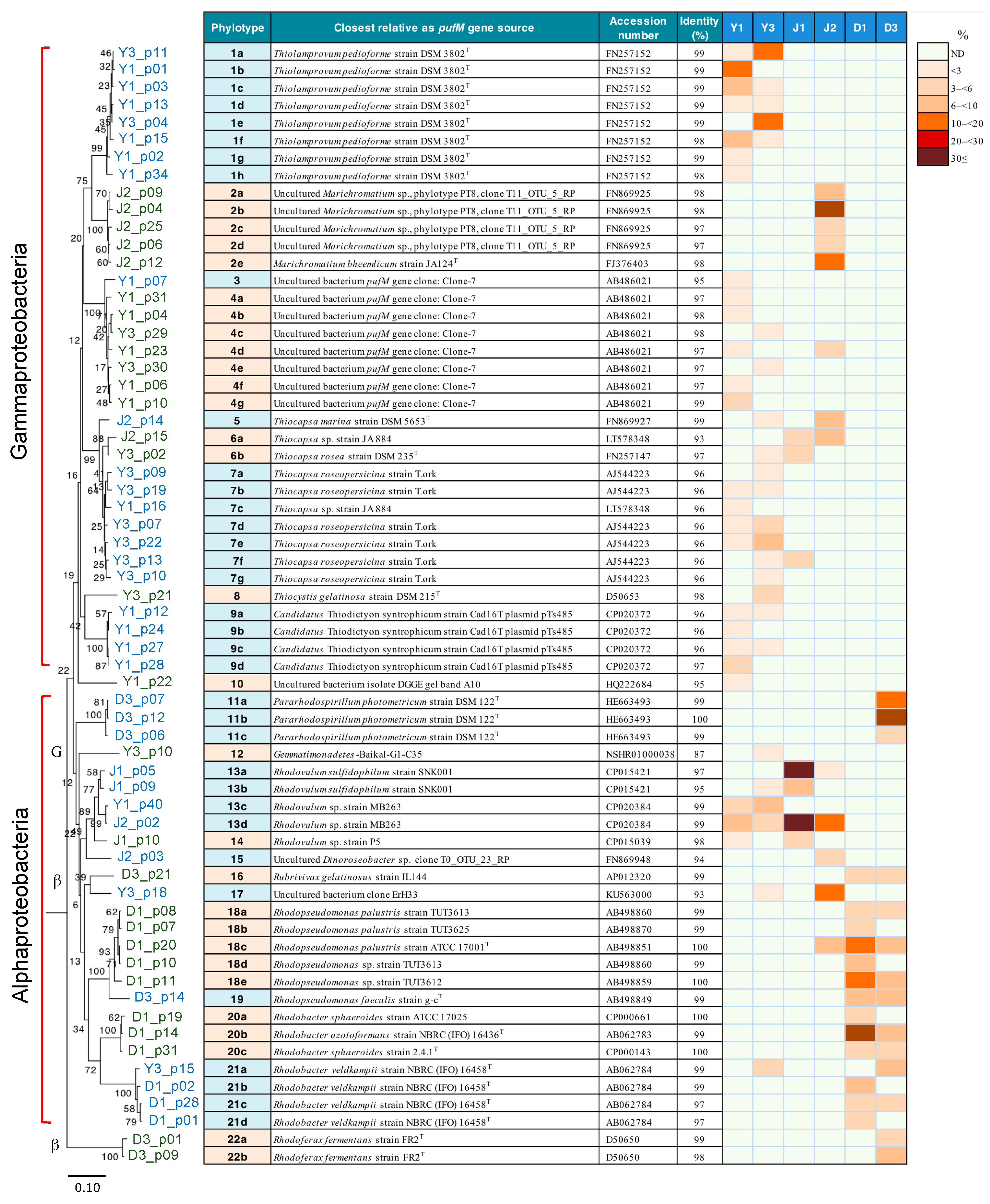
3.6. pufM Gene Clone Phylotyping
3.7. Phylogenetic Identification of Isolates
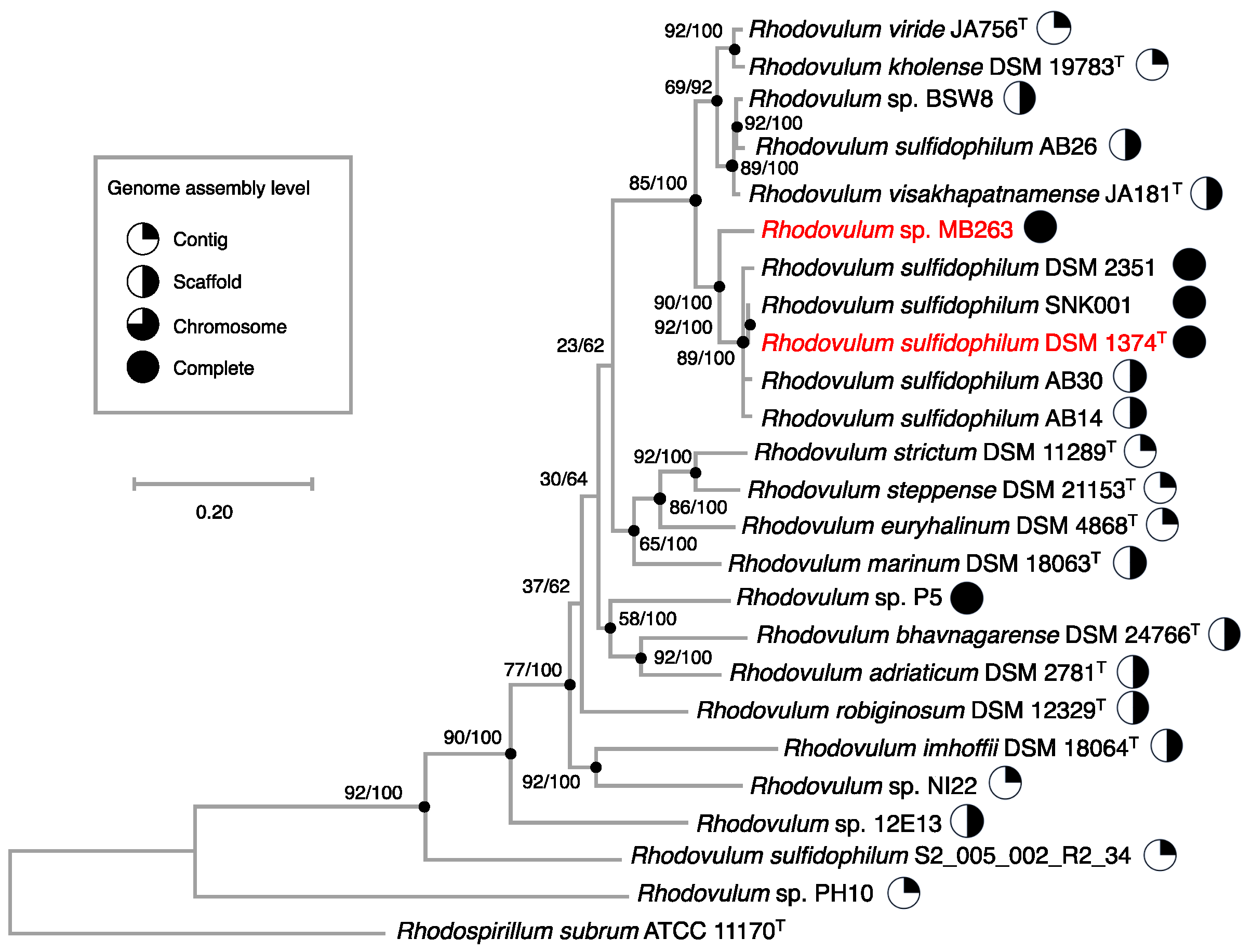
3.8. Genomic Analysis of Rhodovulum Strains
3.9. Phylogenomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Imhoff, J.F. Diversity of anaerobic anoxygenic phototrophic purple bacteria. In Modern Topics in the Phototrophic Prokaryotes, Environmental and Applied Aspects; Hallenbeck, P.C., Ed.; Springer: New York, NY, USA, 2017; pp. 47–85. [Google Scholar]
- Takahashi, M.; Ichimura, S. Vertical distribution and organic matter production of photosynthetic sulfur bacteria in Japanese lakes. Limnol. Oceanogr. 1968, 13, 644–655. [Google Scholar] [CrossRef]
- Matsuyama, M.; Shirouzu, E. Importance of photosynthetic sulfur bacteria, Chromatium sp. as an organic matter producer in Lake Kaiike. Jpn. J. Limnol. 1978, 39, 103–111. [Google Scholar] [CrossRef]
- Overmann, J.; Beauty, J.T.; Hall, K.J.N.; Pfennig, N.; Northcote, T.G. Characterization of a dense, purple sulfur bacterial layer in a meromictic salt lake. Limnol. Oceanogr. 1991, 36, 846–859. [Google Scholar] [CrossRef]
- Schanz, F.; Fischer-Romero, C.; Bachofen, R. Photosynthetic production and photoadaptation of phototrophic sulfur bacteria in Lake Cadagno (Switzerland). Limnol. Oceanogr. 1998, 43, 1262–1269. [Google Scholar] [CrossRef]
- Nakajima, Y.; Okada, H.; Oguri, K.; Suga, H.; Kitazato, H.; Koizumi, Y.; Fukui, M.; Ohkouchi, N. Distribution of chloropigments in suspended particulate matter and benthic microbial mat of a meromictic lake, Lake Kaiike, Japan. Environ. Microbiol. 2003, 5, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Caumette, P. Phototrophic sulfur bacteria and sulfate-reducing bacteria causing red waters in a shallow brackish coastal lagoon (Prévost Lagoon, France). FEMS Microbiol. Lett. 1986, 38, 113–124. [Google Scholar] [CrossRef]
- Tank, M.; Blümel, M.; Imhoff, J.F. Communities of purple sulfur bacteria in a Baltic Sea coastal lagoon analyzed by pufLM gene libraries and the impact of temperature and NaCl concentration in experimental enrichment cultures. FEMS Microbiol. Ecol. 2011, 78, 428–438. [Google Scholar] [CrossRef]
- Nicholson, J.A.M.; Stolz, J.F.; Pierson, B.K. Structure of a microbial mat at Great Sippewissett Marsh, Cape Cod, Massachusetts. FEMS Microbiol. Ecol. 1987, 45, 343–364. [Google Scholar] [CrossRef]
- Thiel, V.; Tank, M.; Neulinger, S.C.; Gehrmann, L.; Dorador, C.; Imhoff, J.F. Unique communities of anoxygenic phototrophic bacteria in saline lakes of Salar de Atacama (Chile): Evidence for a new phylogenetic lineage of phototrophic Gammaproteobacteria from pufLM gene analyses. FEMS Microbiol. Ecol. 2010, 74, 510–522. [Google Scholar] [CrossRef]
- Korponai, K.; Szabó, A.; Somogyi, B.; Boros, E.; Borsodi, A.K.; Jurecska, L.; Vörös, L.; Felföldi, T. Dual bloom of green algae and purple bacteria in an extremely shallow soda pan. Extremophiles 2019, 23, 467–477. [Google Scholar] [CrossRef]
- Sletten, O.; Singer, R.H. Sulfur bacteria in red lagoons. J. Water Pollut. Control Fed. 1971, 43, 2118–2122. [Google Scholar]
- Cooper, D.E.; Rands, M.B.; Woo, C.P. Sulfide reduction in fellmongery effluent by red sulfur bacteria. J. Water Pollut. Control Fed. 1975, 47, 2088–2100. [Google Scholar] [PubMed]
- Wenke, T.L.; Vogt, J.C. Temporal changes in a pink feedlot lagoon. Appl. Environ. Microbiol. 1981, 41, 381–385. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, J.A.; Miller, W.G.; Lathrop, J.R.; Silva, C.J.; Bullard, G.L. Induction of purple sulfur bacterial growth in dairy wastewater lagoons by circulation. Lett. Appl. Microbiol. 2009, 49, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Belila, A.; Abbas, B.; Fazaa, I.; Saidi, N.; Snoussi, M.; Hassen, A.; Muyzer, G. Sulfur bacteria in wastewater stabilization ponds periodically affected by the red-water phenomenon. Appl. Microbiol. Biotechnol. 2013, 97, 379–394. [Google Scholar] [CrossRef]
- Belila, A.; Fazaa, I.; Hassen, A.; Ghrabi, A. Anoxygenic phototrophic bacterial diversity within wastewater stabilization plant during red water phenomenon. Int. J. Environ. Sci. Technol. 2013, 10, 837–846. [Google Scholar] [CrossRef]
- Dungan, R.S.; Leytem, A.B. Detection of purple sulfur bacteria in purple and non-purple dairy wastewaters. J. Environ. Qual. 2015, 44, 1550–1555. [Google Scholar] [CrossRef]
- Okubo, Y.; Futamata, H.; Hiraishi, A. Characterization of phototrophic purple nonsulfur bacteria forming colored microbial mats in a swine wastewater ditch. Appl. Environ. Microbiol. 2006, 72, 6225–6233. [Google Scholar] [CrossRef]
- Hiraishi, A.; Kitamura, H. Distribution of phototrophic purple nonsulfur bacteria in activated sludge systems and other aquatic environments. Bull. Jpn. Soc. Sci. Fish. 1984, 50, 1929–1937. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y. Isolation and characterization of Rhodovulum strictum sp. nov. and some other members of purple nonsulfur bacteria from colored blooms in tidal and seawater pools. Int. J. Syst. Bacteriol. 1995, 45, 319–326. [Google Scholar] [CrossRef]
- Hiraishi, A. Isoprenoid quinones as biomarkers of microbial populations in the environment. J. Biosci. Bioeng. 1999, 88, 449–460. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y.; Ishihara, J.; Mori, T. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 1996, 42, 457–469. [Google Scholar] [CrossRef]
- Achenbach, L.A.; Carey, J.; Madigan, M.T. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microbiol. 2001, 67, 2922–2926. [Google Scholar] [CrossRef] [PubMed]
- Hirose, S.; Nagashima, K.V.P.; Matsuura, K.; Haruta, S. Diversity of purple phototrophic bacteria, Inferred from pufM gene, within epilithic Biofilm in Tama River, Japan. Microbes Environ. 2012, 27, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. New dimensions in microbial ecology-functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. Microorganisms 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association; American Water Works Association; Water Pollution Control Federation. Standard Methods for the Examination of Water and Wastewater, 19th ed.; American Public Health Association: Washington, DC, USA, 1995. [Google Scholar]
- Clayton, R.K. Toward the isolation of a photochemical reaction center in Rhodopseudomonas sphaeroides. Biochim. Biophys. Acta 1963, 75, 312–323. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Hiraishi, A.; Hoshino, Y.; Kitamura, K. Isoprenoid quinone composition in the classification of Rhodospirillaceae. J. Gen. Appl. Microbiol. 1984, 30, 197–210. [Google Scholar] [CrossRef]
- Hiraishi, A.; Hoshino, Y. Distribution of rhodoquinone in Rhodospirillaceae and its taxonomic implications. J. Gen. Appl. Microbiol. 1984, 30, 435–448. [Google Scholar] [CrossRef]
- Yoshida, N.; Hiraishi, A. An improved redox dye-staining method using 5-cyano-2, 3-ditoryl tetrazolium chloride for detection of metabolically active bacteria in activated sludge. Microbes Environ. 2004, 19, 61–70. [Google Scholar] [CrossRef]
- Hiraishi, A.; Okamura, K. Rhodopseudomonas telluris sp. nov., a phototrophic alphaproteobacterium isolated from paddy soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 3369–3374. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Anoxygenic phototrophic bacteria. In Methods in Aquatic Bacteriology; Austin, B., Ed.; John Wiley & Sons: New York, NY, USA, 1988; pp. 207–240. [Google Scholar]
- Biebl, H.; Pfennig, N. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch. Microbiol. 1978, 117, 9–16. [Google Scholar] [CrossRef]
- Hiraishi, A.; Shin, Y.K.; Ueda, Y.; Sugiyama, J. Automated sequencing of PCR-amplified 16S rDNA on “Hydrolink” gels. J. Microbiol. Methods 1994, 19, 145–154. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Hanada, A.; Kurogi, T.; Giang, N.M.; Yamada, T.; Kamimoto, Y.; Kiso, Y.; Hiraishi, A. Bacteria of the candidate phylum TM7 are prevalent in acidophilic nitrifying sequencing-batch reactors. Microbes Environ. 2014, 29, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Hiraishi, A.; Iwasaki, M.; Shinjo, H. Terminal restriction pattern analysis of 16S rRNA genes for the characterization of bacterial communities of activated sludge. J. Biosci. Bioeng. 2000, 90, 148–156. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 1963, 6, 726–738. [Google Scholar]
- Hiraishi, A.; Sakamaki, N.; Miyakoda, H.; Maruyama, T.; Kato, K.; Futamata, H. Estimation of “Dehalococcoides” populations in lake sediment contaminated with low levels of polychlorinated dioxins. Microbes Environ. 2005, 20, 216–226. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P. The Variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Katayama, M.; Ohtsubo, Y.; Misawa, N.; Iioka, E.; Suda, W.; Oshima, K.; Hanaoka, M.; Tanaka, K.; Eki, T.; et al. Complete genome sequence of cyanobacterium Geminocystis sp. strain NIES-3709, which harbors a phycoerythrin-rich phycobilisome. Genome Announc. 2015, 3, e00385-15. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Maruyama, F.; Mitsui, H.; Nagata, Y.; Tsuda, M. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J. Bacteriol. 2012, 194, 6970–6971. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relation to whole genome sequence. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Na, S.-I.; Kim, Y.O.; Yoon, S.-H.; Ha, S.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Imhoff, J.F. Quinones of phototrophic purple bacteria. FEMS Microbiol. Lett. 1984, 25, 85–89. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Neulinger, S.C. Photosynthesis is widely distributed among Proteobacteria as demonstrated by the phylogeny of PufLM reaction center proteins. Front. Microbiol. 2018, 8, 2679. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Feng, F.Y.; Medová, H.; Dean, J.; Koblížek, M. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadates. Proc. Natl. Acad. Sci. USA 2014, 111, 7795–7800. [Google Scholar] [CrossRef]
- Zeng, Y.; Baumbach, J.; Barbosa, E.G.; Azevedo, V.; Zhang, C.; Koblížek, M. Metagenomic evidence for the presence of phototrophic Gemmatimonadetes bacteria in diverse environments. Environ. Microbiol. Rep. 2016, 8, 139–149. [Google Scholar] [CrossRef]
- Nagashima, K.V.; Hiraishi, A.; Shimada, K.; Matsuura, K. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J. Mol. Evol. 1997, 45, 131–136. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Neulinger, S.C. Phylogeny of anoxygenic photosynthesis based on sequences of photosynthetic reaction center proteins and a key enzyme in bacteriochlorophyll biosynthesis, the chlorophyllide reductase. Microorganisms 2019, 7, 576. [Google Scholar] [CrossRef]
- Suresh, G.; Lodha, T.D.; Indu, B.; Sasikala, C.; Ramana, C.V. Taxogenomics resolves conflict in the genus Rhodobacter: A two and half decades pending thought to reclassify the genus Rhodobacter. Front. Microbiol. 2019, 10, 2840. [Google Scholar] [CrossRef]
- Masuda, S.; Hori, K.; Maruyama, F.; Ren, S.; Sugimoto, S.; Yamamoto, N.; Mori, H.; Yamada, T.; Sato, S.; Tabata, S.; et al. Whole-genome sequence of the purple photosynthetic bacterium Rhodovulum sulfidophilum strain W4. Genome Announc. 2013, 1, e00577-13. [Google Scholar] [CrossRef]
- Nagao, N.; Hirose, Y.; Misawa, N.; Ohtsubo, Y.; Umekage, S.; Kikuchi, Y. Complete genome sequence of Rhodovulum sulfidophilum DSM 2351, an extracellular nucleic acid-producing bacterium. Genome Announc. 2015, 3, e00388-15. [Google Scholar] [CrossRef]
- Hansen, T.A.; Veldkamp, H. Rhodopseudomonas sulfidophila, nov. spec., a new species of the purple nonsulfur bacteria. Arch. Mikrobiol. 1973, 92, 45–58. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y. Intrageneric structure of the genus Rhodobacter: Transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int. J. Syst. Bacteriol. 1994, 44, 15–23. [Google Scholar] [CrossRef][Green Version]
- Appia-Ayme, C.; Little, P.J.; Matsumoto, Y.; Leech, A.P.; Berks, B.C. Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum. J. Bacteriol. 2001, 183, 6107–6118. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, W.; Dam, B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 2009, 33, 999–1043. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Brooks, B.; Olm, M.R.; Firek, B.A.; Baker, R.; Thomas, B.C.; Morowitz, M.J.; Banfield, J.F. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat. Commun. 2017, 8, 1814. [Google Scholar] [CrossRef]
- Brown, L.M.; Gunasekera, T.S.; Bowen, L.L.; Ruiz, O.N. Draft genome sequence of Rhodovulum sp. strain NI22, a naphthalene-degrading marine bacterium. Genome Announc. 2015, 3, e01475-14. [Google Scholar] [CrossRef]
- Khatri, I.N.; Korpole, S.; Subramanian, S.; Pinnaka, A.K. Draft genome sequence of Rhodovulum sp. strain PH10, a phototrophic alphaproteobacterium isolated from a soil sample of mangrove of Namkhana, India. J. Bacteriol. 2012, 194, 6363. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kobayashi, M. Waste remediation and treatment using anoxygenic phototrophic bacteria. In Anoxygenic Photosynthetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; pp. 1269–1282. [Google Scholar]
- Hiraishi, A.; Shi, J.L.; Kitamura, H. Effects of organic nutrient strength on the purple nonsulfur bacterial content and metabolic activity of photosynthetic sludge for wastewater treatment. J. Ferment. Bioeng. 1989, 68, 269–276. [Google Scholar] [CrossRef]
- Suzuki, H.; Daimon, M.; Awano, T.; Umekage, S.; Tanaka, T.; Kikuchi, Y. Characterization of extracellular DNA production and flocculation of the marine photosynthetic bacterium Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 2009, 84, 349–356. [Google Scholar] [CrossRef]
| Sample | Sampling Month/Year | Color | Temp. (°C) | pH | COD (mg L−1) | Salinity (‰) | S2− (mg L−1) | ORP (mV) |
|---|---|---|---|---|---|---|---|---|
| Hot spring | ||||||||
| H1 | October/2000 | Red | 35.0 | 7.5 | nd * | nd | nd | nd |
| Tidal flat | ||||||||
| Y1 | May/2007 | Pink | 24.5 | 8.1 | 62 | 21.8 | 6.8 | −170 |
| Y2 | May/2007 | Pink | 24.5 | 8.1 | 44 | 21.8 | 6.2 | −180 |
| Y3 | August/2009 | Pink | 28.7 | 8.0 | 54 | 19.8 | 8.4 | −220 |
| Tide pool | ||||||||
| J1 | September/2002 | Yellow-green | 28.9 | 7.8 | 19 | 28.8 | 9.4 | −280 |
| J2 | September/2004 | Red-brown | 29.5 | 8.2 | 98 | 27.3 | 7.3 | −240 |
| J3 | September/2014 | Yellow-green | 29.4 | 8.5 | 37 | 27.5 | 12 | −320 |
| Ditch | ||||||||
| D1 | May/2004 | Red | 22.3 | 8.9 | 890 | 3.8 | 0 | 23 |
| D2 | August/2012 | Pink-brown | 28.6 | 8.2 | 280 | 1.1 | 0.5 | −56 |
| D3 | August/2002 | Pink-brown | 28.9 | 8.1 | 210 | 2.6 | 0.6 | −93 |
| Feature | Rhodovulum sp. Strain MB263 | Rdv. sulfidophilum Strain DSM 1374T | Rdv. sulfidophilum Strain DSM 2351 |
|---|---|---|---|
| Reference | This study | This study | Nagao et al. [63] |
| Genome size (bp) | 4,162,560 | 4,347,929 | 4,732,772 |
| G+C content (%) | 67.03 | 66.92 | 66.89 |
| Number of: | |||
| CDS | 3725 | 3899 | 4146 |
| rRNA operons | 3 | 3 | 3 |
| tRNA genes | 49 | 50 | 50 |
| Plasmids | 2 | 2 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiraishi, A.; Nagao, N.; Yonekawa, C.; Umekage, S.; Kikuchi, Y.; Eki, T.; Hirose, Y. Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments. Microorganisms 2020, 8, 150. https://doi.org/10.3390/microorganisms8020150
Hiraishi A, Nagao N, Yonekawa C, Umekage S, Kikuchi Y, Eki T, Hirose Y. Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments. Microorganisms. 2020; 8(2):150. https://doi.org/10.3390/microorganisms8020150
Chicago/Turabian StyleHiraishi, Akira, Nobuyoshi Nagao, Chinatsu Yonekawa, So Umekage, Yo Kikuchi, Toshihiko Eki, and Yuu Hirose. 2020. "Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments" Microorganisms 8, no. 2: 150. https://doi.org/10.3390/microorganisms8020150
APA StyleHiraishi, A., Nagao, N., Yonekawa, C., Umekage, S., Kikuchi, Y., Eki, T., & Hirose, Y. (2020). Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments. Microorganisms, 8(2), 150. https://doi.org/10.3390/microorganisms8020150






