Abstract
Iron acquisition pathways have often been considered to be gateways for the uptake of antibiotics into bacteria. Bacteria excrete chelators, called siderophores, to access iron. Antibiotic molecules can be covalently attached to siderophores for their transport into pathogens during the iron-uptake process. P. aeruginosa produces two siderophores and is also able to use many siderophores produced by other bacteria. We investigated the phenotypic plasticity of iron-uptake pathway expression in an epithelial cell infection assay in the presence of two different siderophore–antibiotic conjugates, one with a hydroxamate siderophore and the second with a tris-catechol. Proteomic and RT-qPCR approaches showed that P. aeruginosa was able to sense the presence of both compounds in its environment and adapt the expression of its iron uptake pathways to access iron via them. Moreover, the catechol-type siderophore–antibiotic was clearly more efficient in inducing the expression of its corresponding transporter than the hydroxamate compound when both were simultaneously present. In parallel, the expression of the proteins of the two iron uptake pathways using siderophores produced by P. aeruginosa was significantly repressed in the presence of both conjugates. Altogether, the data indicate that catechol-type siderophores are more promising vectors for antibiotic vectorization using a Trojan-horse strategy.
1. Introduction
Antibiotic resistance is a complex and growing problem for human health. In 2017, the World Health Organization published a list of highly resistant bacteria for which new antibiotics are urgently needed. The most critical pathogens on this list are Gram-negative bacteria, such as Acinetobacter, Pseudomonas, and various enterobacteria (including Klebsiella, Escherichia coli, Serratia, and Proteus). The bottleneck in the development of new antibiotics for Gram-negative bacteria is the need for these drugs to cross the outer membrane. The use of nutrient-importation pathways as gateways for the uptake of antibiotics has often been proposed to bypass the low permeability of the Gram-negative cell wall [1].
Iron is a key nutrient, as this metal is involved in many crucial biological processes, and is therefore essential for bacterial growth and virulence. To access iron, bacteria produce siderophores, small molecules with a very high affinity for ferric iron [2]. These iron chelators are synthesized by bacteria and released into their environment to scavenge iron. The ferric complexes that are formed are recovered by specific transporters [3]. Antibiotics can be covalently linked to siderophores with the idea that they will be transported into the pathogens during the ferri-siderophore uptake process—referred to as a Trojan-horse strategy [1,4,5]. One such siderophore–antibiotic conjugate (Cefiderocol), developed by Shionogi, was approved by the US Food and Drug Administration (FAD) in November 2019 for the treatment of complicated urinary tract infections, showing that Trojan-horse strategies can be successful [6]. Moreover, evolution has also developed natural siderophore–antibiotic conjugates (also called sideromycins), demonstrating the relevance of such a strategy. The archetypes among sideromycins are albomycins [7,8], ferrimycins [9], danomycins [10], and salmycins [11], isolated mainly from streptomycetes or actinomycetes and produced to kill other microorganisms and dominate a given microbiota.
One major bottleneck limiting the development of siderophore–antibiotic conjugates is that all bacteria are equipped with several iron-uptake pathways. Consequently, bacteria can switch off the expression of the targeted iron uptake pathway and express an alternative one, possibly leading to the rapid development of resistance strategies. For example, P. aeruginosa carries genes within its genome for at least 20 different iron-uptake strategies [12]: (i) one ferrous (Fe2+) iron uptake pathway, (ii) three heme acquisition pathways, (iii) ferric iron (Fe3+) uptake pathways by the two siderophores pyoverdine (PVD) and pyochelin (PCH), produced by the pathogen itself, and (iv) at least 10 different “siderophore piracy” strategies to uptake Fe3+ using xenosiderophores (siderophores produced by other bacteria). Indeed, most bacteria can use siderophores produced by other bacteria in multispecies communities and during infections by expressing compatible transporters that are able to capture and import ferric complexes they cannot synthesize themselves [13,14,15,16]. Except for ferrous-iron uptake, bacterial iron-uptake pathways always include an outer membrane transporter (OMT) involved in ferri-siderophore or heme uptake, enzymes involved in iron release from the siderophore or heme, and inner membrane transporters (ATP Binding cassette transporters of proton motive force-dependent permeases). These proteins, especially the OMTs, are always specific for one siderophore or a family of structurally-related siderophores [3,17]. Consequently, bacteria able to access iron via several siderophores carry genes within their genome that encode an OMT specific for each siderophore [3].
In contrast to the large amount of genomic data generated by bacterial genome sequencing, almost no data are available concerning the phenotypic plasticity associated with the expression of these various iron-uptake pathways. Uptake pathways are considered to be generally expressed at a basal level and pathogens only induce the expression of the most efficient pathway(s) for iron acquisition depending on the bacterial environment [16]. It has also been shown that bacteria are often able to sense the presence of xenosiderophores and express the corresponding transporters, allowing them to access iron via these xenosiderophores [16,18,19,20,21,22]. Bacteria are also able to switch from the expression of one iron-uptake pathway to another in response to synthetic iron chelators, as shown for P. aeruginosa cells with an analogue of the siderophore enterobactin (ENT) [23]. Recently, we showed by proteomics and RT-qPCR that tris-catechol type siderophores, such as ENT or vibriobactin (VIB), induce the transcription and expression of the genes involved in their corresponding uptake pathways in P. aeruginosa more efficiently than tris-hydroxamate siderophores, such as ferrichrome [16]. The data also showed that the presence of different iron uptake pathways in the genome of a pathogen involves high phenotypic plasticity in response to diverse environmental stimuli [16]. Such phenotypic plasticity plays a key role in the ability of siderophore-antibiotic conjugates to be transported efficiently into the target bacteria. If the right uptake pathway is not expressed, the drugs have no chance of being efficiently transported into the bacteria by their associated siderophore. Here, we investigated, for the first time, the phenotypic adaptation of P. aeruginosa to the presence of two different siderophore–antibiotic conjugates in an epithelial cell infection assay using proteomics and RT-qPCR. We used the natural sideromycin albomycin (ALBO) (Figure 1a), for which the siderophore component is similar to ferrichrome (FERRI), a siderophore that chelates iron via three hydroxamate groups. For the second conjugate, we used the synthetic compound TCVL6, corresponding to linezolid vectorized by a tris-catechol siderophore (TCV) analogue of ENT (Figure 1a). Both compounds have no antibiotic activity on the pathogen at the concentrations used here [24,25] and could therefore be used as tools to investigate how the bacteria adapts the expression of its various iron-uptake pathways in the presence of such Trojan-horse compounds. We show that both compounds were able to promote P. aeruginosa growth in iron-restricted conditions, ALBO by the FERRI-dependent uptake pathway (Figure 1b) and TCVL6 by the ENT-dependent uptake pathway (Figure 1c). Both compounds were able to induce the transcription and expression of the genes encoding proteins involved in their corresponding uptake pathways (the ENT-dependent uptake pathway for TCVL6 and the FERRI-dependent uptake pathway for ALBO), indicating that P. aeruginosa PAO1 is able to sense the presence of these two compounds and use them to access iron. This occurred in parallel with a decrease in the transcription and expression of the proteins of the endogenous siderophore-dependent iron uptake pathways (PVD and PCH), as well as that of virulence factors. Finally, when bacteria were simultaneously in the presence of both ALBO and TCVL6, the proteins of the ferri-TCVL6 uptake pathway were more highly expressed than those of the ferri-ALBO uptake pathway, indicating that between the two compounds, P. aeruginosa preferentially uses TCVL6 to access iron.
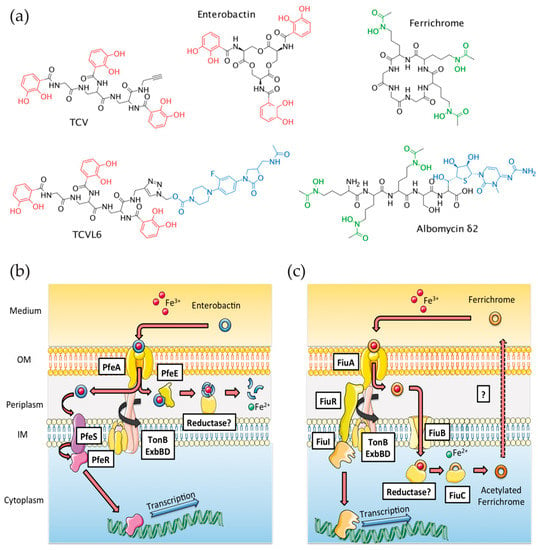
Figure 1.
(a) Structure of ferrichrome (FERRI), albomycin (ALBO), enterobactin (ENT), tris-catechol vector (TCV), and tris-catechol-linezolid vector (TCVL6). (b,c) ENT- and FERRI-dependent iron-uptake pathways in P. aeruginosa, respectively. FERRI and ENT loaded with iron are recognized at the bacterial surface by the outer membrane transporters (OMTs) FiuA and PfeA, respectively, and transported across the outer membrane. Iron release from ENT occurs in the bacterial periplasm and involves hydrolysis of the siderophore by the esterase PfeE and an iron-reduction step by an unidentified reductase [26]. ENT-Fe does not enter the bacterial cytoplasm in P. aeruginosa cells. Concerning FERRI-Fe, once in the bacterial periplasm, the complex is transported further across the inner membrane by the proton motive-dependent permease FiuB and iron is released from FERRI in the bacterial cytoplasm by a mechanism involving acetylation of the siderophore by FiuC and iron reduction by an unknown reductase [14,27]. OM for outer membrane and IM for inner membrane.
2. Materials and Methods
2.1. Chemicals
Enterobactin (ENT) and ferrichrome (FERRI) were obtained from Sigma-Aldrich. Albomycin δ2 (ALBO) was purchased from EMC Microcollection. Pyoverdine (PVD) was purified from P. aeruginosa PAO1 cultures as previously described [28]. TCV was synthesized according to a previously published protocol [29] and TCVL6 was synthesized as described in Supporting information. The protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) was purchased from Sigma-Aldrich. 55FeCl3 was obtained from Perkin Elmer Life and Analytical Sciences (Billerica, MA, USA), at a concentration of 71.1 mM, with a specific activity of 10.18 Ci/g. RPMI was purchased from Thermo-Fisher.
2.2. Bacterial Strains, Plasmids, and Growth Conditions
The P. aeruginosa strains used in this study are all listed in Table S1 in Supplemental Materials. P. aeruginosa strains were grown at 30 °C in LB broth. When P. aeruginosa strains were grown in iron-restricted conditions, CAA medium was used (casamino acid medium, composition: 5 g L−1 low-iron CAA (Difco), 1.46 g L−1 K2HPO4 3H2O, and 0.25 g L−1 MgSO4 7H2O).
2.3. Iron Scavenging from PVD-Fe
Compounds were prepared in solution at 10 mM in DMSO (TCV and TCVL6) or water (FERRI, ALBO, and PVD). The competition assay with PVD-Fe was carried out as described by our team previously [16]. For the kinetic of iron scavenging from PVD-Fe, PVD-Fe at 10 µM in 100 µL of HEPES buffer was incubated at 25 °C in the presence of 100 µM FERRI, ALBO, ENT, or TCVL6 and the kinetic of iron dissociation from PVD followed as described previously [16].
2.4. Growth Assays in Iron-Restricted Conditions
For P. aeruginosa growth assays in microplate, a first overnight culture was carried out at 30 °C in 10 mL of LB broth; afterwards, bacteria were washed and a second overnight culture was carried out in 20 mL CAA medium at 30 °C. Following that, bacteria were washed, resuspended in CAA medium at 0.02 OD at 600 nm, and distributed in the wells of a 96-well plate (Greiner, U-bottomed microplate) in the absence or presence of 10 µM of FERRI, ALBO, ENT, TCV, or TCVL6. The plate was incubated at 30 °C, with shaking, in a Tecan microplate reader (Infinite M200, Tecan) and bacterial growth was monitored at OD600 nm. We calculated the mean of three replicates for each measurement.
2.5. Iron Uptake
ALBO-55Fe complexes were prepared at 55Fe concentrations of 50 µM, with a ALBO:iron (mol:mol) ratio of 20:1. P. aeruginosa strains were first grown overnight at 30 °C in LB broth at 30 °C, and afterwards overnight in CAA medium at 30 °C, and at last, overnight in CAA medium with 10 µM ALBO at 30 °C. The bacteria were then used for 55Fe uptake kinetics as described previously [23], in the absence and presence of 200 µM CCCP (a proton motive force inhibitor [30]).
2.6. Infection Assay
A549 (ATCC® CCL-185™) human pulmonary epithelial cells were grown as previously described [16] in RPMI 1640 medium (Gibco) supplemented with 10% vol/vol FBS (Gibco) [16]. A549 cells infections were also carried out exactly, as we described previously [16], with P. aeruginosa PAO1 cells grown in LB medium and with a multiplicity of infection of 50; 10 µM FERRI, ALBO, ENT, TCV, or TCVL6 were added to the A549 cells at the same moment as P. aeruginosa cells. After 3 h of infection at 37 °C, 5% CO2, the plates were washed with cold PBS 1X buffer and the A549 cells and bacteria still adhering to the bottom of the plate harvested, and prepared for RT-qPCR and proteomic analyses, as described previously [16].
2.7. Quantitative Real-Time PCR on Bacteria Grown in the Presence of TCVL6
This RT-qPCR assay was carried out as described previously for TCV in Gasser et al.’s study [23] on PAO1 cells grown in CAA medium, in the presence of 10 µM TCVL6.
2.8. Quantitative Real-Time PCR on Bacteria Infecting Epithelial Cells
A549 cells were infected as described above, in the absence or presence of FERRI, ENT, TCV, ALBO or TCVL6. The sample preparation and RT-qPCR analyses were carried out as previously described by our team [16]. The primers used are given in Table S2 in Supplementary Materials as well as the uvrD mRNA used as an internal control to normalize the transcript levels for a given gene in a given strain. The data are expressed as log2 of the ratio (fold-change) relative to the conditions without conjugates or siderophores.
2.9. Label-Free Proteomic Analysis on Bacteria Infecting Epithelial Cells
A549 cells were infected as described above, in the absence and presence of conjugates, FERRI, ENT, or TCV. The pellets of A549 cells and bacteria (five independent biological replicates) in PBS were prepared for proteomic analyses, as described by our team previously [16]. The resulting peptides were submitted to nanoLC-MS/MS analyses (750 ng injected) on a nanoACQUITY Ultra-Performance-LC systems hyphenated to a Q-Exactive Plus mass spectrometer, as previously described [16]. The raw data obtained were converted into “.mfg” files with MSConvert software (ProteomeWizard, version 3.0.6090) and analyzed as previously described [16].
3. Results
3.1. Ability of ALBO and TCVL6 to Compete for Iron with PVD
The efficient uptake of siderophore–antibiotic conjugates into bacteria requires that various conditions be met. Among them, the siderophore moiety must have a high affinity for iron and be able to compete for this metal with other siderophores that may be present in the bacterial environment, such as the siderophores produced by the pathogens themselves. P. aeruginosa produces two siderophores, PVD and PCH, with PVD having a higher affinity for iron than PCH (Ka of 1030.8 M−1 for PVD vs. 1018 M−2 for PCH [31,32]. Apo PVD also has specific spectral properties, with a characteristic absorbance peak at 400 nm (pH 7.0) and fluorescence emission at 447 nM, whereas its fluorescence is quenched when complexed with iron [33]. As we have described previously, these spectral properties can be used to compare the ability of various chelators to scavenge iron [16]. Here we used them to monitor the competition between PVD and the compounds FERRI, ALBO, TCV, and TCVL6 for iron (Figure 2). PVD-Fe (non-fluorescent) at 10 µM was incubated in the presence of increasing concentrations of apo FERRI, ALBO, TCV, and TCVL6 (Figure 2a). The complete removal of iron from PVD was only observed for the catechol-type compounds (TCV and TCVL6) at concentrations of approximately 20 µM (two-fold excess compared to PVD-Fe; Figure 2a). Neither FERRI nor ALBO were able to completely remove iron from PVD at the concentrations tested, (70–80% of the PVD was still in its ferric form, Figure 2a), indicating that these compounds compete less efficiently for the scavenging of iron than catechol-type siderophores or conjugates. We also monitored the kinetics of PVD-Fe dissociation by incubating 10 µM PVD-Fe with 100 µM FERRI, ALBO, TCV, or TCVL6 (Figure 2b) and confirmed that catechol-type compounds clearly compete more efficiently for iron with PVD than hydroxamate molecules. These data are consistent with the affinities of these families of siderophores for iron: Ka of 1049 M−1 for ENT [34], a tricatechol compound like TCV, 1029 M−1 for FERRI [35], and 1030.8 M−1 for PVD [31]. Furthermore, they show that the presence of the antibiotic moiety does not significantly affect the chelation properties of the vector moiety.
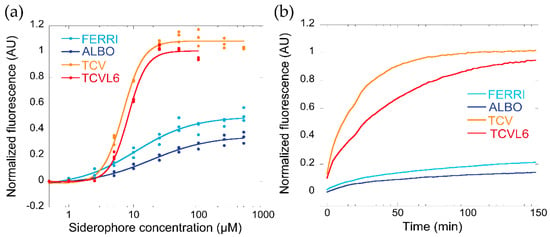
Figure 2.
(a) Ability of FERRI, ALBO, TCV, and TCVL6 to scavenge iron from PVD-Fe complexes. Ten micromoles of PVD-Fe in 100 mM HEPES pH 7.4 were incubated with increasing concentrations of FERRI, ALBO, TCV, or TCVL6 for 48 h (until equilibrium was reached), as described previously [16]. Metal-free PVD is fluorescent, with absorbance typically at 447 nm, and PVD-Fe is not [33]. Apo PVD formation was thus followed by monitoring its fluorescence at 447 nm (excitation at 400 nm). The data were normalized using the formula (FMEASURED—FPVD-Fe)/(FPVD—FPVD-Fe), FMEASURED being the fluorescence measured for each experimental condition, FPVD-Fe the fluorescence of 10 µM PVD-Fe, and FPVD the fluorescence of 10 µM PVD. (b) Kinetics of PVD-Fe dissociation in the presence of FERRI, ALBO, TCV, or TCVL6. Ten micromoles of PVD-Fe in 100 mM HEPES buffer pH 7.4 was incubated with 100 µM FERRI, ALBO, TCV, or TCVL6, as described in Materials and Methods. The kinetics of apo PVD formation were followed by monitoring the fluorescence emission at 447 nm (excitation at 400 nm), as previously described [16].
3.2. ALBO and TCVL6 Both Transport Iron into P. aeruginosa Cells
Previous studies have shown that FERRI and ENT are both able to transport Fe into bacteria via the OMT FiuA for FERRI and PfeA for ENT [13,14]. Nothing has been described in the literature about the ability of ALBO to transport iron into P. aeruginosa cells. Consequently, we carried out an 55Fe uptake assay on iron-starved P. aeruginosa cells that were unable to produce PVD or PCH (∆pvdF∆pchA strain [23]) with ALBO as a siderophore (Figure 3a). The ∆pvdF∆pchA mutant was used to avoid any iron uptake via the siderophores produced by the bacteria itself. We observed an uptake rate of 150 pmol 55Fe transported per OD600 nm per min, showing that ALBO is able to transport iron into P. aeruginosa cells. Uptake was completely abolished in the presence of carbonyl cyanide m-chlorophenyl hydrazine (CCCP), a proton motive force inhibitor [30], indicating that such uptake is not due to diffusion via porins across the outer membrane but is TonB-dependent. TonB is an inner-membrane protein that spans into the periplasm and provides the energy to the ferri-siderophore OMTs for uptake across the outer membrane [36,37]. The deletion of fiuA (FERRI OMT) resulted in 40% inhibition of 55Fe uptake at 2 h. These data show that ferric iron-loaded ALBO uptake occurs via FiuA and at least one other OMT.
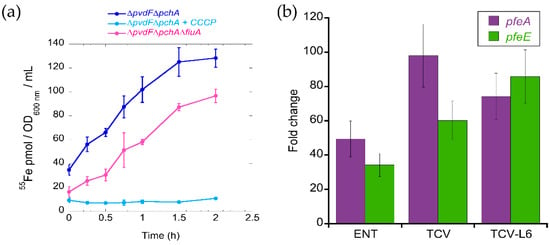
Figure 3.
(a) 55Fe uptake in P. aeruginosa strains using ALBO as a siderophore. ∆pvdF∆pchA and its isogenic mutant ∆pvdF∆pchA grown in iron-restricted casamino acid medium (CAA) medium, in the presence of 10 µM ALBO to induce any needed OMT, were incubated with 500 nM ALBO-55Fe and the kinetics of 55Fe uptake measured as previously described [23]. As a control, the experiment was repeated in the presence of 200 μM carbonyl cyanide m-chlorophenyl hydrazine (CCCP) protonophore for ∆pvdF∆pchA cells. Errors bars were calculated from three independent biological replicates. (b) Analysis of changes in the transcription of the pfeA and pfeE genes. RT-qPCR was performed on RNA from P. aeruginosa PAO1 cells grown in CAA medium, with or without supplementation with 10 µM ENT, TCV, or TCVL6 conjugate. The data are normalized relative to the reference gene uvrD. The results show the ratio between the values obtained in the presence of ENT, TCV or TCVL6 over those obtained in the absence of the compounds. The data are representative of three independent experiments performed in triplicate (n = 3). pfeA encodes the OMT of ENT and pfeE the esterase involved in ENT hydrolysis.
For TCVL6, no 55Fe uptake assays could be carried out because of small precipitation of the conjugates generating a significant background noise in the radioactivity signal. Consequently, we used an indirect approach to show that TCVL6 is able to cross the bacterial outer membrane. Indeed, previous proteomic and RT-qPCR analyses of P. aeruginosa grown in iron-restricted medium have shown that TCV is able to enter P. aeruginosa cells via the OMT PfeA, as the presence of this compound induces the expression of PfeA and PfeE, two proteins of the ENT-Fe uptake pathway [23]. Such induction involves an interaction in the periplasm between TCV and the inner membrane sensor PfeS of the two-component system PfeS/PfeR [18,23]. We used the same approach here. P. aeruginosa PAO1 cells were grown in the presence of TCVL6 and the transcription of pfeA and pfeE analyzed by RT-qPCR (Figure 3b). We observed a strong induction of the transcription of the pfeA and pfeE genes in the presence of TCVL6 with an efficiency equivalent to that for the vector TCV, without the antibiotic component, and higher efficiency than that for ENT. Such induction of pfeA and pfeE transcription is a proof that TCVL6 is able to cross the outer membrane of P. aeruginosa cells and interact with the two-component system PfeS/PfeR in the periplasm.
We also evaluated the ability of ALBO and TCVL6 to provide bacteria with iron by carrying out growth assays on P. aeruginosa cells unable to produce PVD or PCH (∆pvdF∆pchA strain) in iron-restricted CAA medium, in the presence or absence of 10 µM ENT, TCV, TCVL6, FERRI, or ALBO (Figure 4). The same assays were also carried out with various OMT mutants to identify the transporter involved in the uptake. As these growth assays were carried out using strains unable to produce siderophores, all iron present in the media was scavenged by ENT, TCV, TCVL6, FERRI, or ALBO. Consequently, bacteria only grew if they expressed the OMT necessary to import the ferric forms of these compounds. The ∆pvdF∆pchA strain grew in the presence of all tested compounds (Figure 4), showing that P. aeruginosa is able to access iron by using them as siderophores and that neither ALBO nor TCVL6 had antibiotic activity at the concentrations tested. Growth was still observed in the presence of ENT, TCV, and TCVL6 with the ∆pvdF∆pchA∆pfeA and ∆pvdF∆pchA∆pirA mutants, but not with ∆pvdF∆pchA∆pfeA∆pirA cells, indicating that both PfeA and PirA are involved in iron uptake by these three compounds. Both transporters had to be deleted to stop the bacteria from growing in the presence of the catechol compounds. Concerning FERRI and ALBO, we observed growth for ∆pvdF∆pchA and ∆pvdF∆pchA∆fiuA, showing that although FiuA is able to transport iron into bacteria (Figure 3b), at least one other unidentified OMT can perform the same function. In conclusion, ENT, TCV, and TCVL6 are able to transport iron into P. aeruginosa cells via the OMTs PfeA and PirA and FERRI and ALBO by FiuA and another unidentified OMT.
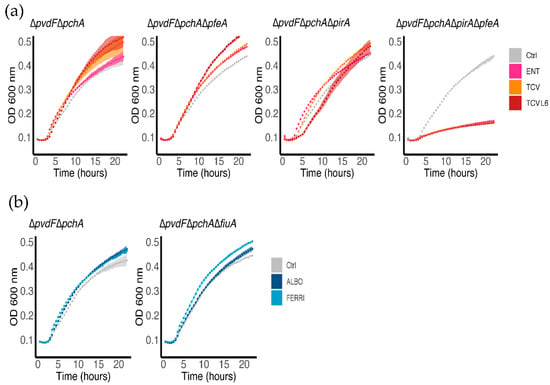
Figure 4.
Bacterial growth in the presence of ENT, TCV, TCVL6, FERRI, or ALBO. Various strains (∆pvdF∆pchA and its corresponding pfeA, pirA and fiuA deletion mutants) were grown in iron-restricted CAA medium, with or without 10 µM ENT, TCVor TCVL6 (a), or FERRI or ALBO (b) at 37 °C. For the Ctrl (control) only, the solvent (DMSO) used to solubilize the conjugates has been added to the cells. The growth curves are the mean of three independent biological triplicates.
3.3. The Presence of Siderophore–Antibiotic Conjugates Induces the Transcription and Expression of their Corresponding OMTs in P. aeruginosa Cells Infecting Epithelial Cells
It is well documented that bacteria are able to sense the presence of xenosiderophores in their environment, consequently leading to induction of the transcription and expression of their corresponding OMTs [16,18,19,20,21,22]. We have previously shown that the presence of either TCV or ENT in planktonic cultures of P. aeruginosa induces the transcription and expression of the OMT pfeA and the presence of FERRI, the OMT fiuA [16,23]. Proteomic and RT-qPCR approaches have shown that P. aeruginosa is also able to sense the presence of xenosiderophores and subsequently adapt its phenotype in even more complex systems, such as an epithelial infection assay [16]. We used the same approach here to investigate whether P. aeruginosa PAO1 senses the presence of ALBO and TCVL6 in an epithelial cell infection assay.
We first used a proteomic approach to identify the bacterial proteins, especially the OMTs, showing differential expression in the presence of ALBO or TCVL6 (Figure 5) and then, RT-qPCR to confirm the proteomic results and compare the levels of transcription of the different genes (Figure 6). Lung epithelial cells (A549 cells, a relevant infection model) were infected with P. aeruginosa PAO1 cells grown overnight in LB (conditions under which the iron-uptake pathways are poorly expressed [16]). The bacteria/epithelial cell ratio used resulted in 40 to 50% viability of the cells 10 h after infection, as we previously described in Perraud et al., 2020 [16]. Only the bacteria associated with the epithelial cells were harvested for proteomic analysis, after 3 h of infection, when we started to observe apoptosis of the epithelial cells. The sample preparation and proteomic workflow used allowed the identification of 1300 proteins from P. aeruginosa in the presence of a large number of proteins from the epithelial cells.
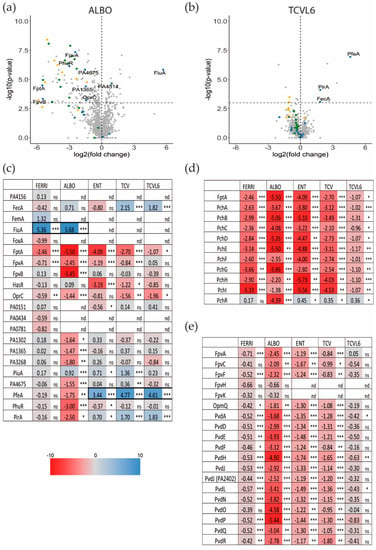
Figure 5.
Analysis of changes in the expression of proteins involved in iron-uptake pathways in P. aeruginosa cells during A549 epithelial cell infection, in the presence or absence of ALBO, TCVL6, FERRI, ENT, or TCV. (a,b) Proteomic analyses were performed on P. aeruginosa PAO1 cells after a 3-h infection of A549 epithelial cells in RPMI medium, in the presence or absence of 10 µM ALBO or TCVL6. Median values measured in the infection assay supplemented with either 10 µM ALBO or TCVL6 were plotted against those measured in the infection assay in the absence of supplementation with siderophore–antibiotic compounds. The median values represent the median of the relative intensity of each protein normalized against all proteins detected by shotgun analysis (n = 5). In blue are OMTs, in yellow, proteins of the pyochelin (PCH) pathway and in green proteins of the PVD pathway. (c–e) Heat maps of OMTs (c), and various proteins involved in the PVD (e) and PCH (d) pathways. The darker the shade of blue, the higher the induced expression of the protein. The darker the shade of red, the higher the repressed expression of the proteins. Only the proteins for which a change in the level of expression was observed are shown. The nd note indicates that the protein has not been identified, ns indicates that the p-value is higher than 0.05; the *, ** and *** symbols indicates a p-value lower than 0.05, 0.01 and 0.001 respectively. All siderophore or siderophore–antibiotic compounds were added at 10 µm.
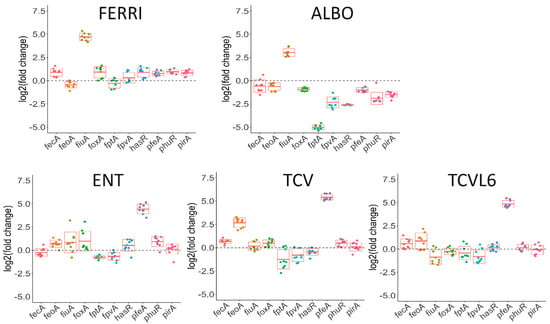
Figure 6.
Analysis of changes in the transcription of genes involved in iron-uptake pathways in P. aeruginosa cells during A549 epithelial cell infection, in the presence or absence of FERRI, ALBO, ENT, TCV, or TCVL6. RT-qPCR analyses were performed, similar to the proteomic analyses, on P. aeruginosa PAO1 cells after a 3-h infection of A549 epithelial cells in RPMI medium, in the presence of absence of 10 µM FERRI, ALBO, ENT, TCV, or TCVL6. The data were normalized relative to the reference gene uvrD and are representative of three independent experiments, each performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of the siderophores or conjugates over those obtained in their absence. fecA encodes the OMT of ferri-citrate, feoA, the ferrous transporter, and fiuA, the OMT of FERRI, foxA, that of ferrioxamine B, fptA, that of PCH, fpvA, that of PVD, hasR and phuR, that of heme, pfeA and pirA, that of ENT and piuA, that of an unknown siderophore.
The presence of ALBO or FERRI induced the transcription and expression of FiuA (log2 fold change of 5.68 for proteomics and 3.03 for RT-qPCR for ALBO) and no other OMT, suggesting that the other unidentified OMT that is involved in iron acquisition by ALBO (Figure 5 and Figure 6) must be expressed constitutively at sufficiently high levels to allow uptake, regardless of the growth conditions. Supporting this hypothesis, previous data have shown no induction of the expression of another OMT when bacteria unable to produce PVD and PCH, and deleted for fiuA, were grown in the presence of FERRI [27]. In addition, in the case of ALBO, the antibiotic moiety did not interfere with the molecular mechanisms involving the sigma/anti-sigma system FiuI/FiuR [21] and required for the detection of these ferri-hydroxamate compounds by P. aeruginosa.
Proteomic and RT-qPCR data (Figure 5 and Figure 6) showed the presence of either TCVL6, TCV, or ENT to strongly induce the expression of PfeA (log2 fold change of 4.61 for proteomics and 4.71 for RT-qPCR for TCVL6). The expression of PfeE (gene number PA2689, esterase hydrolyzing ENT Figure 1c) was also induced, with a log2 fold change of 2.60, 2.58 and 0.91 in the proteomic data in the presence of TCVL6, TCV, and ENT. There was also a very low effect on PirA transcription or expression, by ENT, TCV, and TCVL6 (Figure 5 and Figure 6) with a log2 fold change of 1.83 for TCVL6, 1.70 for TCV and 0.70 for ENT. All three molecules (ENT, TCV, and TCVL6) had a similar effect on the P. aeruginosa phenotype, confirming that TCVL6 is able to enter the bacterial periplasm, like ENT and TCV, and bind to PfeS, the sensor of the two-component system involved in the regulation of the expression of the ENT-dependent iron-uptake pathway in P. aeruginosa cells [18,23], and showing that the linezolide component linked to TCV does not affect this detection mechanism.
The data also show that all five compounds strongly repress the expression of the various proteins of the PCH pathway, such as the OMT FptA (mean log2 fold change of −3.16 for the proteomic data) as well as the enzymes involved in PCH biosynthesis (mean log2 fold change of −2.92 for all enzymes) (Figure 5d). The expression of the proteins of the PVD pathway was more strongly repressed by ALBO than the other compounds tested, with FERRI and TCVL6 having almost no effect on the transcription and expression of these genes (Figure 5e). As most of the proteins in the Fur regulon appeared to also be under-expressed in the presence of ALBO relative to the other compounds tested (Figure S1 in Supplemental Materials), this phenotype could be due to low-level iron contamination of ALBO that probably repressed the expression of all proteins having their corresponding genes transcription regulated by the transcriptional regulator Fur [38]. However, the presence of small amounts of iron should also have as consequence a growth stimulation when ∆pvdF∆pchA cells are grown in the presence of 10 µM ALBO compared to the growth kinetic in the absence of conjugates (Figure 4a). The lack of growth stimulation goes against this hypothesis of iron traces associated to ALBO. At this stage, it is difficult to conclude about the effect of ALBO on the transcription and expression of the proteins of the PVD pathway.
In conclusion, P. aeruginosa is able to detect the two siderophore conjugates used in this study (ALBO and TCVL6) in its growth environment, even in a complex system such as an epithelial infection assay, and clear phenotypic adaption is observed. Both compounds were able to induce the transcription and expression of their corresponding OMT in P. aeruginosa cells: FiuA for ALBO and PfeA for TCVL6. The phenotypic adaptation of P. aeruginosa in this infection assay also involved repression of the transcription and expression of the proteins of the PCH and PVD pathways.
3.4. Siderophore–Antibiotic Conjugates also Affect the Expression of Virulence Factors
Transcription of the genes of the PVD pathway is regulated by two sigma factors, FpvI and PvdS, the second also regulating the transcription of virulence factors such as exotoxin A (ToxA) [39,40], a major virulence factor of P. aeruginosa and Piv (also called PrpL) endoprotease [41,42]. The alkaline protease AprA is regulated by OxyR and is thus dependent on redox stress, which can be induced by high iron concentrations [43]. Thus, we also performed proteomic analysis of the expression of all virulence factors expressed by P. aeruginosa cells during the infection of A549 cells (Figure 7a). However, all secreted virulence factors could not be detected with the protocol used to harvest the P. aeruginosa cells after infection. Consequently, we used RT-qPCR to evaluate the infected samples for the transcription of the genes encoding virulence factors for which their transcription is regulated or not by PvdS or OxyR (Figure 7b), as well as several other virulence factors, such as AmrZ, a transcriptional regulator that controls the expression of many genes involved in environmental adaptation [44], PhzA2, a flavin-containing monooxygenase involved in phenazine biosynthesis [45,46], and, finally, secreted virulence factors such as ExoY, a protease injected into eukaryotic cells by the type III secretion system [47,48,49,50] and the LasB elastase [51].
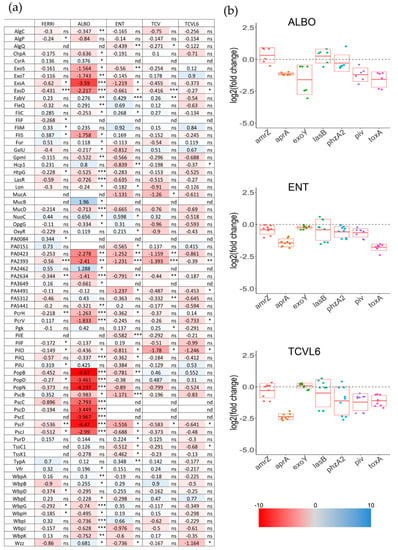
Figure 7.
(a) Analysis of changes in the expression of virulence factors in P. aeruginosa cells during A549 epithelial cell infection, in the presence or absence of FERRI, ALBO, ENT, TCV, or TCVL6. Proteomic analyses were performed on P. aeruginosa PAO1 cells as in Figure 5. Median values measured in the infection assay supplemented with either 10 µM FERRI, ALBO, ENT, TCV, or TCVL6 were plotted against those measured in the infection assay in the absence of supplementation with siderophore or siderophore–antibiotic compounds. The median values represent the median of the relative intensity of each protein normalized against all proteins detected by shotgun analysis (n = 5). The darker the shade of blue, the higher the induced expression of the protein. The darker the shade of red, the higher the repressed expression of the proteins. The nd note indicates that the protein has not been identified, ns indicates that the p-value is higher than 0.05; the *, ** and *** symbols indicates a p-value lower than 0.05, 0.01 and 0.001 respectively. (b) Analysis of the changes in the transcription of genes encoding virulence factors in P. aeruginosa cells during A549 epithelial cell infection, in the presence or absence of ENT, TCVL6, or ALBO. RT-qPCR analyses were performed on the same infection assays as in Figure 5. The data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of siderophores or conjugates over those obtained in their absence. amrZ encodes a transcriptional regulator controlling many genes involved in environmental adaptation [44], aprA, an alkaline protease [43], exoY [50], lasB, an elastase [51], phzA2, a flavin-containing monooxygenase involved in phenazine biosynthesis [45,46], piv, a lysyl-endopeptidase (also called PrpL) [41,42] and toxA, an exotoxin [39,40].
Proteomic analyses showed repression of the expression of many virulence factors, mostly in the presence of ALBO, but also FERRI, ENT, and TCV, and, to a lower extent, TCVL6 (Figure 7). The lower effect of TCVL6 can be explained by the slightly lower solubility of this compound. Indeed, in some cases, a small amount of precipitate of TCVL6 could be observed. In addition, RT-qPCR showed different transcriptional profiles of the tested virulence factors in the presence of ENT, TCVL6, and ALBO. The presence of ENT mostly repressed the transcription of aprA and toxA, as the transcription of these virulence factors is regulated by PvdS. TCVL6 also repressed the transcription of aprA (more strongly than ENT) and toxA, as well as the transcription of piv and phzA2. Finally, ALBO repressed the transcription of aprA, exoY, piv, and toxA.
3.5. A Tris-Catechol Conjugate More Efficiently Induces the Transcription of its OMT than a Hydroxamate Conjugate in P. aeruginosa Cells Infecting Epithelial Cells
We have previously shown that P. aeruginosa cells infecting epithelial A549 cells increase the transcription and expression of catechol siderophore-dependent iron-uptake pathways, and not those of hydroxamate siderophores, in the presence of a mixture of four different siderophores [16]. Here, the infection assay was repeated with a mixture of either TCV and FERRI or TCVL6 and ALBO and analyzed by RT-qPCR (Figure 8). The presence of a mixture of FERRI and TCV in the infection assay strongly induced the transcription of pfeA (ENT OMT), with a log2 fold change of 5; fiuA (FERRI OMT) transcription being almost not induced (Figure 8a). In addition, we observed repression in the range of a 0.95–1.8 log2 fold change for fptA, fpvA, and hasR transcription (the genes encoding the OMTs of PCH, PVD, and the HasA hemophore, respectively). In the presence of a mixture of ALBO and TCVL6, only pfeA transcription was strongly induced (log2 fold change of 5.2), that of fiuA almost not, and that of fptA, fpvA, and hasR repressed, with a of 0.5–0.9 log2 fold change. As the various genes involved in iron acquisition by a given siderophore are always in an operon, a change in the transcription and expression of the gene encoding an OMT implies a similar change for the transcription and expression of the other genes present in the operon. In conclusion, the data show that conjugates with a catechol-type siderophore moiety have more chances to induce the transcription and expression of their corresponding OMT, and consequently, their corresponding uptake pathway than hydroxamate vectors.
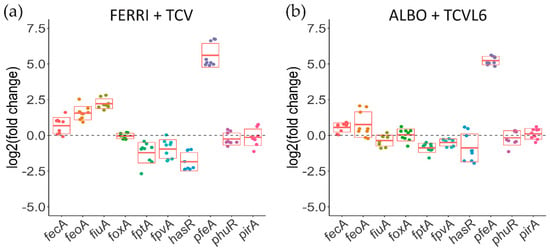
Figure 8.
Analysis of the changes in the transcription of genes encoding OMTs involved in iron-uptake pathways in P. aeruginosa cells during A549 epithelial cell infection, in the presence or absence of a mixture of either FERRI and TCV (a) or ALBO and TCVL6 (b). RT-qPCR analyses were performed on P. aeruginosa PAO1 cells after 3 h of incubation with A549 epithelial cells in RPMI medium, with or without a mixture of two compounds: either FERRI and TCV or ALBO and TCVL6 (each of the compounds being present at a concentration of 10 µM). The data were normalized relative to the reference gene uvrD and are representative of three independent experiments, each performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of the conjugates over those obtained in their absence. fecA encodes the OMT of ferri-citrate, feoA, the ferrous transporter, and fiuA, the OMT of FERRI, foxA, that of ferrioxamine B, fptA, that of PCH, fpvA, that of PVD, hasR and phuR, that of heme, pfeA and pirA, that of ENT, and piuA, that of an unknown siderophore.
4. Discussion
Introducing antibiotics into Gram negative bacteria, such as P. aeruginosa, via a Trojan-horse strategy using siderophores as vectors is a promising approach. However, there are still several bottlenecks in its development. The high antibiotic activity of natural sideromycins clearly shows that the strategy works and the fact that cefiderocol has been approved by the FDA for use in the treatment of urinary infections is a sign that antibiotic vectorization by siderophores should be explored further by pharmaceutical companies. However, developing such compounds requires extensive knowledge of the molecular mechanisms and regulatory network involved in bacterial siderophore-dependent iron acquisition. Most bacteria carry genes encoding several iron uptake pathways in their genome, allowing very high phenotypic plasticity, depending on the presence or not of xenosiderophores or sideromycins. Such phenotypic plasticity of P. aeruginosa has been investigated in various growth media and in the presence of several xenosiderophores [16]. The data show that if siderophores produced by other bacteria (xenosiderophores) are present, bacteria induce the expression of the uptake pathway corresponding to the xenosiderophore(s) (corresponding OMT and enzymes necessary to release the iron from the siderophore) and, in parallel, repress those corresponding to the endogenous siderophores [16].
Here, we show that this also occurs in the presence of the two siderophore–antibiotic conjugates ALBO and TCVL6 in a complex system (an epithelial cell infection assay) with rapid kinetics (3-h infection with the bacteria having been previously grown in iron-rich media). Both compounds have no antibiotic activities on P. aeruginosa on the concentrations used. The bacteria sense the presence of both conjugates in their environment and adapt their phenotypes to access iron via the sideromycins that are present. This occurs via auto-loop regulating systems: the two-component system PfeS/PfeR for TCVL6 and the sigma and anti-sigma factors FiuI/FiuR for ALBO. Comparison of the transcription induction levels between FERRI and ALBO and between TCV and TCVL6 for their corresponding OMTs shows that the presence of the antibiotic group on each sideromycin had no inhibitory effect on the regulatory mechanism: the presence of the conjugates was sensed by P. aeruginosa with the same efficiency as the vector or corresponding siderophore alone.
Such induction of FiuA and PfeA expression occurred in parallel with a decrease in the transcription and expression of the proteins involved in PVD and PCH biosynthesis and ferri-PVD and ferri-PCH uptake. In such a context, the bacteria use the conjugates to get access to iron and less their own siderophores. Moreover, the phenotypic effect induced by the presence of the siderophore conjugates was not only limited to the expression or not of the iron uptake pathways but also that of virulence factors, especially those regulated by PvdS, the sigma factor that regulates the transcription of the genes involved in PVD production The ability of siderophore conjugates to repress the transcription and expression of virulence factors is clearly an asset that has to also be considered in such a Trojan-horse strategy using siderophores as vectors.
When the infection assay was carried out in the simultaneous presence of both sideromycins, ALBO and TCVL6 (present in equivalent amounts), the catechol-type vector was clearly more efficient in inducing the expression of its corresponding transporters, PfeA, than that for ferrichrome, because of its strong affinity for iron. Catechol-type compounds are clearly more efficient in competing for iron with PVD than hydroxamate molecules (Figure 2), consistent with the affinities of such families of siderophores for iron: Ka of 1049 M−1 for ENT [34], 1029 M−1 for FERRI [35], and 1030.8 M−1 for PVD [31]. Because of their higher affinity for iron, tris-catecholate vectors scavenge iron more efficiently in the bacterial environment than hydroxamate compounds (Figure 2). Probably, only the iron-loaded forms of the siderophores or conjugates are able to induce the transcription of the proteins of their corresponding uptake pathways. Consequently, catecholate vectors have a higher chance to induce the transcription of their corresponding uptake pathways in the bacteria via the auto-loop regulating systems.
5. Conclusions
The ability of siderophore conjugates to be detected by the pathogen in its growth environment, even in a complex system, such as an epithelial infection assay, is a great advantage in Trojan-horse strategies, because it ensures the expression of the needed uptake pathway and suggests that such induction should occur as well in the host during an infection. Moreover, the data show that conjugates with a catechol-type siderophore moiety have a greater chance to induce their corresponding OMT, and consequently, their corresponding uptake pathway, than hydroxamate vectors and are potentially more efficient Trojan-horse vectors.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1820/s1, Figure S1: Differential production of iron-regulated proteins in the presence of ALBO based on the Palma, Worgall, and Quadri (2003) and Ochsner et al., (2002) studies [5,6]. Table S1: Strains and plasmids used in this study, Table S2: Primers used for RT-qPCR analysis.
Author Contributions
Conceptualization, I.J.S. and L.E.-S.; methodology, Q.P., G.L.A.M. and L.E.-S.; validation, I.J.S. and L.E.-S.; formal analysis, P.C.; investigation, Q.P., M.M., P.C., F.H., N.Z., and V.G.; data curation, P.C. and Q.P.; writing—original draft preparation, I.J.S. and Q.P.; writing—review and editing, I.J.S. and L.E.-S.; visualization, Q.P. and I.J.S.; supervision, I.J.S and L.E.-S.; project administration, I.J.S and L.E.-S.; funding acquisition, I.J.S. and L.E.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Centre National de la Recherche Scientifique and grants from the associations Vaincre la Mucoviscidose and Association Gregory Lemarchal. In addition, these results were generated as part of the work of the Translocation Consortium (www.translocation.com), supported by the Innovative Medicines Joint Undertaking under Grant Agreement no. 115525, through financial contributions from the European Union’s Seventh Framework Program (FP7/2007-2013) and contributions in kind from EFPIA companies. The equipment at the IPHC was partly funded by the French Proteomics Infrastructure (ProFI; ANR-10-INSB-08-03). P.Q. was funded by Vaincre la Mucoviscidose and Association Gregory Lemarchal and MM by the Translocation Consortium.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tillotson, G.S. Trojan Horse Antibiotics—A Novel Way to Circumvent Gram-Negative Bacterial Resistance? Infect. Dis. 2016, 9, 45–52. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2011, 27, 637–657. [Google Scholar] [CrossRef]
- Schalk, I.J.; Mislin, G.L.A.; Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 2012, 69, 37–66. [Google Scholar] [CrossRef] [PubMed]
- Mislin, G.L.A.; Schalk, I.J. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 2014, 6, 408–420. [Google Scholar] [CrossRef]
- Górska, A.; Sloderbach, A.; Marszałł, M.P. Siderophore-drug complexes: Potential medicinal applications of the “Trojan horse” strategy. Trends Pharmacol. Sci. 2014, 35, 442–449. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef]
- Gause, G.F. Recent studies on albomycin, a new antibiotic. Br. Med. J. 1955, 2, 1177–1179. [Google Scholar] [CrossRef]
- Benz, G.; Schroeder, T.; Kurz, J.; Wuensche, C.; Karl, W.; Steffens, G.; Pfitzner, J.; Schmidt, D. Konstitution der Desferriform der Albomycine δ1, δ2, ε. Angew. Chem. 1982, 21, 1322–1335. [Google Scholar]
- Sackmann, W.; Reusser, P.; Neipp, L.; Kradolfer, F.; Gross, F. Ferrimycin A, a new iron-containing antibiotic. Antibiot. Chemother. 1962, 12, 34–45. [Google Scholar]
- Selzer, G.B. Antibiotics: Isolation, separation, and purification. J. Pharm. Sci. 1980, 69, 754. [Google Scholar] [CrossRef]
- Vertesy, L.; Aretz, W.; Fehlhaber, H.-W.; Kogler, H. Salmycins A-D, antibiotics from Streptomyces violaceus DSM 8286 having a siderophore-aminoglycoside structure. Helv. Chim. Acta 1995, 78, 46–60. [Google Scholar]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Young, L.; Neshat, S. Enterobactin-mediated iron transport in Pseudomonas aeruginosa. J. Bacteriol. 1990, 172, 6991–6996. [Google Scholar] [PubMed]
- Hannauer, M.; Barda, Y.; Mislin, G.L.; Shanzer, A.; Schalk, I.J. The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and a recycling of the modified desferrichrome. J. Bacteriol. 2010, 192, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.F.; Elde, N.C. Buried Treasure: Evolutionary Perspectives on Microbial Iron Piracy. Trends Genet. 2015, 31, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Perraud, Q.; Cantero, P.; Roche, B.; Gasser, V.; Normant, V.P.; Kuhn, L.; Hammann, P.; Mislin, G.L.A.; Ehret-Sabatier, L.; Schalk, I.J. Phenotypic Adaption of Pseudomonas aeruginosa by Hacking Siderophores Produced by Other Microorganisms. Mol. Cell. Proteom. 2020, 19, 589–607. [Google Scholar] [CrossRef]
- Schalk, I.J.; Guillon, L. Fate of ferrisiderophores after import across bacterial outer membranes: Different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 2013, 44, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Dean, C.R.; Neshat, S.; Poole, K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1996, 178, 5361–5369. [Google Scholar]
- Visca, P.; Leoni, L.; Wilson, M.J.; Lamont, I.L. Iron transport and regulation, cell signalling and genomics: Lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 2002, 45, 1177–1190. [Google Scholar]
- Michel, L.; Gonzalez, N.; Jagdeep, S.; Nguyen-Ngoc, T.; Reimmann, C. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 2005, 58, 495–509. [Google Scholar]
- Llamas, M.A.; Sparrius, M.; Kloet, R.; Jimenez, C.R.; Vandenbroucke-Grauls, C.; Bitter, W. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 1882–1891. [Google Scholar] [PubMed]
- Llamas, M.A.; Imperi, F.; Visca, P.; Lamont, I.L. Cell-surface signaling in Pseudomonas: Stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 2014, 38, 569–597. [Google Scholar] [CrossRef] [PubMed]
- Gasser, V.; Baco, E.; Cunrath, O.; August, P.S.; Perraud, Q.; Zill, N.; Schleberger, C.; Schmidt, A.; Paulen, A.; Bumann, D.; et al. Catechol siderophores repress the pyochelin pathway and activate the enterobactin pathway in Pseudomonas aeruginosa: An opportunity for siderophore-antibiotic conjugates development. Environ. Microbiol. 2016, 18, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Pramanik, A.; Gwinner, T.; Koberle, M.; Bohn, E. Sideromycins: Tools and antibiotics. Biometals Int. J. Role Metal Ions Biol. Biochem. Med. 2009, 22, 3–13. [Google Scholar] [CrossRef]
- Wencewicz, T.A.; Mollmann, U.; Long, T.E.; Miller, M.J. Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan Horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. Biometals Int. J. Role Metal Ions Biol. Biochem. Med. 2009, 22, 633–648. [Google Scholar] [CrossRef]
- Perraud, Q.; Moynié, L.; Gasser, V.; Munier, M.; Godet, J.; Hoegy, F.; Mély, Y.; Mislin, G.L.A.; Naismith, J.H.; Schalk, I.J. A key role for the periplasmic PfeE esterase in iron acquisition via the siderophore enterobactin in Pseudomonas aeruginosa. ACS Chem. Biol. 2018, 13, 2603–2614. [Google Scholar] [CrossRef]
- Normant, V.; Josts, I.; Kuhn, L.; Perraud, Q.; Fritsch, S.; Hammann, P.; Mislin, G.L.A.; Tidow, H.; Schalk, I.J. Nocardamine-Dependent Iron Uptake in Pseudomonas aeruginosa: Exclusive Involvement of the FoxA Outer Membrane Transporter. ACS Chem. Biol. 2020. [Google Scholar] [CrossRef]
- Demange, P.; Wendenbaum, S.; Linget, C.; Mertz, C.; Cung, M.T.; Dell, A.; Abdallah, M.A. Bacterial siderophores: Structure and NMR assigment of pyoverdins PaA, siderophores of Pseudomonas aeruginosa ATCC 15692. Biol. Metals 1990, 3, 155–170. [Google Scholar]
- Baco, E.; Hoegy, F.; Schalk, I.J.; Mislin, G.L. Diphenyl-benzo[1,3]dioxole-4-carboxylic acid pentafluorophenyl ester: A convenient catechol precursor in the synthesis of siderophore vectors suitable for antibiotic Trojan horse strategies. Org. Biomol. Chem. 2014, 12, 749–757. [Google Scholar] [CrossRef]
- Clément, E.; Mesini, P.J.; Pattus, F.; Abdallah, M.A.; Schalk, I.J. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 2004, 43, 7954–7965. [Google Scholar]
- Albrecht-Gary, A.M.; Blanc, S.; Rochel, N.; Ocacktan, A.Z.; Abdallah, M.A. Bacterial iron transport: Coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 1994, 33, 6391–6402. [Google Scholar]
- Brandel, J.; Humbert, N.; Elhabiri, M.; Schalk, I.J.; Mislin, G.L.A.; Albrecht-Garry, A.-M. Pyochelin, a siderophore of Pseudomonas aeruginosa: Physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans 2012, 41, 2820–2834. [Google Scholar] [PubMed]
- Folschweiller, N.; Gallay, J.; Vincent, M.; Abdallah, M.A.; Pattus, F.; Schalk, I.J. The interaction between pyoverdin and its outer membrane receptor in Pseudomonas aeruginosa leads to different conformers: A time-resolved fluorescence study. Biochemistry 2002, 41, 14591–14601. [Google Scholar] [PubMed]
- Loomis, L.; Raymond, K.N. Solution equilibria of enterobactin complexes. Inorg. Chem. 1991, 30, 906–911. [Google Scholar]
- Anderegg, G.; L’Eplattenier, F.; Schwarzenbach, G. Hydroxamatkomplexe III. Eisen(III)-Austausch zwischen Sideraminen und Komplexonen. Diskussion der Bildungskonstanten der Hydroxamatkomplexe. Helv. Chim. Acta 1963, 46, 1409–1422. [Google Scholar] [CrossRef]
- Postle, K.; Larsen, R.A. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 2007, 20, 453–465. [Google Scholar] [PubMed]
- Celia, H.; Noinaj, N.; Buchanan, S.K. Structure and Stoichiometry of the Ton Molecular Motor. Int. J. Mol. Sci. 2020, 21, 375. [Google Scholar] [CrossRef]
- Cornelis, P.; Matthijs, S.; Van Oeffelen, L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 2009, 22, 15–22. [Google Scholar] [CrossRef]
- Pollack, M. The role of exotoxin A in Pseudomonas disease and immunity. Rev. Infect. Dis. 1983, 5 (Suppl. 5), S979–S984. [Google Scholar] [CrossRef]
- Chaudry, G.J.; Holmans, P.L.; Clowes, R.C.; Draper, R.K. Pseudomonas aeruginosa exotoxin A. Target Diagn. Ther. 1992, 7, 405–423. [Google Scholar]
- Thibodeaux, B.A.; Caballero, A.R.; Dajcs, J.J.; Marquart, M.E.; Engel, L.S.; O’Callaghan, R.J. Pseudomonas aeruginosa protease IV: A corneal virulence factor of low immunogenicity. Ocul. Immunol. Inflamm. 2005, 13, 169–182. [Google Scholar] [CrossRef]
- Oh, J.; Li, X.-H.; Kim, S.-K.; Lee, J.-H. Post-secretional activation of Protease IV by quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Laarman, A.J.; Bardoel, B.W.; Ruyken, M.; Fernie, J.; Milder, F.J.; van Strijp, J.A.G.; Rooijakkers, S.H.M. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J. Immunol. 2012, 188, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Muriel, C.; Arrebola, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Jalvo, B.; Pfeilmeier, S.; Blanco-Romero, E.; Baena, I.; Malone, J.G.; Rivilla, R.; et al. AmrZ is a major determinant of c-di-GMP levels in Pseudomonas fluorescens F113. Sci. Rep. 2018, 8, 1979. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef]
- Recinos, D.A.; Sekedat, M.D.; Hernandez, A.; Cohen, T.S.; Sakhtah, H.; Prince, A.S.; Price-Whelan, A.; Dietrich, L.E.P. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc. Natl. Acad. Sci. USA 2012, 109, 19420–19425. [Google Scholar] [CrossRef]
- Duong, F.; Bonnet, E.; Géli, V.; Lazdunski, A.; Murgier, M.; Filloux, A. The AprX protein of Pseudomonas aeruginosa: A new substrate for the Apr type I secretion system. Gene 2001, 262, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Wilson, M.J.; Cunliffe, H.E.; Lamont, I.L.; Vasil, M.L. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001, 69, 5385–5394. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef]
- Yahr, T.L.; Vallis, A.J.; Hancock, M.K.; Barbieri, J.T.; Frank, D.W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 1998, 95, 13899–13904. [Google Scholar] [CrossRef]
- Wolz, C.; Hellstern, E.; Haug, M.; Galloway, D.R.; Vasil, M.L.; Döring, G. Pseudomonas aeruginosa LasB mutant constructed by insertional mutagenesis reveals elastolytic activity due to alkaline proteinase and the LasA fragment. Mol. Microbiol. 1991, 5, 2125–2131. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).