Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Transjugation Assays
2.3. Whole Genome Sequencing (WGS)
2.4. Parenthood Analysis
2.5. Identification of Non-Homologous Genes Acquisition
3. Results
3.1. Selection of T. thermophilus Strains for Parenthood Analysis
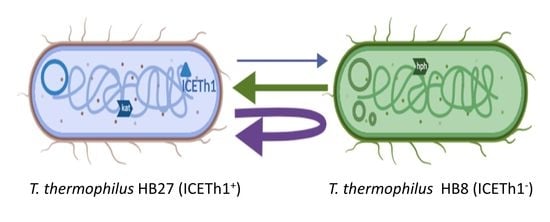
3.2. Transjugation between T. thermophilus HB7 and HB8 Strains
3.3. Selection of HB8-Derived Transjugants
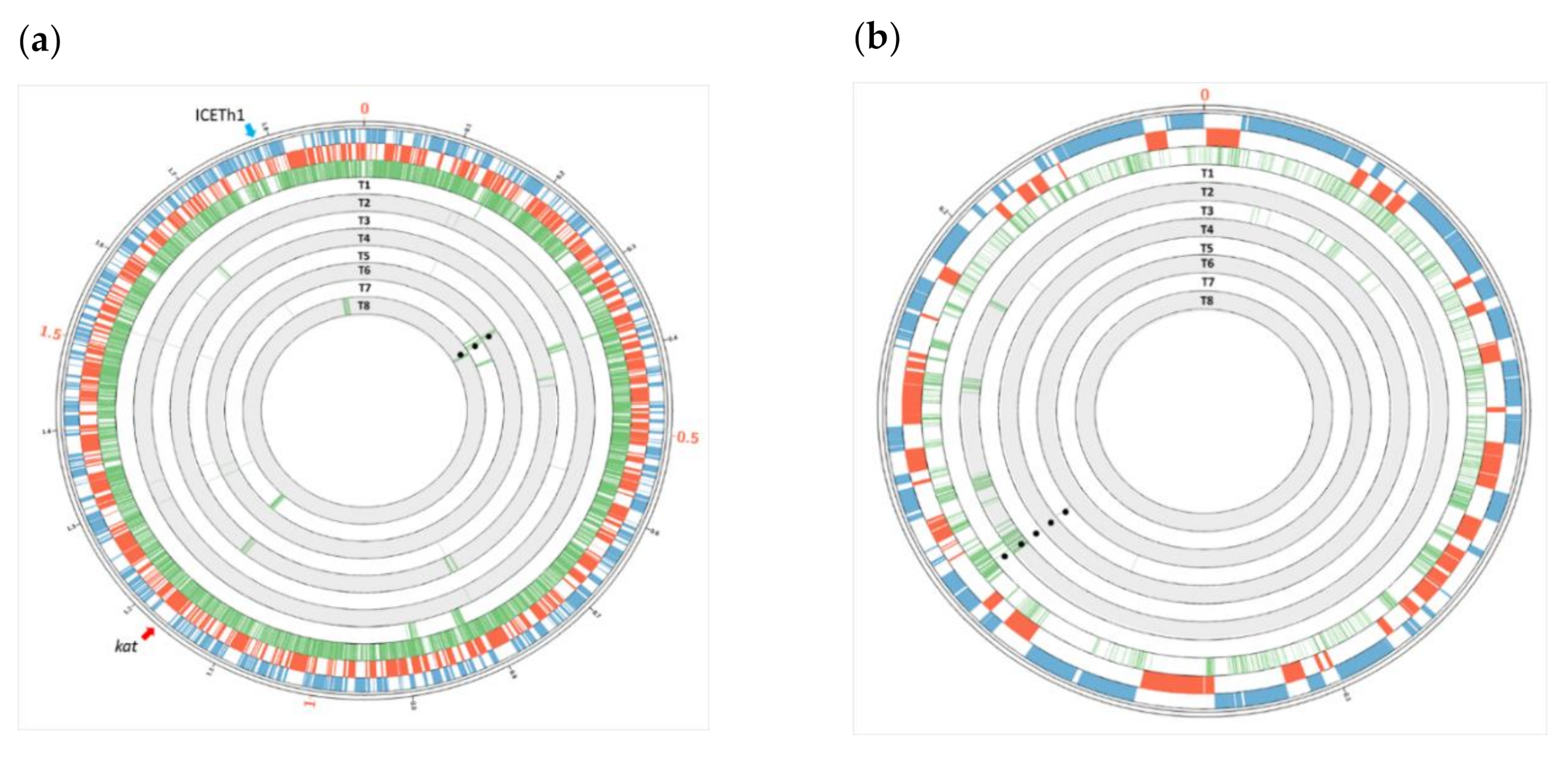
3.4. Single Nucleotide Polymorphisms (SNPs) Reveal Parenthood of Genes
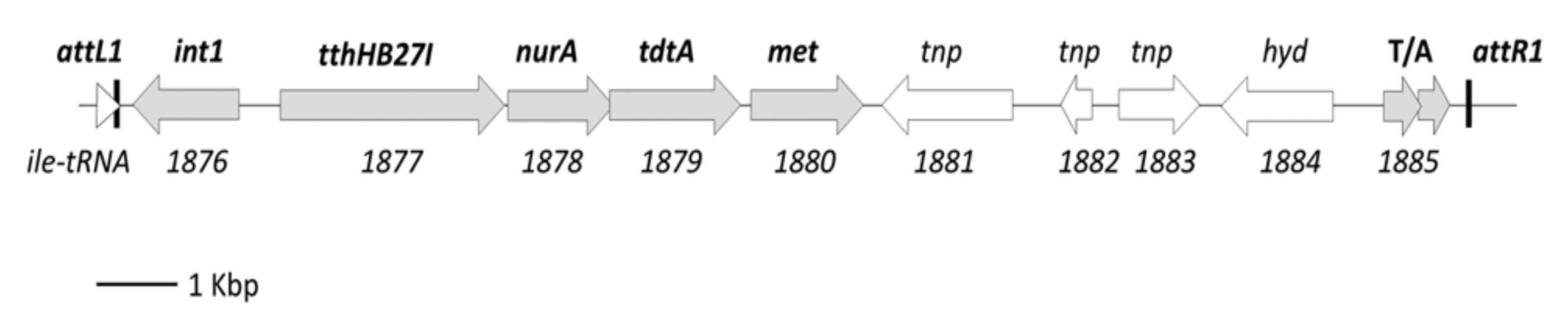
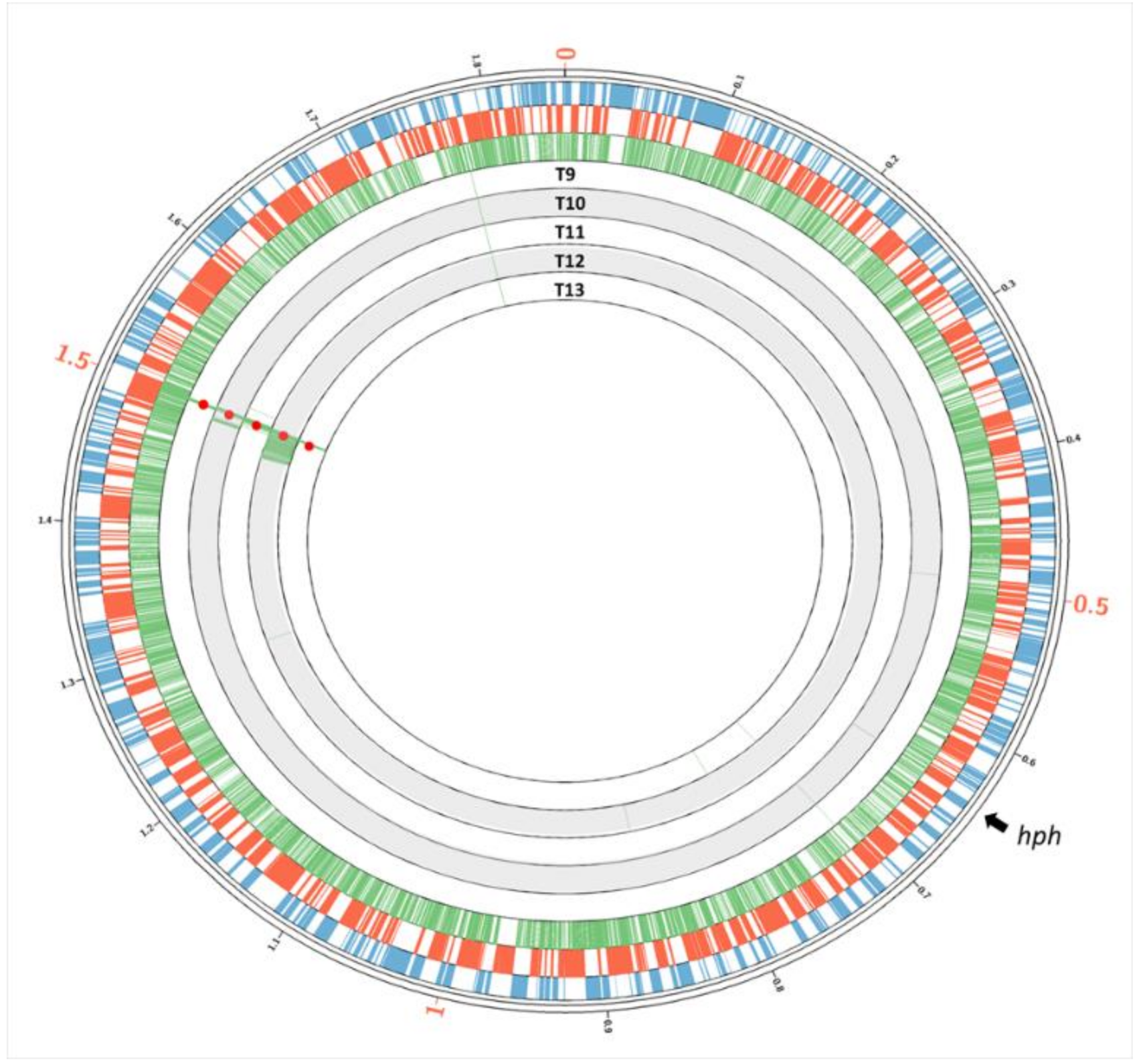
3.5. Transfer of Parental-Specific Genes
4. Discussion
4.1. Detection of Retrotransfer
4.2. Mosaicity of the Progeny
4.3. A Putative Mechanism for Retro-Transfer
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Originality-Significance Statement
References
- Garcia-Aljaro, C.; Balleste, E.; Muniesa, M. Beyond the canonical strategies of horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017, 38, 95–105. [Google Scholar] [CrossRef]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents, phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef]
- Blesa, A.; Berenguer, J. Contribution of vesicle-protected extracellular DNA to horizontal gene transfer in Thermus spp. Int. Microbiol. 2015, 18, 177–187. [Google Scholar]
- Roier, S.; Zingl, F.G.; Cakar, F.; Durakovic, S.; Kohl, P.; Eichmann, T.O.; Klug, L.; Gadermaier, B.; Weinzerl, K.; Prassl, R.; et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016, 7, 10515. [Google Scholar] [CrossRef] [PubMed]
- Cabezón, E.; Ripoll-Rozada, J.; Pena, A.; De la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Broda, L. The formation of Hfr strains in Escherichia coli K12. Genet. Res. 1967, 9, 35–47. [Google Scholar] [CrossRef]
- Hochhut, B.; Marrero, J.; Waldor, M.K. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a const in found in Vibrio cholerae O139. J. Bacteriol. 2000, 182, 2043–2047. [Google Scholar] [CrossRef]
- Thoma, L.; Muth, G. The conjugative DNA-transfer apparatus of Streptomyces. Int. J. Med. Microbiol. 2015, 305, 224–229. [Google Scholar] [CrossRef]
- Blesa, A.; Berenguer, J. Alternative ways to exchange DNA: Unconventional conjugation among bacteria. In Horizontal Gene Transfer; Villa, T., Viñas, M., Eds.; Springer: Cham, Germany, 2019; pp. 77–96. [Google Scholar]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef]
- Li, H. Random chromosome partitioning in the polyploid bacterium Thermus thermophilus HB27. G3 (Bethesda) 2019, 9, 1249–1261. [Google Scholar] [CrossRef]
- Henne, A.; Bruggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Baquedano, I.; Quintans, N.G.; Mata, C.P.; Castón, J.R.; Berenguer, J. The transjugation machinery of Thermus thermophilus, Identification of TdtA, an ATPase involved in DNA donation. PLoS Genet. 2017, 13, e1006669. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Cesar, C.E.; Averhoff, B.; Berenguer, J. Non-canonical cell-to-cell DNA transfer in Thermus spp. is insensitive to Argonaute-mediated interference. J. Bacteriol. 2015, 197, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Baquedano, I.; Mencia, M.; Blesa, A.; Burrus, V.; Berenguer, J. ICETh1 and ICETh2, two interdependent mobile genetic elements in Thermus thermophilus transjugation. Environ. Microbiol. 2020, 22, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Krefft, D.; Zylicz-Stachula, A.; Mulkiewicz, E.; Papkov, A.; Jezewska-Frackowiak, J.; Skowron, P.M. Two-stage gene assembly/cloning of a member of the TspDTI subfamily of bifunctional restriction endonucleases, TthHB27I. J. Biotechnol. 2015, 194, 67–80. [Google Scholar] [CrossRef]
- Baquedano, I. ICEth1 and ICEth2, Two Mobile Genetic Elements Coordinated in Thermus thermophilus Transjugation. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2019; p. 188. [Google Scholar]
- Blesa, A.; Quintans, N.G.; Baquedano, I.; Mata, C.P.; Castón, J.R.; Berenguer, J. Role of archaeal HerA protein in the biology of the bacterium Thermus thermophilus. Genes 2017, 8, 130. [Google Scholar] [CrossRef]
- Dordet-Frisoni, E.; Sagne, E.; Baranowski, E.; Breton, M.; Nouvel, L.X.; Blanchard, A.; Marenda, M.S.; Tardy, F.; Sirand-Pugnet, P.; Citti, C. Chromosomal transfers in mycoplasmas, when minimal genomes go mobile. mBio 2014, 5, e01958. [Google Scholar] [CrossRef]
- Bruggemann, H.; Chen, C. Comparative genomics of Thermus thermophilus, Plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 2006, 124, 654–661. [Google Scholar] [CrossRef]
- Cava, F.; Laptenko, O.; Borukhov, S.; Chahlafi, Z.; Blas-Galindo, E.; Gomez-Puertas, P.; Berenguer, J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 2007, 64, 630–646. [Google Scholar] [CrossRef]
- Gray, T.A.; Derbyshire, K.M. Blending genomes, distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol. Microbiol. 2018, 108, 601–613. [Google Scholar] [CrossRef]
- Blesa, A.; Berenguer, J. Cell-to-cell DNA transfer among Thermus species. Bio-protocol. 2016, 6, 22006. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic, a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen, A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, S.; De Graaf, A.O.; Karimi, M.; Van der Reijden, B.A.; Hellstrom-Lindberg, E.; Jansen, J.H.; Dugas, M. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci. Rep. 2017, 7, 43169. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit, a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff, SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos, an information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Lasa, I.; Castón, J.R.; Fernández-Herrero, L.A.; De Pedro, M.A.; Berenguer, J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol. 1992, 6, 1555–1564. [Google Scholar] [CrossRef]
- Nakamura, A.; Takakura, Y.; Kobayashi, H.; Hoshino, T. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J. Biosci. Bioeng. 2005, 100, 158–163. [Google Scholar] [CrossRef]
- Ohtani, N.; Tomita, M.; Itaya, M. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 2010, 192, 5499–5505. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Tomita, M.; Itaya, M. The third plasmid pVV8 from Thermus thermophilus HB8, isolation, characterization, and sequence determination. Extremophiles 2012, 16, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Sánchez, M.; Sacristán-Horcajada, E.; González-de la Fuente, S.; Peiro, R.; Berenguer, J. Into the Thermus mobilome, Presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A. Horizontal gene transfer in Thermus thermophilus: Mechanisms and barriers. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2016; p. 194. [Google Scholar]
- Dordet Frisoni, E.; Marenda, M.S.; Sagne, E.; Nouvel, L.X.; Guerillot, R.; Glaser, P.; Blanchard, A.; Tardy, F.; Sirand‐Pugnet, P.; Baranowski, E.; et al. ICEA of Mycoplasma agalactiae, a new family of self-transmissible integrative elements that confers conjugative properties to the recipient strain. Mol. Microbiol. 2013, 89, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Barany, F.; Danzitz, M.; Zebala, J.; Mayer, A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes, comparison with the isoschizomeric TaqI enzymes. Gene 1992, 112, 3–12. [Google Scholar] [CrossRef]
- Bellanger, X.; Morel, C.; Gonot, F.; Puymege, A.; Decaris, B.; Guedon, G. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol. Microbiol. 2011, 81, 912–925. [Google Scholar] [CrossRef]
| Strain | Genotype/Resistance | Use | Reference |
|---|---|---|---|
| CK1 | HB27 derivative, TTC1211(gdh1)::kat/KanR | Parental | [21] |
| CK2 | HB27 derivative, TTC1211(gdh1)::kat, ΔpilA4/KanR | Competence deficient | [22] |
| CH81 | HB8 derivative, TTHA0672::hph/HygR | Parental | This work |
| PH81 | HB8 derivative, TTHB198::hph/HygR | Parental | This work |
| Strain | % Align vs. HB27 Reference | % Align vs. HB8 Reference |
|---|---|---|
| CK1 | 98.45 | 87.53 |
| C/PH81 | 79.46 | 99.70 |
| T1 1 | 98.54 | 87.90 |
| T2 1 | 98.76 | 87.89 |
| T3 1 | 98.14 | 86.49 |
| T4 1 | 97.87 | 85.39 |
| T5 1 | 98.80 | 87.22 |
| T6 2 | 98.93 | 87.09 |
| T7 2 | 98.73 | 85.96 |
| T8 2 | 98.48 | 85.45 |
| T9 3 | 82.19 | 99.54 |
| T10 3 | 84.76 | 99.53 |
| T11 3 | 82.22 | 99.51 |
| T12 3 | 83.63 | 99.52 |
| T13 3 | 81.46 | 99.19 |
| Strain | SNPs vs. HB27 Reference | Nº of Chromosome Regions Involved 1 | SNPs vs. pTT27 Reference | Nº of pTT27 Regions Involved 1 | HB8-Specific Genes Detected |
|---|---|---|---|---|---|
| CK1 | 0 2 | - | 0 2 | - | - |
| P/CH81 | 14,818 | - | 2167 | - | - |
| T1 | 109 | 6 | 31 | 1 1 | ISTh4 |
| T2 | 6 | 6 | 283 | 5 1 | ISTh4 |
| T3 | 65 | 6 | 51 | 6 1 | TTHA0285 TTHA0286 TTHB073 |
| T4 | 92 | 6 | 0 | 1 1 | - |
| T5 | 5 | 5 | 5 | 2 1 | - |
| T6 | 39 | 5 1 | 0 | 0 | - |
| T7 | 114 | 11 1 | 0 | 0 | - |
| T8 | 50 | 2 1 | 0 | 0 | TTHA498 |
| Strain | HB8 Reference | Chromosome Regions Involved 1 | pTT27 (HB8) Reference | pTT27 Regions Involved 1 | HB27-Specific Genes Detected |
|---|---|---|---|---|---|
| CH8 | 0 | - | 0 | - | - |
| T9 | 16 | 2 1 | 0 | 0 | tdtA |
| T10 | 51 | 3 1 | 0 | 0 | TTC0952 |
| T11 | 36 | 2 1 | 0 | 0 | |
| T12 | 168 | 2 1 | 0 | 0 | TTC0398 TTC0857 |
| T13 | 14 | 2 1 | 0 | 0 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blesa, A.; Baquedano, I.; González-de la Fuente, S.; Mencía, M.; Berenguer, J. Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms 2020, 8, 2051. https://doi.org/10.3390/microorganisms8122051
Blesa A, Baquedano I, González-de la Fuente S, Mencía M, Berenguer J. Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms. 2020; 8(12):2051. https://doi.org/10.3390/microorganisms8122051
Chicago/Turabian StyleBlesa, Alba, Ignacio Baquedano, Sandra González-de la Fuente, Mario Mencía, and José Berenguer. 2020. "Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus" Microorganisms 8, no. 12: 2051. https://doi.org/10.3390/microorganisms8122051
APA StyleBlesa, A., Baquedano, I., González-de la Fuente, S., Mencía, M., & Berenguer, J. (2020). Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms, 8(12), 2051. https://doi.org/10.3390/microorganisms8122051