Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review
Abstract
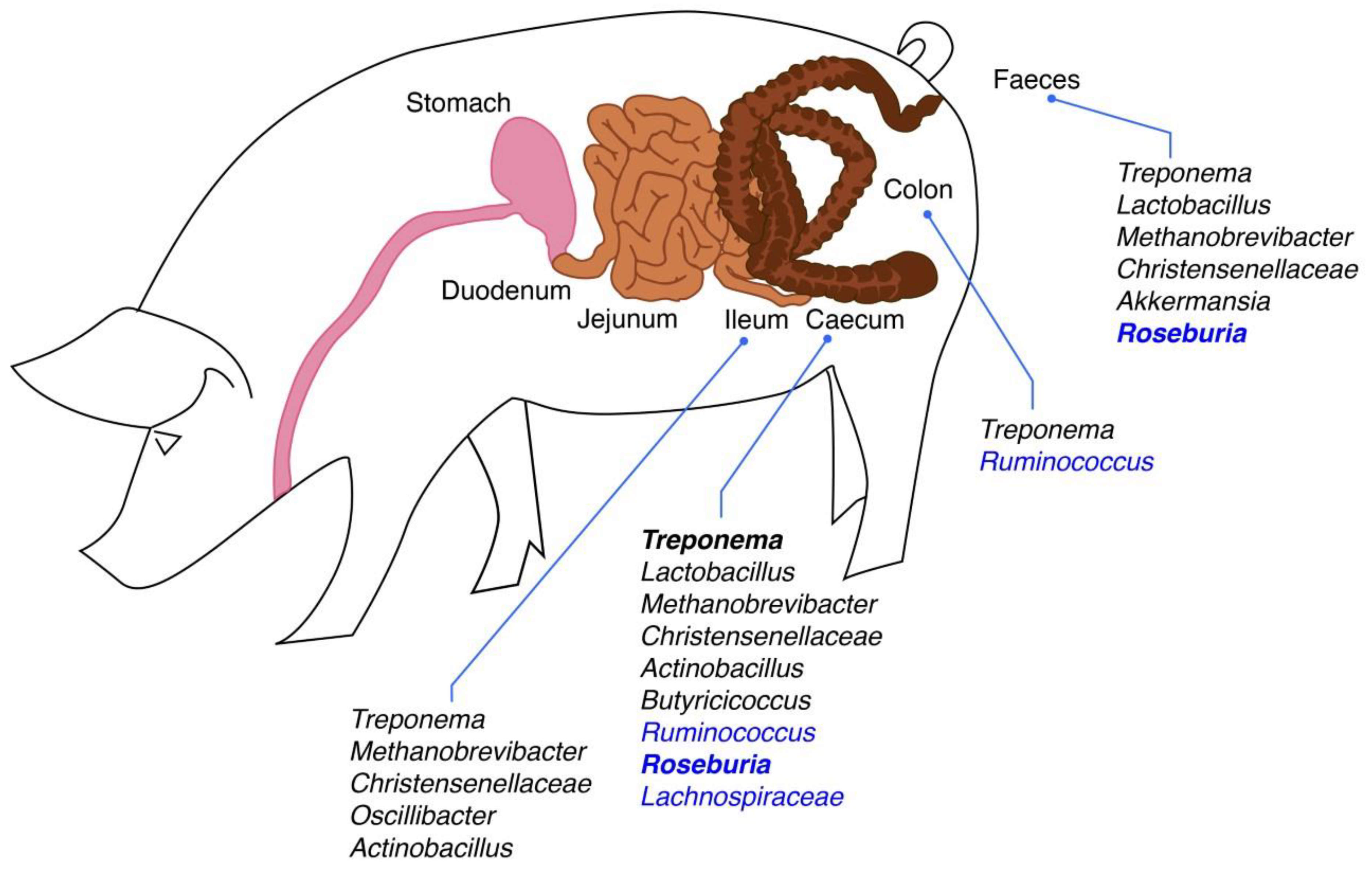
1. Introduction
2. Evidence for a Link between the Pig Intestinal Microbiota and Pig Growth, Body Weight and Body Composition
3. Evidence for a Link between the Pig Intestinal Microbiota and Feed Efficiency
4. Potential Mechanisms by Which the Gut Microbiome Impacts Growth and FE in Pigs
4.1. Fermentation of Dietary Substrates
4.2. Modulation of Inflammation
5. Applying Knowledge on FE- and Growth-Associated Microbiota to Improve Production Traits in Pigs
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cullen, J.; Lawlor, P.G.; Gardiner, G.E. Microbiological services delivered by the pig gut microbiome. In Understanding Gut Microbiomes as Targets for Improving Pig Gut Health; Bailey, M., Stokes, C., Eds.; Burleigh Dodds Science Publishing Ltd.: Cambridge, UK, 2020; in press. [Google Scholar]
- Deusch, S.; Tilocca, B.; Camarinha-Silva, A.; Seifert, J. News in livestock research-use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Comp. Struct. Biotechnol. J. 2015, 13, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Tröscher-Mußotter, J.; Tilocca, B.; Stefanski, V.; Seifert, J. Analysis of the bacterial and host proteins along and across the porcine gastrointestinal tract. Proteomes 2019, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Han, G.G.; Lee, J.Y.; Jin, G.D.; Park, J.; Choi, Y.H.; Chae, B.J.; Kim, E.B.; Choi, Y.J. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Appl. Microbiol. Biotechnol. 2017, 101, 5903–5911. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016, 6, 27427. [Google Scholar] [CrossRef]
- Lu, D.; Tiezzi, F.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Maltecca, C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome 2018, 6, 4. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Woo, G.K.S.; Fung, A.M.Y.; Ngan, A.H.Y.; Song, Y.; Liu, C.; Summanen, P.; Finegold, S.M.; Yuen, K. Bacteraemia caused by Anaerotruncus colihominis and emended description of the species. J. Clin. Pathol. 2006, 59, 748–752. [Google Scholar] [CrossRef]
- Yekani, M.; Baghi, H.B.; Naghili, B.; Vahed, S.Z.; Sóki, J.; Memar, M.Y. To resist and persist: Important factors in the pathogenesis of Bacteroides fragilis. Microb. Pathog. 2020, 149, 104506. [Google Scholar] [CrossRef]
- Togo, A.H.; Diop, A.; Dubourg, G.; Khelaifia, S.; Richez, M.; Armstrong, N.; Maraninchi, M.; Fournier, P.E.; Raoult, D.; Million, M. Anaerotruncusmassiliensis sp. nov., a succinate-producing bacterium isolated from human stool from an obese patient after bariatric surgery. New Microbes New Infect. 2019, 29, 100508. [Google Scholar] [CrossRef]
- Zhong, X.; Harrington, J.M.; Millar, S.R.; Perry, I.J.; O’Toole, P.W.; Phillips, C.M. Gut microbiota associations with metabolic health and obesity status in older adults. Nutrients 2020, 12, 2364. [Google Scholar] [CrossRef] [PubMed]
- Torres-Pitarch, A.; Gardiner, G.E.; Cormican, P.; Rea, M.; Crispie, F.; O’Doherty, J.V.; Cozannet, P.; Ryan, T.; Lawlor, P.G. Effect of cereal soaking and carbohydrase supplementation on growth, nutrient digestibility and intestinal microbiota in liquid-fed grow-finishing pigs. Sci. Rep. 2020, 10, 1023. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Doré, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P. Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health. Appl. Microbiol. Biotechnol. 2019, 103, 7287–7315. [Google Scholar] [CrossRef]
- Torres-Pitarch, A.; Gardiner, G.E.; Cormican, P.; Rea, M.; Crispie, F.; O’Doherty, J.V.; Cozannet, P.; Ryan, T.; Cullen, J.; Lawlor, P.G. Effect of cereal fermentation and carbohydrase supplementation on growth, nutrient digestibility and intestinal microbiota in liquid-fed grow-finishing pigs. Sci. Rep. 2020, 10, 13716. [Google Scholar] [CrossRef]
- Holman, D.B.; Brunelle, B.W.; Trachsel, J.; Allena, H.K. Meta-analysis to define a core microbiota in the swine gut. mSystems 2017, 2, e00004-17. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Maltecca, C.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Fix, J.; Tiezzi, F. Heritability and genome-wide association of swine gut microbiome features with growth and fatness parameters. Sci. Rep. 2020, 10, 10134. [Google Scholar] [CrossRef]
- Fang, S.; Xiong, X.; Su, Y.; Huang, L.; Chen, C. 16S rRNA gene-based association study identified microbial taxa associated with pork intramuscular fat content in feces and cecum lumen. BMC Microbiol. 2017, 17, 162. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Tiezzi, F.; Howard, J.; Huang, Y.J.; Gray, K.A.; Schillebeeckx, C.; McNulty, N.P.; Maltecca, C. Gut microbiome composition differences among breeds impact feed efficiency in swine. Microbiome 2020, 8, 110. [Google Scholar] [CrossRef]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Maltecca, C.; Lu, D.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Tiezzi, F. Predicting growth and carcass traits in swine using microbiome data and machine learning algorithms. Sci. Rep. 2019, 9, 6574. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curião, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D.; et al. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 2017, 83, e00380-17. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curião, T.; Metzler-Zebeli, B.U.; Wilkinson, T.; Magowan, E.; Berry, D.P.; Reyer, H.; Prieto, M.L.; Buzoianu, S.G.; Harrison, M.; et al. Porcine feed efficiency-associated intestinal microbiota and physiological traits: Finding consistent cross-locational biomarkers for residual feed intake. mSystems 2019, 4, e00324-18. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Cai, G.; Ye, J.; Yang, M.; Ding, R.; Wang, X.; Zheng, E.; Fu, D.; Li, S.; Zhou, S.; et al. A global comparison of the microbiome compositions of three gut locations in commercial pigs with extreme feed conversion ratios. Sci. Rep. 2018, 8, 4536. [Google Scholar] [CrossRef]
- Vigors, S.; O’ Doherty, J.V.; Sweeney, T. Colonic microbiome profiles for improved feed efficiency can be identified despite major effects of farm of origin and contemporary group in pigs. Animal 2020, in press. [Google Scholar] [CrossRef]
- Quan, J.; Wu, Z.; Ye, Y.; Peng, L.; Wu, J.; Ruan, D.; Qiu, Y.; Ding, R.; Wang, X.; Zheng, E.; et al. Metagenomic characterization of intestinal regions in pigs with contrasting feed efficiency. Front. Microbiol. 2020, 11, 32. [Google Scholar] [CrossRef]
- Tan, Z.; Yang, T.; Wang, Y.; Xing, K.; Zhang, F.; Zhao, X.; Ao, F.; Chen, S.; Liu, J.; Wang, C. Metagenomic analysis of cecal microbiome identified microbiota and functional capacities associated with feed efficiency in landrace finishing pigs. Front. Microbiol. 2017, 8, 1546. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Ma, X.; Gao, S.; He, L.; Wu, W.; et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015, 5, 9938. [Google Scholar] [CrossRef]
- Rosewarne, C.P.; Cheung, J.L.; Smith, W.J.M.; Evans, P.N.; Tomkins, N.W.; Denman, S.E.; Cuív, P.Ó.; Morrison, M. Draft genome sequence of Treponema sp. strain JC4, a novel spirochete isolated from the bovine rumen. J. Bacteriol. 2012, 194, 4130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, H.; Huang, X.; Fang, S.; He, M.; Zhao, Y.; Wu, Z.; Yang, M.; Zhang, Z.; Chen, C.; Huang, L. Unravelling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 2017, 8, 1555. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Angelakis, E.; Maraninchi, M.; Henry, M.; Giorgi, R.; Valero, R.; Vialettes, B.; Raoult, D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int. J. Obes. 2013, 37, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Nørskov-Lauritsen, N. Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin. Microbiol. Rev. 2014, 27, 214–240. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sung, C.Y.J.; Lee, N.; Ni, Y.; Pihlajamäki, J.; Panagiotou, G.; El-Nezami, H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef] [PubMed]
- Valeriano, V.D.V.; Balolong, M.P.; Kang, D.K. Probiotic roles of Lactobacillus sp. in swine: Insights from gut microbiota. J. Appl. Microbiol. 2016, 122, 554–567. [Google Scholar] [CrossRef]
- Vigors, S.; Sweeney, T.; O’Shea, C.J.; Kelly, A.K.; O’Doherty, J.V. Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal 2016, 10, 1848–1855. [Google Scholar] [CrossRef]
- Songer, G.; Francisco, A.U. Clostridial enteric infections in pigs. J. Vet. Diagn. Investig. 2005, 17, 528–536. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host Front. Cell Infect Microbiol. 2015, 5, 84. [Google Scholar]
- Weishaar, R.; Wellmann, R.; Camarinha-Silva, A.; Rodehutscord, M.; Bennewitz, J. Selecting the hologenome to breed for an improved feed efficiency in pigs-A novel selection index. J. Anim. Breed. Genet. 2020, 137, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Machado, D.; Andrade, J.C.; Mendo, S.; Gomes, A.M.; Freitas, A.C. Evolving trends in next-generation probiotics: A 5W1H perspective. Crit. Rev. Food Sci. Nutr. 2020, 60, 11. [Google Scholar] [CrossRef] [PubMed]
- Krzyściak, W.; Pluskwa, K.K.; Jurczak, A.; Kościelniak, D. The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1361–1376. [Google Scholar] [CrossRef] [PubMed]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [PubMed]
- Hols, P.; Ledesma-García, L.; Gabant, P.; Mignolet, J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019, 27, 690–702. [Google Scholar] [CrossRef]
- Sommer, F.; Nookaew, I.; Sommer, N.; Fogelstrand, P.; Bäckhed, F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015, 16, 62. [Google Scholar] [CrossRef]
- Grubbs, J.K.; Fritchen, A.N.; Huff-Lonergan, E.; Gabler, N.K.; Lonergan, S.M. Selection for residual feed intake alters the mitochondria protein profile in pigs. J. Proteomics. 2013, 80, 334–345. [Google Scholar] [CrossRef]
- Reyer, H.; Oster, M.; Magowan, E.; Muráni, E.; Sauerwein, H.; Dannenberger, D.; Kuhla, B.; Ponsuksili, S.; Wimmers, K. Feed-efficient pigs exhibit molecular patterns allowing a timely circulation of hormones and nutrients. Physiol. Genomics 2018, 50, 726–734. [Google Scholar] [CrossRef]
- Vincent, A.; Louveau, I.; Gondret, F.; Tréfeu, C.; Gilbert, H.; Lefaucheur, L. Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 2015, 93, 2745–2758. [Google Scholar] [CrossRef]
- Reyer, H.; Oster, M.; McCormack, U.M.; Muráni, E.; Gardiner, G.E.; Ponsuksili, S.; Lawlor, P.G.; Wimmers, K. Host-microbiota interactions in ileum and caecum of pigs divergent in feed efficiency contribute to nutrient utilization. Microorganisms 2020, 8, 563. [Google Scholar] [CrossRef]
- Metzler-Zebeli, B.U.; Lawlor, P.G.; Magowan, E.; McCormack, U.M.; Curião, T.; Hollmann, M.; Ertl, R.; Aschenbach, J.R.; Zebeli, Q. Finishing pigs that are divergent in feed efficiency show small differences in intestinal functionality and structure. PLoS ONE 2017, 12, e0174917. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Broom, L.; Kogut, M. Gut immunity: Its development and reasons and opportunities for modulation in monogastric production animals. Anim. Health Res. Rev. 2018, 19, 46–52. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.; Tan, J.; Macia, L.; Mackay, C.R. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 2017, 278, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Kaiko, G.E.; Stappenbeck, T.S. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014, 35, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef] [PubMed]
- Mauch, E.D.; Young, J.M.; Serão, N.V.L.; Hsu, W.L.; Patience, J.F.; Kerr, B.J.; Weber, T.E.; Gabler, N.K.; Dekkers, J.C.M. Effect of lower-energy, higher-fiber diets on pigs divergently selected for residual feed intake when fed higher-energy, lower-fiber diets. J. Anim. Sci. 2018, 96, 1221–1236. [Google Scholar] [CrossRef]
- Barea, R.; Dubois, S.; Gilbert, H.; Sellier, P.; van Milgen, J.; Noblet, J. Energy utilization in pigs selected for high and low residual feed intake. J. Anim. Sci. 2010, 88, 2062–2072. [Google Scholar] [CrossRef]
- Montagne, L.; Loisel, F.; Le Naou, T.; Gondret, F.; Gilbert, H.; Le Gall, M. Difference in short-term responses to a high-fiber diet in pigs divergently selected for residual feed intake. J. Anim. Sci. 2014, 92, 1512–1523. [Google Scholar] [CrossRef]
- Harris, A.J.; Patience, J.F.; Lonergan, S.M.; Dekkers, J.C.M.; Gabler, N.K. Improved nutrient digestibility and retention partially explains feed efficiency gains in pigs selected for low residual feed intake. J. Anim. Sci. 2012, 90, 164–166. [Google Scholar] [CrossRef]
- Vigors, S.; O’Doherty, J.V.; Kelly, A.K.; O’Shea, C.J.; Sweeney, T. The effect of divergence in feed efficiency on the intestinal microbiota and the intestinal immune response in both unchallenged and lipopolysaccharide challenged ileal and colonic explants. PLoS ONE 2016, 11, e0148145. [Google Scholar] [CrossRef] [PubMed]
- Metzler-Zebeli, B.U.; Lawlor, P.G.; Magowan, E.; Zebeli, Q. Interactions between metabolically active bacteria and host gene expression at the cecal mucosa in pigs of diverging feed efficiency. J. Anim. Sci. 2018, 96, 2249–2264. [Google Scholar] [CrossRef]
- Li, X.; Shimizu, Y.; Kimura, I. Gut microbial metabolite short-chain fatty acids and obesity. Biosci. Microbiota Food Health 2017, 36, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Gondret, F.; Louveau, I.; Mourot, J.; Duclos, M.J.; Lagarrigue, S.; Gilbert, H.; van Milgen, J. Dietary energy sources affect the partition of body lipids and the hierarchy of energy metabolic pathways in growing pigs differing in feed efficiency. J. Anim. Sci. 2014, 92, 4865–4877. [Google Scholar] [CrossRef] [PubMed]
- Zaibi, M.S.; Stocker, C.J.; O’Dowd, J.; Davies, A.; Bellahcene, M.; Cawthorne, M.A.; Brown, A.J.; Smith, D.M.; Arch, J.R. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett. 2010, 584, 2381–2386. [Google Scholar] [CrossRef] [PubMed]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, J.R.; Clausen, M.R.; Mortensen, P.B. Oxidation of short and medium chain C2-C8 fatty acids in Sprague -Dawley rat colonocytes. Gut 1997, 40, 400–405. [Google Scholar] [CrossRef]
- Shimizu, H.; Masujima, Y.; Ushiroda, C.; Mizushima, R.; Taira, S.; Ohue-Kitano, R.; Kimura, I. Dietary short-chain fatty acid intake improves the hepatic metabolic condition via FFAR3. Sci. Rep. 2019, 9, 16574. [Google Scholar] [CrossRef]
- Saika, A.; Nagatake, T.; Kunisawa, J. Host- and microbe-dependent dietary lipid metabolism in the control of allergy, inflammation, and immunity. Front. Nutr. 2019, 6, 36. [Google Scholar] [CrossRef]
- Lochmiller, R.L.; Deerenberg, C. Trade-offs in evolutionary immunology: Just what is the cost of immunity? Oikos 2000, 88, 87–98. [Google Scholar] [CrossRef]
- Rauw, W.M. Immune response from a resource allocation perspective. Front. Genet. 2012, 3, 267. [Google Scholar] [CrossRef] [PubMed]
- Frosali, S.; Pagliari, D.; Gambassi, G.; Landolfi, R.; Pandolfi, F.; Cianci, R. How the intricate interaction among toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J. Immunol. Res. 2015, 2015, 489821. [Google Scholar] [CrossRef]
- Mani, V.; Harris, A.J.; Keating, A.F.; Weber, T.E.; Dekkers, J.C.M.; Gabler, N.K. Intestinal integrity, endotoxin transport and detoxification in pigs divergently selected for residual feed intake. J. Anim. Sci. 2013, 91, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.L.; An, H.J.; Kim, J.; Canfield, D.R.; Torres, J.; Lebrilla, C.B.; Solnick, J.V. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology 2009, 137, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Day, C.J.; Semchenko, E.A.; Korolik, V. Glycoconjugates play a key role in Campylobacter jejuni infection: Interactions between host and pathogen. Front. Cell. Infect. Microbiol. 2012, 2, 9. [Google Scholar] [CrossRef]
- Quintana-Hayashi, M.P.; Mahu, M.; De Pauw, N.; Boyen, F.; Pasmans, F.; Martel, A.; Premaratne, P.; Fernandez, H.R.; Teymournejad, O.; Vande Maele, L.; et al. The levels of Brachyspira hyodysenteriae binding to porcine colonic mucins differ between individuals, and binding is increased to mucins from infected pigs with de novo MUC5AC synthesis. Infect. Immun. 2015, 83, 1610–1619. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 252, 1–8. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Nakanishi, N.; Tashiro, K.; Kuhara, S.; Hayashi, T.; Sugimoto, N.; and Tobe, T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 2019, 155, 521–530. [Google Scholar] [CrossRef]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 2018, 24, 296–307. [Google Scholar] [CrossRef]
- De Ridder, L.; Maes, D.; Dewulf, J.; Pasmans, F.; Boyen, F.; Haesebrouck, F.; Méroc, E.; Butaye, P.; Van der Stede, Y. Evaluation of three intervention strategies to reduce the transmission of Salmonella Typhimurium in pigs. Vet. J. 2013, 197, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Luethy, P.M.; Huynh, S.; Ribardo, D.A.; Winter, S.E.; Parker, C.T.; Hendrixson, D.R. Microbiota-derived short-chain fatty acids modulate expression of Campylobacter jejuni determinants required for commensalism and virulence. mBio 2017, 8, e00407-17. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543. [Google Scholar] [CrossRef] [PubMed]
- Marsh, A.J.; Azcarate-Peril, M.A. Hologenomics: The interaction between host, microbiome and diet. Reference Module in Food Science. 2020, 212–228. [Google Scholar] [CrossRef]
- Rychlik, J.L.; May, T. The effect of a methanogen, Methanobrevibacter smithii, on the growth rate, organic acid production, and specific ATP activity of three predominant ruminal cellulolytic bacteria. Curr. Microbiol. 2000, 40, 76–180. [Google Scholar] [CrossRef]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Fecal Microbiota Transplantation Workgroup. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef]
- Canibe, N.; O’Dea, M.; Abraham, S. Potential relevance of pig gut content transplantation for production and research. J. Anim. Sci. Biotechnol. 2019, 10, 55. [Google Scholar] [CrossRef]
- McCormack, U.M.; Curião, T.; Wilkinson, T.; Metzler-Zebeli, B.U.; Reyer, H.; Ryan, T.; Calderon-Diaz, J.A.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; et al. Fecal microbiota transplantation in gestating sows and neonatal offspring alters lifetime intestinal microbiota and growth in offspring. mSystems 2018, 3, e00134-17. [Google Scholar] [CrossRef]
- McCormack, U.M.; Curião, T.; Metzler-Zebeli, B.U.; Wilkinson, T.; Reyer, H.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; Gardiner, G.E.; Lawlor, P.G. Improvement of feed efficiency in pigs through microbial modulation via fecal microbiota transplantation in sows and dietary supplementation of inulin in offspring. Appl. Environ. Microbiol. 2019, 85, e01255-19. [Google Scholar] [CrossRef]
- Hu, J.; Ma, L.; Nie, Y.; Chen, J.; Zheng, W.; Wang, X.; Xie, C.; Zheng, Z.; Wang, Z.; Yang, T.; et al. A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe 2018, 24, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Metzler-Zebeli, B.U.; Canibe, N.; Montagne, L.; Freire, J.; Bosi, P.; Prates, J.A.M.; Tanghe, S.; Trevisi, P. Resistant starch reduces large intestinal pH and promotes fecal lactobacilli and bifidobacteria in pigs. Anim. Int. J. Anim. Biosci. 2017, 13, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Biagi, G.; Piva, A.; Moschini, M.; Vezzali, E.; Roth, F.X. Performance, intestinal microflora, and wall morphology of weanling pigs fed sodium butyrate. J. Anim. Sci. 2007, 85, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.L.; Sun, H.; Wu, J.; Niu, H.H.; Feng, J. Effects of sodium butyrate on growth performance, haematological and immunological characteristics of weanling piglets. J. Anim. Physiol. Anim. Nutr. 2014, 98, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Sun, J.; Li, M.; Xu, Q.; Zhang, X.; Tang, Z.; Chen, J.; Zhen, J.; Sun, Z. The effects of dietary sodium butyrate supplementation on the growth performance, carcass traits and intestinal microbiota of growing-finishing pigs. J. Appl. Microbiol. 2020, 128, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Hirschberg, S.; Gisevius, B.; Duscha, A.; Haghikia, A. Implications of diet and the gut microbiome in neuroinflammatory and neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 3109. [Google Scholar] [CrossRef]
- Tilocca, B.; Burbach, K.; Heyer, C.M.E.; Hoelzle, L.E.; Mosenthin, R.; Stefanski, V.; Camarinha-Silva, A.; Seifert, J. Dietary changes in nutritional studies shape the structural and functional composition of the pigs’ fecal microbiome-from days to weeks. Microbiome 2017, 5, 144. [Google Scholar] [CrossRef]
| Trait (Methodology for Structural and Functional Analysis of Gut Microbiome) | Sample Type (Total Number of Pigs) | Age of Pigs | α- and β-Diversity Effects in Animals with Better Production Traits | Genera/Species More Abundant in Animals with Better Production Traits, i.e., Heavier, Lower Back Fat (Leaner) | Genera/Species Less Abundant in Animals with Better Production Traits, i.e., Heavier, Lower Back Fat (Leaner) | Functional Analysis (↓ = Bacterial Pathways Less Abundant in Animals with Better Production Traits; ↑ = Pathways More Abundant in Animals with Better Production Traits) 1 | Ref |
|---|---|---|---|---|---|---|---|
| Body weight (16S rRNA gene sequencing (V4) on Illumina MiSeq + PICRUSt predictions) | Faeces (n = 18) | Day 63 of age | Higher α-diversity (ACE, Chao1, and observed OTU indices) for heavier pigs. β-diversity effects at phylum but not genus level | Anaerococcus Sediminibacterium Butyrivibrio | Lactococcus Anaerotruncus Eubacterium Bilophila Bacteroides Prevotella Corynebacterium | ↑dioxin, xylene, benzoate degradation ↓Nucleotide-binding oligomerization domain-like receptor (NLR) signalling ↓Alanine, aspartate, glutamate metabolism ↓Novobiocin biosynthesis | [5] |
| Back fat (16S rRNA gene sequencing (V4) on Illumina MiSeq + metagenomic shotgun sequencing on Illumina HiSeq 2000) | Jejunum (n = 8) | ~Day 300 of age | α-diversity not measured No β-diversity differences | Actinobacillus succinogenes Cellulosilyticum lentocellum Fusobacterium nucleatum Haemophilus influenzae Mannheimia succiniciproducens | Clostridium phytofermentans Lactobacillus johnsonii Escherichia fergusonii | ↑DNA repair and recombination ↑Pyrimidine metabolism ↑Secretion system ↑Lysozyme and chitinase degradation ↑Folate biosynthesis ↑Tropane, piperidine, pyridine alkaloid biosynthesis ↓Nucleotide excision and mismatch repair ↓Pyruvate, propanoate, cysteine, methionine metabolism | [6] |
| Ileum (n = 8) | ~Day 300 of age | α-diversity not measured No β-diversity differences | Actinobacillus succinogenes Cellulosilyticum lentocellum Bacteroides dorei Bacteroides fragilis Bacteroides helcogenes Bacteroides salanitronis Bacteroides thetaiotaomicron Bacteroides vulgatus Bacteroides xylanisolvens Barnesiella viscericola Prevotella dentalis Prevotella denticola Prevotella ruminicola | Chamaesiphon minutus Clostridium difficile Clostridium sordellii Escherichia coli Sphaerochaeta coccoides Vibrio harveyi | ↑Glycoside hydrolase ↑Glycan degradation ↑Pyrimidine metabolism ↑Chaperones and folding catalysts ↑Bacterial toxins ↑Glycosaminoglycan degradation ↑Glycosphingolipid biosynthesis ↑Lysosome ↑Protein digestion and absorption ↓Tropane, piperidine, pyridine alkaloid biosynthesis ↓Chitin and peptidoglycan cleaving ↓Antigen processing and presentation ↓Influenza A ↓MAPK signalling pathway ↓Endocytosis | ||
| Caecum (n = 8) | ~Day 300 of age | α-diversity not measured β-diversity differences between high- and low-fatness pigs | Bacteroidesdorei Bacteroides fragilis Bacteroides helcogenes Bacteroides salanitronis Bacteroides thetaiotaomicron Megamonas hypermegale Megasphaera elsdenii Prevotella ruminicola Treponema succinifaciens | Escherichia coli Eubacteriumrectale Oscillibacter valericigenes Parabacteroides distasonis Roseburia hominis Roseburia intestinalis | ↑Chaperones and folding catalysts ↑Glycosaminoglycan degradation ↑Glycosphingolipid biosynthesis ↑Glycoside hydrolase ↑Adipocytokine signalling ↑Cellular antigens ↑Protein digestion and absorption ↓Lipopolysaccharide biosynthesis ↓Cytoskeleton proteins ↓Two-component system ↓Transporters ↓Benzoate degradation ↓Flagellar assembly ↓Bacterial chemotaxis |
| Trait (Methodology for Structural and Functional Analysis of Gut Microbiome) | Sample Type (Total Number of Pigs) | Age of Pigs | α- and β-Diversity Effects in More Feed-Efficient Animals | Genera/Species More Abundant in More Feed-Efficient Animals | Genera/Species Less Abundant in More Feed-Efficient Animals | Functional Analysis (↓ = Bacterial Pathways Less Abundant in More Feed-Efficient Animals; ↑ = Pathways More Abundant in More Feed-Efficient Animals) 1 | Ref |
|---|---|---|---|---|---|---|---|
| Residual feed intake (16S rRNA gene sequencing (V3-V4) on Illumina MiSeq + PICRUSt predictions) | Faeces (n = 32 at each time point) | Weaning (~Day 27 of age) | No effect | Bacteroides | No effect | [23] | |
| D42 pw 2 | Streptococcus | No effect | |||||
| Day 138 pw | 3 UN_Christensenellaceae Cellulosilyticum UN_vadinBB60 Bacteroides | Clostridium sensu stricto 1 Can. Saccharimonas | ↑RE 4 processing ↓Biosynthesis of 2° metabolites | ||||
| Caecum (n = 30) | Day 139 pw | No effect | Actinobacillus porcinus | Solobacterium | ↓Thiamine metabolism | ||
| Ileum (n = 24) | Day 139 pw | No effect | Oscillibacter Methanobrevibacter Treponema berlinense | Rhodococcus Methanosphaera Clostridiales | ↑Bacterial invasion ↑Biosynthesis of amino acids (phenylalanine, tyrosine, tryptophan, valine, leucine, isoleucine) ↑Metabolism of C5-branched dibasic acid, terpenoids, polyketides ↑RE processing ↓PTS 5 | ||
| Residual feed intake (16S rRNA gene sequencing (V3-V4) on Illumina MiSeq + PICRUSt predictions) (3 different geographic locations (AT 7, NI 8, ROI 9, with 2 batches in ROI (ROI1, ROI2)) | Faeces (n = 100) | Day 70 of age | No effect | UN_S24.7 (AT) Mucispirillum (NI, AT) | UN_S24.7 (NI) | No common effects | [24] |
| Day 134 of age | No effect | Roseburia (NI) Treponema brennaborense (AT) Methanobrevibacter (ROI1 and 2) UN_BS11.gut.group (ROI2) UN_RF16 (ROI1, AT) Alistipes (ROI2) Sedimentibacter (ROI2) UN_Family XIII (ROI2) Oribacterium (NI) Pseudobutyrivibrio (NI) Papillibacter (ROI2) 10 UNID_vadinBB60 (ROI2) UN_RF12 gut group (ROI1) UN_WCHB1.25 (ROI1) Leeia (ROI1) UN_Succinivibrionaceae (ROI) UN_TA18 (ROI2) Desulfovibrio (AT) UN_RF9 (AT) Akkermansia (ROI1) | Mogibacterium (NI) Ruminococcaceae IS 6 (ROI1) UN_Ruminococcaceae (NI) UN_Erysipelotrichaceae (NI) Alloprevotella (ROI2) UN_Bacteroidetes (ROI1) Can. Saccharimonas (ROI2) UN_Defluviitaleaceae (ROI1) Lachnospira (ROI1) Marvinbryantia (ROI2) Oribacterium (ROI1) Faecalibacterium(ROI2) Ruminococcus(NI) Subdoligranulum (ROI2) Asteroleplasma (ROI2) Erysipelotrichaceae IS (ROI1) Allisonella (ROI1) Mitsuokella (ROI1) Selenomonas (ROI2) UN_Veillonellaceae (ROI2) | ↑Biosynthesis of unsaturated fatty acids (only common results shown, i.e., these followed the same trend in both ROI batches) | |||
| Ileum (n = 76) (NI pigs not sampled) | Day 134 of age | Higher α-diversity (Shannon and Simpson indices) (ROI2) No effect for β-diversity | Dietzia (ROI2) Leucobacter (ROI2) Rothia (ROI2) Saccharopolyspora (ROI2) UN_S24.7 (ROI2) UN_Gastranaerophilales (ROI2) Oceanobacillus (ROI2) Paucisalibacillus (ROI2) Dolosicoccus (ROI2) Globicatella (ROI2) Enterococcus (ROI2) Clostridium sensu stricto 1(ROI1) Blautia (ROI2) Pseudobutyrivibrio (ROI2) Subdoligranulum (ROI2) UN_vadinBB60 (ROI2) Turicibacter (ROI1) Solobacterium (ROI2) Dialister (ROI2) Mitsuokella(ROI2) p1088 a5 gut group (ROI2) Desulfovibrio (ROI2) Actinobacillus (ROI2) Spirochaeta (ROI2) | UN_vadinBB60 (ROI1) Helicobacter (ROI1) | No common effects | ||
| Caecum (n = 76) (NI pigs not sampled) | Day 134 of age | Higher α-diversity (Shannon and Simpson indices) (ROI2) No effect for β-diversity | Microbacterium (ROI2) UN_Coriobacteriaceae (ROI2) Paludibacter (ROI2) UN_RF16 (ROI1, ROI2) dgA 11 gut group (ROI2) RC9 gut group (ROI2) UN_4c0d (AT) UN_Gastranaerophilales (ROI2) UN_Erysipelotrichaceae (ROI2) Asteroleplasma (ROI2) UN_Family XIII (ROI2) Peptococcus (ROI2) Ruminococcaceae IS (ROI2) Papillibacter (ROI2) UN_vadinBB60 (ROI2) UN_RF12 gut group (ROI2) Sutterella (ROI1) Noviherbaspirillum (ROI2) UN_GR WP33 58 (ROI2) Helicobacter (ROI2) Ruminobacter (ROI1) | Butyrivibrio (ROI2) Oscillospira (ROI1) Streptococcus (ROI2) UN_Veillonellaceae (ROI1) | ↑Inositol phosphate metabolism ↓Porphyrin/chlorophyll metabolism (only common results shown, i.e., these followed the same trend in both ROI batches) | ||
| Feed conversion ratio ((16S rRNA gene sequencing (V4–V5) on Illumina HiSeq+ PICRUSt predictions)) | Ileum (n = 24) | Day 141–142 of age | No effect | Streptococcus Ruminococcaceae spp. Christensenellaceae R7 | Roseburia Prevotellaceae spp. Rikenellaceae RC9 | ↓Infectious diseases | [25] |
| Caecum (n = 24) | Day 141–142 of age | Higher α-diversity (Shannon index) | Eubacteriumhallii Clostridium sensu stricto1 Lachnospiraceae sp. Dialister Rikenellaceae RC9 Prevotellaceae sp. Ruminiclostridium Treponema Ruminococcus p-1088-a5 gut group | Bacteroides Parabacteroides Eubacterium coprostanoligenes Sutterella Lachnoclostridium | ↑Carbohydrate metabolism ↑Lipid metabolism | ||
| Colon (n = 24) | Day 141–142 of age | Higher α-diversity (Shannon and Chao1 indices) No effect for β-diversity | Ruminococcaceae spp. Ruminococcus Prevotella Rikenellaceae RC9 Anaerotruncus Eubacterium coprostanoligenes Lawsonia Dielma Treponema Hydrogenoanaerobacterium Ruminiclostridium | Bacteroides Streptococcus Lachnospiraceae sp. Macrococcus Ruminiclostridium Streptococcus Prevotellaceae spp. | ↑Metabolism of amino acids ↑Signalling molecules and interaction ↑Metabolism of cofactors and vitamins ↑Digestive system ↑Glycan biosynthesis and metabolism ↑Folding, sorting, degradation ↑Immune system ↑Metabolism of terpenoids and polyketides | ||
| Residual feed intake 2 farms ((16S rRNA gene sequencing (V3–V4) on Illumina MiSeq + PICRUSt predictions)) | Colon (n = 40) | Day 105–141 of age | No effect | Ruminococcus flavefaciens | Collinsella | No effect | [26] |
| Feed conversion ratio (Metagenomic shotgun sequencing on Illumina platform) | Caecum (n = 6 pigs from [22]) | Day 141–142 of age | β-diversity differences No effect for α-diversity | Lactobacillus johnsonii Acetatifactor muris Firmicutes bacterium CAG:110 Firmicutes bacterium CAG:124 Firmicutes bacterium CAG:475 Treponema bryantii Treponema berlinense Treponema succinifaciens Treponema socranskii Treponema saccharophilum Treponema brennaborense Butyricicoccus_porcorum Clostridiales_bacterium Bacteroidales WCE2004 Methanogenic archaeon Clostridia bacterium Alistipes sp. Eubacterium sp. CAG:180 Verrucomicrobia Faecalibacterium sp. Bacteroides sp. Clostridium Fibrobacter Ruminococcus Roseburia Butyrivibrio Desulfurispirillum Paludibacter Blautia Methanobrevibacter | Prevotella sp. Bacteroides sp. Escherichia coli Prevotella stercorea Prevotella bryantii Prevotella ruminicola Prevotella timonensis Succinivibrio dextrinosolvens Succinatimonas sp. Clostridium sp. | ↑Metabolism of protein, nucleotide, cofactors, vitamins ↑Monosaccharide and energy transportation ↑13 Antibiotic resistance genes ↓12 Antibiotic resistance genes ↓Genetic information processing ↓Amino sugar and nucleotide sugar metabolism ↓Transport system | [27] |
| Feed conversion ratio (Metagenomic shotgun sequencing on Illumina HiSeq 2500) | Caecum (n = 8) | Day 166 of age | β-diversity differences α-diversity not measured | Acetivibrio ethanolgignens Lactobacillus ruminis Lactobacillus amylophilus Lactobacillus sp. Butyricicoccus sp. UN_Butyricicoccus sp. Clostridium bolteae Ruminococcus sp. Clostridium clostridioforme Clostridium saccharolyticum Christensenella sp. Erysipelotrichaceae sp. Roseburia intestinalis Coprococcus spp. Lachnospiraceae sp. Roseburia hominis Eubacterium siraeum Butyrate producing bacterium | Prevotella | ↑ABC transporters ↑Bacterial chemotaxis ↑D-arginine and D-ornithine metabolism ↑Flagellar assembly ↑Lysine degradation ↑Phenylalanine metabolism ↑Sulphur relay system ↑Synthesis and degradation of ketone bodies ↑Two-component system ↓Nitrogen metabolism ↓Glycan degradation | [28] 11 |
| Residual feed intake (16S rRNA gene sequencing (V4) on Illumina MiSeq and metagenomic shotgun sequencing on Illumina HiSeq 2500) | Faeces (n = 18) | Day 140 of age | Not determined | Aequorivita sublithincola Ammonifex degensii Akkermansia Bacteroidales bacterium CF Brachyspira pilosicoli Butyrate-producing bacterium Clostridium cellulosi Clostridium clariflavum Coriobacterium glomerans Desulfatibacillum alkenivorans Desulfovibrio desulfuricans Desulfovibrio vulgaris Eggerthella lenta Ethanoligenens harbinense Heliobacterium modesticaldum Lactobacillus casei Methylotenera versatilis methao valericigenes Paenibacillus graminis Paenibacillus sabinae Paenibacillus sp. Paenibacillus stellifer Pelobacter propionicus Podospora anserina Propionibacterium propionicum Pseudomonas fluorescens Pseudomonas mendocina Rikenellaceae bacterium M3 Streptococcus gordonii Symbiobacterium thermophilum Syntrophobotulus glycolicus Tepidanaerobacter acetatoxydans Thermacetogenium phaeum Treponema denticola | Bordetella avium Prevotella Faecalibacterium | ↑Nitrogen metabolism ↑Amino acid metabolism ↑Transport system ↑PPAR signalling pathway ↑Amino acid metabolism ↑Fatty acid metabolism ↑Adipocytokine signalling pathway ↓Metabolism and transport of monosaccharide ↓Signal transduction ↓Bacterial PTS ↓Phosphonate and phosphinate metabolism | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardiner, G.E.; Metzler-Zebeli, B.U.; Lawlor, P.G. Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review. Microorganisms 2020, 8, 1886. https://doi.org/10.3390/microorganisms8121886
Gardiner GE, Metzler-Zebeli BU, Lawlor PG. Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review. Microorganisms. 2020; 8(12):1886. https://doi.org/10.3390/microorganisms8121886
Chicago/Turabian StyleGardiner, Gillian E., Barbara U. Metzler-Zebeli, and Peadar G. Lawlor. 2020. "Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review" Microorganisms 8, no. 12: 1886. https://doi.org/10.3390/microorganisms8121886
APA StyleGardiner, G. E., Metzler-Zebeli, B. U., & Lawlor, P. G. (2020). Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review. Microorganisms, 8(12), 1886. https://doi.org/10.3390/microorganisms8121886






