Abstract
The GacS histidine kinase is the membrane sensor of the major upstream two-component system of the regulatory Gac/Rsm signal transduction pathway. This pathway governs the expression of a wide range of genes in pseudomonads and controls bacterial fitness and motility, tolerance to stress, biofilm formation, and virulence or plant protection. Despite the importance of these roles, the ligands binding to the sensor domain of GacS remain unknown, and their identification is an exciting challenge in this domain. At high population densities, the GacS signal triggers a switch from primary to secondary metabolism and a change in bacterial lifestyle. It has been suggested, based on these observations, that the GacS signal is a marker of the emergence of nutritional stress and competition. Biochemical investigations have yet to characterize the GacS signal fully. However, they portray this cue as a low-molecular weight, relatively simple and moderately apolar metabolite possibly resembling, but nevertheless different, from the aliphatic organic acids acting as quorum-sensing signaling molecules in other Proteobacteria. Significant progress in the development of metabolomic tools and new databases dedicated to Pseudomonas metabolism should help to unlock some of the last remaining secrets of GacS induction, making it possible to control the Gac/Rsm pathway.
1. Introduction
The ability of an organism to colonize and adapt to an ecological niche is highly dependent on its capacity to perceive and analyze its environment. Two-component systems (TCSs, a detailed list of all abbreviations used in this article is available in Figure S1) are powerful tools, widely encountered in bacteria, that fulfill these functions [,]. The structures and subtleties of molecular functioning in the wide diversity of TCS families have recently been reviewed in detail by Jacob-Dubuisson et al. [] and Zschiedrich et al. []. Briefly, TCSs consist of a sensor kinase embedded in the bacterial (inner) membrane and a cytosolic cognate response regulator. After coming into contact with its biotic or abiotic environmental cue, the sensor (known as S) undergoes autophosphorylation, and activates the response regulator (also known as the response activator, or A) by phosphotransfer. The GacS/GacA TCS is highly conserved in many Gram-negative bacteria, but that of the Pseudomonas genus has been promoted as an experimental model [,,,,]. Pseudomonas spp. have considerable potential for adaptation to fluctuating environmental conditions and different hosts, thanks to the plasticity of their large genome [,]. The type species of the genus, P. aeruginosa, is an opportunistic pathogen of humans and animals that can also cause necrosis in plants, but most Pseudomonas species are commonly found associated with plants [,]. Some are saprotrophs that may have beneficial effects on plant growth and health, such as P. chlororaphis and P. fluorescens, whereas others are redoubtable phytopathogens, such as P. syringae and P. viridiflava [,,]. The GacS/GacA TCS supports the diversity of behavior in this genus and its hosts, by playing major ecological (e.g., control of fitness, host virulence or protection), physiological (e.g., control of swimming and swarming motilities, stress tolerance, biofilm formation) and metabolic (e.g., control of secondary metabolites and extracellular enzyme production) roles in pseudomonads [,,,,,]. Thus, the GacS/GacA TCS appears to be a master sensor of the microenvironment, operating in both pathogenic and beneficial pseudomonads [,,,].
The GacS/GacA TCS has been studied since the early 1990s. These studies were initiated essentially by Professor Dieter Haas and his team at the Department of Fundamental Microbiology of the University of Lausanne []. This is well illustrated by the long list of references from his laboratory cited in this review. These researchers were the first to assign the acronym “Gac”, for “global activator antibiotic and cyanide synthesis”, to the response regulator GacA, which was described in the “Pseudomonas fluorescens” model strain CHA0 []. This biocontrol strain has since been reassigned to the more recently described species Pseudomonas protegens, which characteristically produces the antimicrobial compounds pyoluteorin and 2,4-diacetylphloroglucinol (DAPG) []. During the same epoch, Estelle Hrabak and David Willis of the Department of Plant Pathology of the University of Wisconsin, discovered the existence of the membrane sensor kinase LemA (for “lesion manifestation”) [,]. This kinase was identified in the phytopathogen P. syringae pv. syringae B278a, and was implicated in bean leaf damage. The connection between LemA (now called GacS) and GacA resulting in the formation of a TCS—which was non-intuitive given that gacA is not clustered next to the corresponding sensor kinase gene—was demonstrated in the P. syringae pv. syringae model, by the same team []. Subsequent studies led to (i) the detection of GacS and GacA analogs in many other model strains and species, and (ii) detailed characterization of the links between the TCS constituents and their downstream regulatory network leading to the activation or inhibition of target genes [,]. This downstream regulatory cascade involves the Rsm proteins. RsmA and its paralogs are RNA-binding proteins that modulate translation initiation or alter the stability of target messenger RNAs. For example, RsmA acts as a global posttranscriptional regulator, controlling the expression of more than 500 genes in P. aeruginosa, many of which are involved in virulence or biofilm formation and are downregulated []. GacA upregulates the expression of several small non-coding RNAs (sRNAs), the number of which depends on the Pseudomonas species considered []. These sRNAs are posttranslational regulators, as they can sequester the regulatory protein RsmA and other RsmA-like proteins, depending on the strain, thereby blocking their activity [,,,,,,]. Thus, the activation of GacS suppresses the inhibitory or stimulating effects exerted by Rsm proteins on the expression of their target genes. Based on these findings, it became clear that GacS/GacA TCS functioning needed to be integrated into a more global cellular mechanism, which the authors referred to as the Gac/Rsm signal transduction (regulatory) pathway [,,].
Almost 20 years ago, Stephan Heeb and Dieter Haas [] concluded one of the first reviews on the GacS/GacA TCS with a statement that: “the current challenges are to discover the signal molecule(s) specific to the GacS sensor and to identify additional components in this complex regulatory pathway”. Today, many of the major elements involved in the functioning of the Gac/Rsm regulon have been identified, but the precise nature of the signal activating GacS remains a mystery. This major gap in our knowledge is particularly penalizing, because knowledge of the GacS signal is essential if we are to understand how various strong physiological and ecological adaptations are triggered in numerous bacteria, with a view to controlling these mechanisms, through anti-virulence and smart biocontrol treatments in particular. This review follows three axes for deciphering the structure and function of the GacS signal. First, the architecture and functioning of the GacS sensor kinase, to date the only known pedestal for the GacS signal, were considered. Then, the candidate cues, already suspected to play the role of the GacS signal on the basis of screening for demonstrated or presumed functions in bacterial metabolism and communication, adaptation to the host and lifestyle selection, were evaluated. Finally, the progress made and novel strategies for biochemical characterization of the GacS signal were summarized, leading to establish a plausible profile for the elusive GacS signal.
2. GacS Is a Membrane Sensor Centralizing Multiple Environmental Stimuli
2.1. Architecture and Functioning of the GacS Sensor
The GacS sensor is a homodimeric protein in which each of the monomers consists of an N-terminal transmembrane α-helix, followed by a periplasmic detector domain linked to a second transmembrane α-helix connected to a large cytoplasmic region [] (Figure 1). The first structural and functional insights into the periplasmic sensor domain were obtained by Zuber et al. [] in P. protegens CHA0. These findings were then clarified by Ali-Ahmad et al. [] in the P. aeruginosa model, in a study based on nuclear magnetic resonance (NMR) spectroscopy and GacS mutants. This study also compared structural sequence alignments of GacS sensor domains with the most closely related kinase sensory domains DcuS from Escherichia coli and CitA from Klebsiella pneumoniae. Determination of the NMR solution structure of the GacS sensor domain highlighted the presence of a flexible major loop well conserved in the bacterial models studied. This loop has a dynamic conformation in solution, suggesting that ligand binding may induce a conformational change. At the top of the β-sheet facing this loop, three basic amino-acyl residues form an interacting network defining a pocket that could act as a putative ligand-binding site. In P. aeruginosa, the functional region consists of the histidyl-97, -124 and -133 residues, which define a positively charged binding-pocket. However, the histidyl-133 residue is not conserved in the DcuS and CitA structural homologs, suggesting that the ligand binding site in GacS is not located in exactly the same place as those of DcuS and CitA []. Finally, in silico analyses of more than 200 sequence homologs of the GacS periplasmic sensor domain led to the establishment of a phylogenic tree revealing early divergence between P. aeruginosa and P. fluorescens within the Pseudomonas group []. These findings raise questions about the conserved function between the GacS proteins of different species, and about the similarity of the signals recognized by the binding site. These elements will be addressed in Section 4.
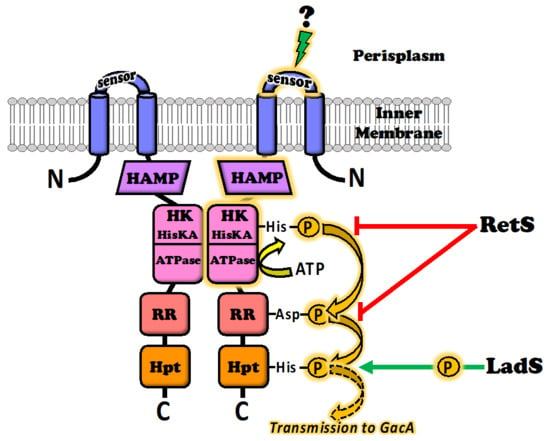
Figure 1.
Architecture and functioning of the GacS sensor kinase. GacS is capable of collecting a pool of membrane stimuli: the central environmental source signal (still unknown) is perceived by the periplasmic sensor domain and causes a conformational change that is mechanically transmitted by the HAMP domain to the histidine kinase (HK) domain, leading to autophosphorylation of the key histidyl residue. The phosphorelay involves transmission by the response signal receiver (RR) and histidine phosphotransfer (Hpt) domains, to the cytosolic GacA response regulator. The GacS signal route (marked in amber) is modulated by other environmental cues provided by two other membrane kinase sensors: LadS stimulates GacS activity by shuttling additional phosphate groups into the Hpt domain (green arrow), whereas RetS inhibits GacS activity by siphoning phosphates from the GacS HK, dephosphorylating the RR domain, and/or blocking access to the key histidyl residue of the HK domain (red arrows).
The periplasmic domain of GacS plays a crucial role in the sensing and binding of extracellular cues, but the cytoplasmic part of the molecule is also important for the transmission of information to trigger the GacA regulatory response (Figure 1). The GacS cytoplasmic region contains a HAMP phosphatase domain; such domains are present in histidine kinases, adenylate cyclases, methyl-accepting proteins and phosphatases (hence HAMP). The HAMP domain transmits conformational changes from the periplasmic ligand-binding domains to the cytoplasmic part of the protein, more precisely to a histidine kinase (HK) domain. The deletion of this HAMP domain thus results in a strong signal-independent activation of the Gac/Rsm cascade []. The HK domain harbors the histidine kinase A, plus a histidine kinase-like ATPase. Its role is to translate the received signal into a phosphorylation event leading to signal transmission. The HK domain can, thus, bind ATP and has histidine kinase activity, leading to self-phosphorylation on a conserved histidyl residue to generate the activated form of the molecule. Interestingly, the HK domain is targeted by ATPase inhibitors from the benzothiazole family []. These compounds therefore inhibit the functioning of GacS data transduction and have been considered as a means of controlling TCS activity and associated virulence []. Autophosphorylation of the first histidyl residue of the HK domain initiates an aspartyl phosphorelay involving a response signal receiver (RR or REC) domain, followed by a second histidyl residue phosphorelay involving a histidine phosphotransfer (Hpt) domain. Finally, phosphate groups are transferred to a conserved aspartyl residue in the recipient domain of the GacA transcriptional regulator [,,]. The HK and RR domains of GacS also act together in structural interactions of GacS with GacA, and the HAMP domain of GacS is responsible for GacS dimerization. GacS autophosphorylation presumably occurs in trans, within a GacS homodimer [].
2.2. Modulators of GacS Sensor Activity
In pseudomonads, GacS activity is modulated by at least two other inner membrane sensors, called LadS for “loss of adherence sensor” and RetS for “regulator of exopolysaccharide and type III secretion system” [,,,]. LadS and RetS are also sensor kinases with an N-terminal periplasm-facing signal-receiving domain. However, unlike GacS, they are considered to be orphan sensor kinases, because they are not genetically associated with a cognate response regulator. The LadS and RetS sensors do not directly affect the phosphorylation status of the GacS response regulator (i.e., the GacA protein) []. They compensate for the absence of a dedicated response regulator by interacting directly with the GacS sensor, to transmit environmental stimuli and integrate their inputs into the Gac/Rsm regulatory pathway. LadS and RetS provide a certain degree of flexibility, because they have counterbalancing effects, with one enhancing GacS signaling, whereas the other attenuates this signaling (Figure 1). LadS stimulates GacS activity by shuttling additional phosphate groups into the Hpt domain of GacS, enhancing the GacS-GacA phosphorelay [,]. Conversely, RetS counteracts GacS activity by at least three different mechanisms recently elucidated in the P. aeruginosa model. In the first two of these mechanisms, RetS captures phosphate groups from the GacS HK domain, preventing the initial phosphorelay following autophosphorylation from occurring, or dephosphorylates the GacS RR domain. Both these mechanisms prevent the efficient transfer of phosphate groups from GacS to GacA []. In the third mechanism, the HK domains of RetS and GacS establish a non-enzymatic tight bond, blocking access to the histidyl residue of the GacS HK domain, thereby preventing its phosphorylation [,]. According to the authors, this last mechanism results from the reversible unfolding of a short helical region (helix-cracking) of the RetS HK domain triggered by ligand binding to the periplasmic sensory domain of RetS []. Finally, at least on other membrane histidine kinase, PA1611, can also participate in the GacS/LadS/RetS network, by preventing the inhibitory effects of RetS on GacS activation [,].
Attempts to identify the signals likely to activate the LadS and RetS membrane sensors have been met with more success than those for the GacS sensor. However, LadS and RetS are characterized by a periplasmic sensor domain different from that of GacS. They carry an N-terminal seven-transmembrane domain region with diverse intracellular signaling modules extracellular domain 2 (7TMR-DISMED2) [,]. Broder et al. [] showed that calcium binds to the 7TMR-DISMED2 binding pocket, activating the LadS kinase in P. aeruginosa. These authors suggested that calcium is used to sense the intracellular environment of the eukaryote host, to improve the chances of successful long-term host tissue colonization. By contrast, the ligand that binds to the sensor domain of RetS has yet to be fully identified, although the crystal structure of the RetS periplasmic domain revealed the presence of a carbohydrate-binding module []. Like LadS, RetS may be responsible for sensing the prevailing conditions in the host. Indeed, RetS can sense unknown cues related to skin cell lysis in P. aeruginosa []. Moreover, a recent study showed that a component of the mucus coating epithelial cells, mucin glycans, activates RetS via its carbohydrate-binding site in the same model []. RetS also appears to be temperature sensitive. Indeed, high temperatures (close to 35 °C) affect antibiotic and cyanide production in P. protegens CHA0, possibly via a mechanism involving a change in membrane fluidity, leading to a stronger RetS-GacS interaction []. Thus, RetS acts as a temperature-sensitive sensor, inducing a switch in cellular metabolism in conditions of temperature stress for environmental pseudomonad strains, or on contact with warm-blooded host organisms for strains pathogenic to humans and animals.
3. Candidate GacS Signal Molecules Identified on the Basis of Their Demonstrated or Speculated Roles
Our ignorance of the identity of the GacS signal is not an isolated case: in many TCSs, the environmental or physiological stimulus triggering the activation of the sensor kinase and, thus, the intracellular regulation cascade, remains unclear [,]. The few TCS stimuli identified to date include biotic and abiotic factors, with diverse structures. The affinities of these signals for their sensor kinases range from submicromolar to millimolar values, reflecting the physiological need to trigger a response at a particular signal concentration []. The putative stimuli identified encompass simple and complex ions, mineral and organic molecules, including gases, but also solar radiation and physical phenomena acting at the nanoscale, such as the molecular agitation generated by heating. For example, the TodS/TodT TCS detects recalcitrant aromatic compounds in the cell environment, leading to expression of the toluene dioxygenase catabolic pathway [], whereas DesR/DesK senses decreases in temperature and increases membrane fluidity through Δ5-lipid desaturation [], and the sensing of blue light by LovK/LovR triggers a mechanism favoring bacterial cell attachment in response to the circadian photocycle []. In the face of the profusion of possible sensor kinase-activating candidates, most attempts to identify the GacS signal have been guided by the presumed or demonstrated role of the Gac/Rsm regulon in the model bacterium studied.
3.1. The GacS Sensor Kinase as the Trigger of a Metabolic Switch
Pseudomonads are known to have a considerable potential for metabolic adaptation, whether from oligotrophic to rich niches and vice versa, thanks to their highly versatile metabolism [,,,,,,]. They can assimilate diverse compounds, ranging from highly assimilable sugars, and organic and amino acids to much more recalcitrant molecules, such as hydrocarbons and xenobiotics, depending on the strain considered [,,,]. The whole metabolism of Pseudomonas is centered on the tricarboxylic acid cycle (TCA cycle, also known as the Krebs cycle), which is itself connected to the glyoxylate cycle and the oxygen and nitrogen oxide respiratory pathways [,,,]. Exceptionally, under anaerobic conditions, most pseudomonads can ferment pyruvate and arginine to generate acetate and lactate plus ornithine, respectively, a strategy that is used for long-term survival and redox balance rather than for growth [,]. The primary metabolism pathway of pseudomonads is associated with another based on the production of a wide range of secondary compounds, including cyanide, antibiotics (e.g., DAPG, phenazines, pyoluteorin), cyclolipopeptides, siderophores and phytohormones [,,,]. The synthesis of secondary metabolites is costly in terms of energy and is therefore affected by basal metabolism and used as an alternative by bacteria in conditions of competitive and nutritional stress [,,]. Secondary metabolites are thought to confer a selective advantage on the organisms producing them, in conditions in which these organisms cannot rely on their full growth potential to compete with other organisms in natural environments [,]. By contrast, during culture in synthetic media, these metabolites are not essential to the survival of their producers and are generally produced only during stationary phase, when cell population density is very high, and bacterial growth is restricted [,,].
The Gac/Rsm regulon encodes the components of the master regulatory pathway for secondary metabolite production in pseudomonads [,,]. The switch between primary and secondary metabolism is controlled by the RsmA (for “regulator of secondary metabolism”) and RsmA-like intracellular proteins. In E. coli, the RsmA homolog is called CsrA, for “carbon storage regulator”, revealing the connection of this protein to the regulation of basal metabolism [,]. A similar connection is observed in P. fluorescens, in which energy and carbon metabolism are influenced by Rsm proteins, and an rsmA/rsmE double mutant has been show to display slow growth []. RsmA and RsmE prevent the translation of transcripts for proteins involved in secondary metabolism, by preventing the ribosome from binding to target mRNAs. This repression is relieved by Rsm sRNAs, which are produced in large amounts under positive regulation by GacS/GacA, which act to neutralize the effects of RsmA and RsmE by titration [,]. RsmA can also have a positive effect on the translation of mRNAs encoding proteins involved in the induction of lipase and rhamnolipid synthesis, probably through mRNA stabilization []. In return, the functioning of Gac/Rsm seems to be affected by core primary metabolism, particularly as concerns the imbalance between the intracellular concentrations of TCAs []. Mutations of the gene encoding pyruvate carboxylase, an enzyme responsible for regenerating the oxaloacetate intensely consumes by the TCA cycle, lead to a downregulation of secondary metabolism in the absence of detectable nutrient limitation. By contrast, a mutation of the gene encoding a fumarase isoenzyme involved in ensuring the supply of malate results in an upregulation of secondary metabolism []. The authors showed that these effects were dependent on the GacS sensor kinase, but not on the RetS or LadS sensors, and that they required the production of RsmX, RsmY, and RsmZ sRNAs. Finally, they observed a strong positive correlation between GacA-dependent sRNA production and the intracellular pools of 2-oxoglutarate, succinate and fumarate []. This correlation can be explained by the amphibolic role of the TCA cycle, in which certain acids may be catabolized to generate energy, or extracted from the cycle to serve as precursors of secondary metabolites. These observations are also supported by the role assigned to thiamine in this switch []. Thiamine is an essential cofactor of TCA cycle enzymes []. It is required by pyruvate dehydrogenase, which supplies acetyl-coA to the cycle, and 2-oxoglutarate dehydrogenase, which oxidizes 2-oxoglutarate to generate succinyl-coA. A thiamine auxotrophic mutant derived from P. protegens CHA0 was found to have low levels of expression for the RsmX, RsmY and RsmZ sRNAs and for the AprA exoprotease and DAPG antibiotic secondary metabolites, when cultured in the presence of trace amounts of thiamine. The addition of excess exogenous thiamine to the medium complemented all the deficiencies of this mutant. Thus, in the presence of thiamine limitation, functions essential for the primary metabolism of the bacterium can occur, but not several key functions of GacS/GacA-dependent secondary metabolism []. These findings led to thiamine and TCA cycle acids being identified and tested as candidate direct or indirect signals activating the GacS sensor. However, none of these molecules seemed to be able to bind to GacS (Table 1).

Table 1.
GacS signal candidates previously studied in pseudomonads.
In Streptomyces, another bacterium producing a wide range of antibiotics, the switch between primary and secondary metabolism is underpinned by a transition from glycolytic to oxidative metabolism [,]. This probably promotes the production of TCA intermediates required for antibiotic synthesis and increases the efficiency of energy extraction from nutrient substrates required for secondary metabolism. Such a transition would be expected to occur in pseudomonads. Indeed, studies of the mode of glucose assimilation in Pseudomonas species have confirmed a preference for organic acids over sugars [,,,]. Glucose catabolism in pseudomonads is not directed by the conventional glycolysis pathway, but by the Entner-Doudoroff pathway, due, in particular, to a lack of phosphofructokinase, a key enzyme of the glycolytic pathway [,]. This results in the prior oxidation of a large proportion of the sugar present by the periplasmic glucose and gluconate dehydrogenases, leading to the production of gluconate and 2-oxogluconate, respectively []. Gluconate and 2-oxogluconate are produced under the control of the Gac/Rsm regulon, as is pyrroloquinoline quinone, a cofactor of glucose dehydrogenase []. Based on its production close to the periplasmic domain of GacS, gluconate has been considered a potential GacS ligand, as has pyrroloquinoline quinone (Table 1). Unfortunately, other compounds involved in the pathway leading to gluconic acid synthesis, such as 2-oxogluconate, 2-oxo-3-deoxygluconate (KDG) and its phosphorylated form 2-oxo-3-deoxy-6-phosphogluconate (KDPG), have yet to be evaluated. Indeed, KDG, a catabolite resulting from pectin breakdown, is known to act as a signal redirecting energy metabolism towards cell wall compounds in other γ-Proteobacteria, such as Dickeya and Pectobacterium spp. In Pseudomonas, KDGP is the central intermediate metabolite generated by the Entner-Doudoroff pathway. This compound binds the cytosolic HexR regulator, which controls glucose-responsive genes and some functions of central carbon metabolism, releasing the repression of target genes exerted by this protein [,].
3.2. The GacS Sensor as a Population Density (Quorum) Sensor
Numerous microorganisms use cell-to-cell communication systems based on both the synthesis and perception of signaling molecules to evaluate population density and synchronize their social behavior [,]. These quorum-sensing (QS) systems act through the following mechanism: each individual constitutively produces diffusible signals; the environmental concentration of signaling molecules is, therefore, directly linked to the density of the emitting population, but also to the level of cell lockdown favoring signal accumulation. Each member of the population is capable of detecting a critical concentration of signals, corresponding to the cellular quorum. The whole population is therefore, informed of the cellular quorum being reached and of the degree of diffusion from the microenvironment, between bacterial sites within and outside the host, for example. QS molecules generally induce their own production. At high concentration, they stimulate the expression of their synthases. Consequently, the production of QS signaling molecules is often optimal at the end of the exponential growth phase []. The best-known QS systems in Gram-negative bacteria produce molecules from the N-acyl-L-homoserine lactone (AHL) family []. In several Pseudomonas species, such as P. aeruginosa, P. chlororaphis and P. syringae, the Gac/Rsm system upregulates the synthesis of AHLs [,,,,]. Such upregulation has also been observed in P. brassicacearum, at least in the DF41 strain [,]. In these bacteria, GacA upregulates the transcription of luxI/luxR analog operons, encoding the enzyme synthesizing AHLs and the corresponding AHL response regulators. In addition, P. aeruginosa strains use at least two AHL-independent QS systems. The first is based on the secretion and detection of 2-heptyl-3-hydroxy-4(1H)-quinolone, which is usually referred to as the Pseudomonas quinone signal (PQS), and its precursor, 2-heptyl-4-quinolone (HHQ) [,]. The second uses 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde QS (IQS) and can integrate environmental stress cues []. The PQS and IQS systems are themselves under AHL-based QS control [], so all of these QS systems are ultimately governed by the GacS/GacA TCS. In other pseudomonads, such as P. protegens and most of the strains of the P. fluorescens-P. putida continuum, no AHLs have not detected, raising questions about the existence of QS systems in these species [,,,,].
At high cell population densities, during the transition from the exponential growth phase to stationary phase, the Gac/Rsm pathway of Pseudomonas spp. is activated by uncharacterized molecules present in their own culture supernatants. These molecules induce the phosphorylation of the GacS sensor kinase and, hence, of the cognate GacA response regulator, leading to sRNA transcription [,,,]. These activators also act as auto-inducers of the Gac/Rsm system via a positive feedback loop. Indeed, very little signal activity is present in the culture supernatants of P. fluorescens and P. aeruginosa gacA mutants [,,]. More generally, mutants with defects of the Gac/Rsm pathway usually display impaired social behavior []. All these traits suggest that the molecules activating GacS are also QS signals acting at the top of the QS signaling hierarchy in AHL-producing pseudomonads, and that the Gac/Rsm system is a QS-like alternative response in the absence of AHLs [,]. AHLs have, therefore, been identified as molecules that, if bound to GacS, could inform all pseudomonads of the nearby presence of bacterial populations producing AHLs, in addition to having a possible self-inducing effect in AHL-producing pseudomonads. Intriguingly, the signals extracted from Pseudomonas cultures with a high cell density appear to be unrelated to well-known QS signals, such as members of the AHL, PQS or auto-inducer-2 (AI-2) families (Table 1). This is also true for another candidate signal, homoserine lactone, the common core of all AHLs, recently identified as the ligand binding to an AHL-sensor in the Rhodococcus erythropolis QS-quencher []. Nevertheless, the corollary of the behavior of the GacS signal as an auto-inducer is that a compound with production stimulated by the Gac/Rsm regulon may be the GacS signal.
3.3. The GacS Sensor as a Promoter of Host Adaptation
Plants and microbes have evolved together over millions of years and collectively form a holobiont, a complex system involving beneficial, neutral and deleterious interactions between partners [,]. In plant-pathogen interactions, the attack on the plant generally begins with a phase of active bacterial multiplication, called primo-invasion, supported by highly assimilable photosynthates released by wounded tissues. A second step involving installation of the disease and its slow progression within the plant then occurs. This phase is less favorable to pathogens, because competition occurs within the dense population of invaders, which is also subjected to the response of the host’s immune defenses [,,,]. The Gac/Rsm regulatory pathway is thought to be involved in the ecological and metabolic transition of pseudomonad cells arising between these two phases of the disease. P. syringae illustrates this assertion well. About 50 pathovars of this pathogen infect almost all economically important crop species, earning P. syringae a place among the most pathogenic bacteria on Earth []. P. syringae is an epiphytic pathogen that infects the internal leaf tissues by swimming through wounds or natural openings, such as the stomata in the leaf surface []. It first secretes immunity-suppressing effectors into host cells via its type III secretion system (T3SS), which is crucial for the success of the attack. The GacS/GacA TCS affects the timing of the deployment of the T3SS and the release of effectors in target cells, although the precise timing of these events depends on the strain []. This has led to suggestions that some of the plant compounds present in the apoplast might be able to induce the Gac/Rsm regulon. It has been shown that the detection of D-fructose, sucrose and trehalose by P. syringae, but also by certain plant growth-promoting rhizobacteria (PGPR), such as P. fluorescens C7R12, induces the expression of key hrpA and hrpN genes involved in T3SS elongation and effector secretion, respectively, in conditions mimicking those of the apoplast [,].
In pseudomonads beneficial to the plant, the GacS/GacA TCS also plays a role in bacterial competition in the rhizosphere [,]. Thus, the GacS sensor is not specifically dedicated to the transition between virulence stages. The role of seed and root exudates as a nutrient source can account for the spectacular increases in the density and activities of some bacterial populations in the rhizosphere relative to bulk soil [,,]. In the PGPR P. protegens, CHA0, exudates released by various monocots and dicots stimulate DAPG antibiotic biosynthesis, which is directly under the control of the GacS/GacA TCS []. Other unidentified compounds present in seed exudates can also stimulate the GacS/GacA TCS of Pseudomonas sp. DSS73. In this biocontrol strain, exudate from sugar beet seed triggers biosynthesis of the antifungal cyclopeptide amphisin. The synthesis of amphisin in DSS73 is strictly dependent on GacS, and even induction by seed exudates requires a functional gacS locus []. Finally, it has also been suggested that polyamines and their precursor, N-acetyl-glutamate, which are present at high concentrations in root exudates, provide bacteria with information about the nutrient composition of the rhizosphere, as in P. chlororaphis, for example, in which the metabolism of these compounds is under the control of the Gac/Rsm network [] (Table 1).
Secondary metabolites, including phenolic compounds and γ-aminobutyric acid (GABA), are also released in large amounts by the plant during its growth and after wounding, revealing the presence of the plant to the soilborne microflora []. Many of these compounds are plant-bacteria signaling molecules [,,]. Single phenolic compounds (i.e., with only one or two aromatic rings), in particular, have been identified as possible candidates for interaction with pseudomonad GacS sensor kinases, because these compounds are already known to interact with GacS analogs encountered in other Proteobacteria phytopathogens. In the soft-rot bacterium Dickeya dadantii, trans-cinnamic and o-coumaric acids, at levels that appear physiologically relevant in plants, have been shown to induce the hrpA and hrpN T3SS genes, and, thus, to suppress plant defenses. The effect of these phenolic compounds is linked to the Gac/Rsm transduction pathway and requires a functional GacS sensor kinase, suggesting that these phenolic compounds are signals detected by GacS in these bacteria []. In Agrobacterium tumefaciens, a pathogen that induces tumorous crown galls in a wide range of dicots, the expression of most virulence (vir) genes is conditioned by the action of the close GacS/GacA homolog, VirA/VirG, with VirA functioning as a histidine sensor kinase, and VirG as a DNA-binding response regulator. VirA recognizes acetosyringone and its structural analogs. It also responds to specific monosaccharides and low pH, by enhancing vir expression []. The cytoplasmic linker domain of VirA has been implicated in pH and phenol sensing, but monosaccharides seem to be sensed by the periplasmic domain, which enhances the sensitivity and maximal response to phenolic compounds []. In P. syringae, some virulence traits are also induced by a mixture of sugar and phenolic compounds: the synthesis of syringomycin and syringopeptin phytotoxins, which are released following plant aggression, is induced by D-fructose and the phenolic sugar arbutin, under the control of the GacS/GacA TCS []. According to the speculative model established by the authors, P. syringae senses these signaling molecules in the plant apoplast via the GacS sensor kinase, and transmits a relay signal to the GacA response regulator.
3.4. The GacS Sensor as a Trigger of Lifestyle Change
Bacteria are only rarely found as single dispersed organisms. They generally live in communities and may colonize the surfaces of minerals and higher organisms, forming biofilms []. During this process, they lose motility and attach to surfaces, forming cellular aggregations or microcolonies that are embedded in extracellular polymers providing protection from the surrounding environment. In various Pseudomonas species capable of infecting both plant and animal hosts, GacS/GacA functions as a master regulator of multiple virulence traits, including those leading to the crucial switches between motile and sessile lifestyles [,,,]. P. syringae regularly switches back and forth between these lifestyles. P. syringae cells live both on mineral and plant surfaces, often in the epilithic and phyllosphere biofilms, respectively []. The apoplast is a potential carbohydrate-rich habitat for the bacterium and is targeted by P. syringae, but heavily defended by the plant. P. syringae therefore has to trigger a succession of lifestyle switches to cause disease, in which it develops motility, toxin and exopolysaccharide production, and the secretion of immunity-suppressing effectors into host cells via its T3SS [,,]. A model has recently been proposed in which the Gac/Rsm operon regulates motility and the T3SS in opposite manners during host infection by P. syringae DC3000. In this model, GacS/GacA TCS is activated on the leaf surface, thereby promoting motility and damping down deployment of the T3SS. It is then deactivated during early apoplast colonization, leading to a decrease in motility and an increase in type III secretion to suppress host immunity []. The authors of this study also suggested that the GacS/GacA TCS may coordinately regulate additional virulence-associated traits, such as toxin production and biofilm formation, in a similar manner []. Thus, the GacS/GacA TCS and ensuing lifestyle switches are triggered by nutrient deficiency, as experienced on the surface of the leaf, and deactivated in conditions of nutrient abundance.
P. aeruginosa is a highly prevalent opportunistic pathogen of animals and humans, particularly immunocompromised individuals and cystic fibrosis patients. However, some studies have shown that although this bacterium does not cause damage in field conditions, it can cause black necrosis when used to inoculate aerial parts of the plant, such as the leaves. These symptoms reflect the action of virulence factors altering both human and plant cells, such as phospholipase C, exotoxin A and T3SS, which is known to be produced under the control of the Gac/Rsm regulon [,,]. The switch from a planktonic to a sessile lifestyle is a survival advantage enabling pathogens, such as P. aeruginosa, to evade stresses and adverse conditions. The formation of a biofilm may decrease the acute virulence of the pathogen, but it provides P. aeruginosa strains with an extremely high capacity to persist against host responses, such as phagocytosis, free radical release by immune cells, and antibiotics used for treatment, in addition to nutrient and oxygen limitation []. P. aeruginosa switches between the motile and sessile lifestyles, modulating its secretion of virulence effectors by a plethora of TCS, QS networks, sRNAs and transcription factors [,,]. The Gac/Rsm regulatory network acts as the decision-making switch [,]. However, the continued functioning and flexibility of Gac/Rsm over time are particularly important during the transition to a sessile lifestyle, because RsmY and RsmZ sRNA levels, and, consequently, RsmA activity, are continually modulated during the process of biofilm development []. The RNA-binding protein RsmA downregulates surface attachment, exopolysaccharide production and biofilm maturation, whilst inducing flagellum-based motility, the production of the T3SS, type 4 pili and other virulence factors, such as exotoxins and proteases []. RsmA also represses diguanylate cyclases, thereby preventing an increase in intracellular 3′,5′-cyclic diguanylic acid (cyclic di-GMP) levels. Cyclic di-GMP is an almost ubiquitous second messenger present in diverse bacteria. Increases in its levels within cells inhibit motility and trigger biofilm formation. The sensing of environmental cues and stress conditions by the envelope sensors of P. aeruginosa triggers the Gac/Rsm cascade. This cascade leads to the generation of the RsmY and RsmZ sRNAs, which counteract the translational repression activity of RsmA, leading to an increase in cyclic di-GMP levels, and, consequently, biofilm formation [,].
During pathogenesis P. aeruginosa cells in the planktonic state are considered to be equipped for the aggressive primo-invasion of the host required for acute infection. By contrast, the formation of a mucoid biofilm is a hallmark of chronic infections and indicative of disease progression and long-term persistence [,]. Calcium has been identified as a first signal by which the Gac/Rsm pathway induces a switch from acute-to-chronic virulence has been identified in P. aeruginosa [] (Section 2.2). This discovery led to the concentrations of ions surrounding the bacteria in the vicinity of the host being considered as important cues, and to the notion of diverse possible roles, including as a potential ligand of the GacS sensor, but without convincing results (Table 1). Another avenue of investigation that has yet to be explored, is the possibility that GacS signal is a typical component of the biofilm initiating its own production or revealing its presence to external bacterial populations, thereby favoring biofilm extension. Such a role has been hypothesized for mannuronic acid and guluronic acid, the monomers of alginate, a major exopolysaccharide component of biofilms. Indeed, alginate production is induced by the Gac/Rsm cascade under conditions of metabolic stress, such as microaerophilia or anaerobiosis, as observed during chronic P. aeruginosa infections and in P. fluorescens mucoid colonies [,]. Moreover, the concentrations of these acids are controlled in the periplasm, the location of GasS signal-binding domain, in which alginates not exported to the microenvironment are degraded. Finally, alginate biosynthesis is costly to the bacterium, due to its requirement of nucleotide sugars and the sequestering of carbon, to the detriment of central metabolism. Biofilm production therefore leads to a switch between cell growth sustained by primary metabolism and the production of exopolysaccharides [,].
The Gac/Rsm signal transduction pathway is also involved in the switch between the planktonic and biofilm lifestyles in non-pathogenic Pseudomonas spp. [,,,]. In PGPR strains, this switch enables the bacteria to proliferate on plant roots and, subsequently, to compete effectively with other rhizosphere organisms in a sessile mode of growth [,]. Genome-wide transcriptome analysis of P. fluorescens SBW25 revealed that a mutation of gacS led to changes in the expression of genes involved in biofilm formation, motility, stress responses and survival []. The spontaneous mutation of Gac/Rsm system genes in P. fluorescens F113 and P. brassicacearum NFM421 enhances motility and root colonization, probably due to improvements in the harvesting and use of rhizosphere exudates [,]. Similar differences were also observed in laboratory conditions, especially in rich media, in which gacS or gacA mutants grow well and may even have a temporary advantage over the wild-type []. By contrast, Gac mutants are generally poor at biofilm formation. They have a lower capacity for biocontrol and are considered to be less competitive than the wild-type against other microbes, particularly in conditions of nutrient limitation. This defect is imputed to the low levels of secondary metabolites produced by Gac mutants, particularly for HCN and antibiotic biosynthesis [,]. Based on these observations, some plant photosynthates and secondary metabolites exuded into the phyllosphere, apoplast or rhizosphere were identified as potential GacS inducers (Table 1). However, the role of each of the molecules studied, particularly in switches to a more efficient lifestyle, is difficult to understand fully, depending on whether the molecules are considered to act as extra- or intracellular signals. This is the case with GABA, a compound produced by numerous bacteria, but also by plant hosts []. Kasumi Takeuchi [] showed that the Gac/Rsm pathway has effects on GABA intracellular accumulation similar to those on secondary metabolism and the production of extracellular enzymes. Curiously, the accumulation of GABA promotes planktonic growth and root colonization, and reduces the formation of biofilms in P. protegens CHA0, entirely contrary to expectations. It appears that GABA should not be considered primarily as an extracellular signal monitoring the plant, but rather as an bacterial intracellular signal inhibiting the Gac/Rsm cascade and biofilm formation, with other intracellular effectors such as cyclic di-GMP and guanosine tetra/pentaphosphate ((p)ppGpp), usually having the opposite effect [,].
4. Strategies for Characterizing the Nature of the GacS Signal
4.1. Massive Extraction and Fine Characterization of Self-Produced GacS Signals
The paradigm according to which the GacS signal is used as a means of communication in pseudomonads is based on observations that (i) Pseudomonas cells growing at high population densities excrete a compound or compounds activating their own GacS sensor kinase, and (ii) these compounds act as a QS-molecule (see Section 3.2). A mutagenesis strategy was therefore envisaged to target the gacS anabolic genes of a Pseudomonas GacS signal emitter and to identify them by comparison with existing gene databases. Provided that the genes concerned are relatively small and clustered, as for the AHL and AI-2 biosynthetic genes, it is possible to establish a genomic library for the isolation of genes involved signal synthesis []. Conversely, if the genes involved in signal design are too numerous and dispersed throughout the genome, the data may not be suitable for full characterization of the signal molecule. This is the case for the cluster of genes directing the unidentified Vfm-QS signal found in Dickeya spp. These genes encode proteins displaying similarities both to enzymes involved in amino-acid activation and enzymes involved in fatty-acid synthesis, but provide no decisive insight into the structure of Vfm signaling molecules []. No mutants with specific defects of structural genes for GacS signal synthesis are currently available []. Unfortunately, it is not possible to obtain any real additional information on the complexity of GacS signal structure by this approach, because it remains possible that the genes involved in the synthesis of the GacS signal or its precursors are essential for cell metabolism and that all mutations affecting GacS signal synthesis are, thus, lethal.
As an alternative to mutagenesis, analytical biochemistry strategies for identifying GacS signals are based on the extraction and characterization of the molecules trapped in the supernatant of the producing bacteria. Thus, supernatants of P. protegens CHA0 cultures grown to late exponential growth phase contain, in addition to the molecules present during the stationary phase, signaling molecules that activate the transcription of the RsmY and RsmZ sRNAs, resulting in the induction of secondary metabolites, such as HCN [,,]. Thanks to this fundamental finding, Christophe Dubuis came very close, during his PhD [], to identifying a GacS signal produced by the CHA0 strain. He first cultured this model strain at high density in GCM synthetic medium []. This medium contains principally glycerol as the basic carbon and energy source, supplemented with casamino acids to satisfy selective nitrogen requirements, buffered with phosphate salts. Casamino acids are a mixture of amino acids derived from fully hydrolyzed casein. The use of GCM medium was based on the hypothesis that none of these amino acids was itself the GacS signal, but that they were likely to support its production. As expected, uninoculated GCM medium did not contain the signal []. Moreover, it has since been shown that, when cultured in this medium, P. aeruginosa rapidly and completely digests amino acids that can be easily integrated into central metabolic pathways until the mid-exponential growth phase, subsequently digesting others that require a more complex catabolism []. Finally, supernatants from the stationary phase mostly lack these amino-acids components []. Dubuis [] subjected a large volume (about 100 L) of culture supernatant to extraction in dichloromethane. The dichloromethane extract was purified by separation on a silica gel column with a linear gradient of methanol. The fractions obtained were then assayed for β-galactosidase activity with P. protegens CHA0 (hcnA’-‘lacZ) or (rsmZ-lacZ) reporter strains, leading to the identification of two different active fractions. The most promising fraction was subjected to chromatography on a Sephadex LH-20 column, with methanol as the solvent. Finally, the constituents of the purest concentrated active fraction were separated by reverse-phase chromatography on a C18 column, by HPLC-MS analysis, or by reverse-phase chromatography followed by GC-MS. The active fraction contained a compound with a molecular mass of m/z 278, in which the MS detection of specific fragments suggested the presence of at least two hydroxyl groups []. The stability of this GacS signal was evaluated by treating the fraction with proteinase K, boiling for 10 min or exposure to pH 2 and pH 12: these treatments did not affect the activity of extracts []. Stability at pH 12 indicates that the signals are not AHLs, because, at this pH, the lactone bond of AHLs is hydrolyzed []. If this stability at extreme pH was confirmed, it would exclude from the list of GacS signal candidates the hydroxycarboxylic acids which spontaneously cyclize to form lactones, with a five- or six-membered ring (γ- and δ-lactones). The absence of absorption at 210 and 254 nm, indicates that the extracted signal is unlikely to contain an aromatic ring or conjugated double bonds. A peptide structure is also unlikely, because the signal provided no evidence of amino groups on MS and was not destroyed by proteinase K []. Pinolenic acid (CAS number 168-33-54-8), which is found in diverse pine nuts, is an unsaturated aliphatic monocarboxylic acid with 18 carbon atoms and the same molecular mass as the GacS signal extracted from the CHA0 strain. However, this compound has no hydroxyl or ketone groups. According to the work of Dubuis, the GacS signal probably has a structure closely resembling that of this compound. By analogy with its QS functions, it could also have a structure similar to the acyl chain part of AHLs, or to the signaling molecules produced by Ralstonia spp. (i.e., the volatile extracellular factor 3-hydroxy-palmitate ester) [], or by Xanthomonas spp. (i.e., the diffusible signal factor cis-11-methyl-2-dodecenoate) [].
Although it was not possible to characterize the active compound present in the CHA0 fraction fully, the GacS induction capacities of the purified fraction were assessed. CHA0 mutants with defects of gacS and gacA produced signals with a strength about 25- to 30-fold weaker than that of the wild-type [], confirming that signaling molecules induce their own synthesis in an auto-induction process. Self-produced GacS signals have been found in various Pseudomonas species, making it possible to compare signaling both within and between species [,]. This work demonstrated the existence of crosstalk responding to the signal extracted from P. protegens CHA0. This crosstalk occurs at variable intensities, between strains from the same species, but also between strains from different Pseudomonas species (e.g., P. protegens CHA0 and P. aeruginosa PAO1) or even different genera (Pseudomonas and Vibrio) [,]. These findings suggest a certain ubiquity in GacS signal recognition and, therefore, structure, at least in Pseudomonas spp. However, the GacS signal may be due to a family of molecules, such as those of the AHL family, for example, accounting for the different intensities recorded in response to the CHA0 signal.
4.2. Metabolomic and Wide Screenings of Potential GacS Signals
The metabolome is the inventory of all the metabolites in a biological system []. Metabolites are the end products of cellular regulatory processes, and their levels can be regarded as the ultimate response of biological systems to genetic or environmental changes []. The GacS signal therefore corresponds to a typical metabolite excreted into the extracellular metabolome. Metabolomic analysis is usually based on techniques such as NMR spectroscopy, matrix-assisted laser desorption-ionization time of flight (MALDI-TOF) and imaging mass spectrometry (IMS). Liquid-state NMR spectroscopy has been used to characterize the medium in which pseudomonad cells are cultured, to identify differences in excreted metabolites and changes in the amount of compounds present in the medium []. It can be used to monitor the metabolic footprint of cultures over the entire growth curve, which is essential when searching for QS signals, for example []. However, the sensitivity of classic NMR analysis is relatively limited for the capture of signaling molecules, which are generally present at very low concentrations in crude extracts. NMR spectroscopy on fractions with GacS activity isolated from P. protegens CHA0 supernatant was inconclusive, probably because the concentration of signal molecules was too low for significance to be achieved in the analysis []. Improvements to NMR spectroscopy, based on techniques such as high-resolution magic-angle spinning (HRMAS), have been used to determine the chemical composition of the cells themselves. Moreover, to promote the separation of metabolites and to increase sensitivity, gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-mass spectrometry (LC-MS) have often been incorporated into NMR studies, for the quantitative profiling of low-molecular weight metabolites in particular [,,]. Finally, MALDI-IMS adds the possibility of analyzing various compounds involved in the interactions of microbial colonies [,]. Metabolomic approaches based on this tool have made it possible to visualize metabolic exchanges within and between microbial species, including the production of secondary metabolites by pseudomonads [,,]. Interpretation of the results obtained with metabolomic tools requires the use of powerful spectral databases (e.g., METLIN, MassBank) and appropriate software (e.g., CFM-ID). Metabolomic databases, such as the Pseudomonas aeruginosa Metabolome Database (PAMDB), the Golm Metabolite Database (GMD), Metabolights, Global Natural Products Social Molecular Networking (GNPS), and the Kyoto Encyclopedia of Genes and Genomes (KEGG), can be used to explore metabolite structures and pathways in addition to significant genes involved in the biosynthesis of compounds [,].
Metabolomic approaches have not yet been used specifically in the search for the GacS signal, but they have been used to study physiological phenomena known to be under the control of the GacS/GacA TCS. The metabolome of wild-type P. protegens CHA0, a gacA-negative mutant, which has lost Gac/Rsm activities, and a retS-negative mutant, which shows strongly enhanced Gac/Rsm-dependent activities, were compared after these strains were grown in a modified GCM medium []. This work highlights some intracellular metabolites, the production of which is controlled by the Gac/Rsm cascade. As example, the production of ppGpp and 3-hydroxyaspartic acid are more than six-fold upregulated in gacA and retS mutants than in the wild-type, respectively []. Other studies have made it possible to identify several metabolic pathways that are stimulated or repressed in P. aeruginosa (i) during changes in environmental metabolic conditions [], (ii) under AHL-dependent QS regulation [,], (iii) during lifestyles adopted by the bacteria under planktonic or biofilm growth [], and (iv) during long-term adaptation of the bacteria in the host [,,]. These studies cannot provide crucial information about the nature of the GacS signal, but they have made it possible to confirm the key role of metabolites already identified as influencing and/or being under the influence of the Gac/Rsm regulon. These metabolites are TCA cycle intermediates, amino acids, polyamines and fatty acids, the concentrations of which are affected during the switch studied [,,].
5. Conclusions
The Gac/Rsm regulon and its master extracellular sensor, GacS/GacA, are powerful elements controlling the expression of the about 10% of the genes in the genomes of various pseudomonads [,,,,]. As a consequence, many studies have attributed diverse roles to the GacS sensor, ranging from bacterial QS to triggering metabolic or lifestyle switches, in response to conditions encountered within or close to the plant, for example. Far from being incompatible, all these functions are tightly interconnected, and can be explained in terms of the nutrient availability. Indeed, the signals activating the GacS/GacA TCS are produced at high cell population densities, triggering the Rsm cascade. This favors the switch from primary to secondary metabolism and from lifestyle, effectively contributing to competition between the GacS signal producer and the native microflora, thereby improving survival or fitness in the ecological niche [,,]. The virulence and biocontrol factors controlled by the Gac/Rsm pathway are highly dependent on population size and can therefore, be regarded as manifestations of social behavior to deal with competition with invaders and the immune responses encountered the host [,,,,,]. Thus, most mutants with defects of the Gac/Rsm pathway have lost part or all of their virulence or have a reduced ability to antagonize fungi and to suppress plant diseases [,]. Serendipitously, these physiological and ecological transitions can easily be mimicked under laboratory conditions. During pure culture, a Pseudomonas strain reaching the late stages of the exponential growth phase produces a critical quantity of GacS signal. This suggests that the nature of the compounds activating GacS is linked to the appearance of nutritional stresses during restricted growth at high cell population density. This observation is supported by the positive regulation of the Gac/Rsm system by ppGpp, an intracellular signal characteristic of nutrient limitation and stress []. Moreover, (p)ppGpp is also known to be integrated into QS signaling networks, which ensure that costly virulence factors are produced at the appropriate cell density and under conditions of nutrient limitation [,]. The hypothesis that the GacS signal monitors metabolic stress might also account for the profusion of potential signal sources proposed in previous studies. During the hostile conditions imposed by competition for nutrients, GacS signals may be produced not only by pseudomonad populations, but also by the indigenous microbial community and/or the eukaryote host. Compounds with a profile of this type would be diffusible catabolites that may or may not be converted into signals by pseudomonads, with concentrations increasing in the microenvironment in a density-dependent manner. Such a mechanism of metabolic transition has recently been described in P. protegens strain Pf5, in which an intermediate catabolite produced during DAPG synthesis is converted into cell-cell communication signals by halogenation, in turn activating the independent pyoluteorin anabolic pathway [].
Structural analysis has shown that the GacS sensor kinase acts as an antenna, receiving an extracellular signal, the impact of which is influenced by subservient membrane modules indicating the presence of the host. The periplasmic GacS ligand probably corresponds to a molecule or family of molecules with a low molecular weight, facilitating passage across the outer membrane of the bacterial wall and binding in the narrow pocket identified in the periplasmic domain of GacS. This ligand may carry one or more negative charges capable of interacting with the positive charges encountered in the ligand-binding site, thus favoring such interactions. For example, TCA cycle components, such as citrate, succinate and fumarate, have been shown to act as ligands of the narrow binding pocket located on the periplasmic sensor domains of their sensor kinases, CitA in K. pneumoniae, DcuS in E. coli, and DctB in Vibrio cholerae, respectively [,]. A screening strategy for competitive inhibitors targeting the periplasmic binding site of GacS may facilitate advances in research, by making it possible to deduce the structure of the GacS signal from that of its inhibitors.
Attempts to purify the GacS signal produced by P. protegens CHA0 showed that it could not be a previously characterized QS molecule, or a small protein or peptide, and that it did not appear to contain aromatic rings []. This last finding rules out plant phenolic candidates, at least in this strain. The same study revealed that the GacS signal was a relatively simple and small molecule, apolar but moderately volatile, as revealed by GC-MS purification. It probably consists of an aliphatic chain with oxygen substituents []. Taking into account all these considerations, aliphatic organic acids are serious candidates for the chemical class of the GacS signal. These compounds are often hydrophobic and, therefore, cell diffusible, depending on the length and degree of unsaturation of their chain. Like other signaling molecules (e.g., AHLs), they can indirectly monitor extra- and intracellular pH changes according to their level of protonation, with pH an abiotic factor known to vary with cell metabolism. Finally, the number and locations of hydrogen substitutions by hydroxyl or ketone groups harbored by the aliphatic chain of the acid would provide the typical features of the molecule necessary to characterize the role and specificity of the signal, as for the N-acyl chain of AHLs.
The increasing development of dynamic and versatile metabolite identification tools may make it possible to identify the family and structure of the GacS signal fully in the near future. It will then be necessary to determine the effective physiological concentration of the GacS signal according to environmental conditions, and its impact on the host and other microorganisms. Finally, these advances should make it possible to establish GacS signal-quenching strategies, similar to those used to control the virulent AHL-based QS communication of certain Gram-negative pathogens [,,,]. Such clinical and agroecological approaches, recently referred to as “bacterial informational wars” [,], might raise a new hope for reducing the Gac/Rsm pathway-based virulence of pathogens. Conversely, the addition of GacS signals or biostimulating molecules (e.g., cheap GacS agonists) could trigger and increase the Gac/Rsm-dependent biocontrol exerted by some PGPRs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1746/s1, Figure S1: List of abbreviations used throughout this review.
Funding
This research was supported by grants from French institutions (Ministère de l’Enseignement Supérieur, de la Recherche et de l’innovation, the Région Normandie and the Evreux Portes de Normandie agglomération, the Pôle de Compétivité Valorial, and FEDER (European Union).
Acknowledgments
This article is dedicated to the memory of Dieter Haas, Head of the Department of Fundamental Microbiology at the University of Lausanne (Switzerland), and expert in the metabolism and Gac/Rsm signal transduction pathway of fluorescent pseudomonads. He passed away on 22 April 2017, at the age of 71. The author thanks Alex Edelman and associates for linguistic support.
Conflicts of Interest
The author declares no conflict of interest.
References
- Bhate, M.P.; Molnar, K.S.; Goulian, M.; William, F.; DeGrado, W.F. Signal transduction in histidine kinases: Insights from new structures. Structure 2015, 23, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Krell, T.; Lacal, J.; Busch, A.; Silva-Jiménez, H.; Guazzaroni, M.E.; Ramos, J.L. Bacterial sensor kinases: Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 2010, 64, 539–559. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Mechaly, A.; Betton, J.-M.; Antoine, R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018, 16, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef] [PubMed]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009, 73, 434–445. [Google Scholar] [CrossRef]
- Heeb, S.; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant-Microbe Interact. 2001, 14, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Marutani, M.; Taguchi, F.; Ogawa, Y.; Hossain, M.M.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol. Genet. Genomics 2008, 279, 313–322. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Malley, M.R.; Chien, C.F.; Peck, S.C.; Lin, N.C.; Anderson, J.C. A revised model for the role of GacS/GacA in regulating type III secretion by Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2020, 21, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Workentine, M.L.; Chang, L.; Ceri, H.; Turner, R.J. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: A bacterial two-hybrid analysis. FEMS Microbiol. Lett. 2009, 292, 50–56. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology 2000, 146, 2345–2350. [Google Scholar] [CrossRef] [PubMed]
- Palleroni, N.J. Introduction to the Family Pseudomonadaceae. In The Prokaryotes, 2nd ed.; Balows, A., Trüper, H.G., Dworkin, M., Harder, W., Schleifer, K.H., Eds.; Springer: New York, NY, USA, 1992; Volume 160, pp. 3071–3085. [Google Scholar]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Ronald, G.; Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]
- Mazurier, S.; Merieau, A.; Bergeau, D.; Decoin, V.; Sperandio, D.; Crépin, A.; Barbey, C.; Jeannot, K.; Vicré-Gibouin, M.; Plésiat, P.; et al. Type III secretion system and virulence markers highlight similarities and differences between human- and plant-associated pseudomonads related to Pseudomonas fluorescens and P. putida. Appl. Environ. Microbiol. 2015, 81, 2579–2590. [Google Scholar] [CrossRef]
- Morris, C.E.; Bardin, M.; Kinkel, L.L.; Moury, B.; Nicot, P.C.; Sands, D.C. Expanding the paradigms of plant pathogen life history and evolution of parasitic fitness beyond agricultural boundaries. PLoS Pathog. 2009, 5, e1000693. [Google Scholar] [CrossRef]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Anderson, A.J.; Kang, B.R.; Kim, Y.C. The Gac/Rsm signaling pathway of a biocontrol bacterium, Pseudomonas chlororaphis O6. Res. Plant Dis. 2017, 23, 212–227. [Google Scholar] [CrossRef]
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364, fnx104. [Google Scholar] [CrossRef]
- Cheng, X.; de Bruijn, I.; van der Voort, M.; Loper, J.E.; Raaijmakers, J.M. The Gac regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 2013, 5, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, K.; Schubert, M.; Allain, F.H.; Haas, D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Borland, S.; Prigent-Combaret, C.; Wisniewski-Dyé, F. Bacterial hybrid histidine kinases in plant-bacteria interactions. Microbiology 2016, 162, 1715–1734. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C.; Reimmann, C. Signal transduction in plant-beneficial rhizobacteria with biocontrol properties. Antonie Van Leeuwenhoek 2002, 81, 385–395. [Google Scholar] [CrossRef]
- Teplitski, M.; Merighi, M.; Gao, M.; Robinson, J. Integration of Cell-to-Cell Signals in Soil Bacterial Communities. In Soil Biology: Biocommunication in Soil Microorganisms; Witzany, G., Ed.; Springer: Berlin, Germany, 2011; Volume 23, Chapter 14; pp. 369–401. [Google Scholar] [CrossRef]
- Heeb, S.; Cámara, M.; Filloux, A.; Williams, P. Professor Dieter Haas (1945–2017). FEMS Microbiol. Rev. 2017, 41, 597–598. [Google Scholar] [CrossRef]
- Laville, J.; Voisard, C.; Keel, C.; Maurhofer, M.; Défago, G.; Haas, D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 1992, 89, 1562–1566. [Google Scholar] [CrossRef]
- Ramette, A.; Frapolli, M.; Fischer, L.M.; Gruffaz, C.; Meyer, J.M.; Défago, G.; Sutra, L.; Moënne, L.Y. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 2011, 34, 180–188. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Willis, D.K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 1992, 174, 3011–3020. [Google Scholar] [CrossRef]
- Willis, D.K.; Hrabak, E.M.; Rich, J.J.; Barta, T.M.; Panopoulos, N.J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation in bean. Mol. Plant-Microbe Interact. 1990, 3, 149–156. [Google Scholar] [CrossRef]
- Rich, J.J.; Kinscherf, T.G.; Kitten, T.; Willis, D.K. Genetic evidence that the gacA gene encodes the cognate response regulator for the LemA sensor in Pseudomonas syringae. J. Bacteriol. 1994, 176, 7468–7475. [Google Scholar] [CrossRef]
- Jimenez, N.P.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The Multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef]
- Sobrero, P.M.; Valverde, C. Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front. Mol. Biosci. 2020, 7, 127. [Google Scholar] [CrossRef]
- Heeb, S.; Blumer, C.; Haas, D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002, 184, 1046–1056. [Google Scholar] [CrossRef]
- Kay, E.; Dubuis, C.; Haas, D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. USA 2005, 102, 17136–17141. [Google Scholar] [CrossRef]
- Kay, E.; Humair, B.; Dénervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; Haas, D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 2011, 91, 63–79. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B.; Wu, H.; Wu, X.; Yan, Q.; Zhang, L.Q. Pleiotropic effects of RsmA and RsmE proteins in Pseudomonas fluorescens 2P24. BMC Microbiol. 2020, 20, 191. [Google Scholar] [CrossRef]
- Zuber, S.; Carruthers, F.; Keel, C.; Mattart, A.; Blumer, C.; Pessi, G.; Bonnefoy, G.C.; Keel, S.U.; Heeb, S.; Reimmann, C.; et al. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 2003, 16, 634–644. [Google Scholar] [CrossRef]
- Ali-Ahmad, A.; Fadel, F.; Sebban-Kreuzer, C.; Ba, M.; Pélissier, G.D.; Bornet, O.; Guerlesquin, F.; Bourne, Y.; Bordi, C.; Vincent, F. Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 11262. [Google Scholar] [CrossRef]
- Goswami, M.; Espinasse, A.; Carlson, E.E. Disarming the virulence arsenal of Pseudomonas aeruginosa by blocking two-component system signaling. Chem. Sci. 2018, 9, 7332–7337. [Google Scholar] [CrossRef]
- Ventre, I.; Goodman, A.L.; Vallet, G.I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004, 7, 745–754. [Google Scholar] [CrossRef]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef]
- Broder, U.N.; Jaeger, T.; Jenal, U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016, 2, 16184. [Google Scholar] [CrossRef]
- Chambonnier, G.; Roux, L.; Redelberger, D.; Fadel, F.; Filloux, A.; Sivaneson, M.; de Bentzmann, S.; Bordi, C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016, 12, e1006032. [Google Scholar] [CrossRef]
- Francis, V.I.; Waters, E.M.; Finton-James, S.E.; Gori, A.; Kadioglu, A.; Brown, A.R.; Porter, S.L. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 2018, 9, 2219. [Google Scholar] [CrossRef] [PubMed]
- Mancl, J.M.; Ray, W.K.; Helm, R.F.; Schubot, F.D. Helix cracking regulates the critical interaction between RetS and GacS in Pseudomonas aeruginosa. Structure 2019, 27, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Pydi, S.P.; Li, Y.; Lin, C.; Kong, W.; Chelikani, P.; Duan, K. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 2017, 3, 162–175. [Google Scholar] [CrossRef]
- Kong, W.; Chen, L.; Zhao, J.; Shen, T.; Surette, M.G.; Shen, L.; Duan, K. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 2013, 88, 784–797. [Google Scholar] [CrossRef]
- Vincent, F.; Round, A.; Reynaud, A.; Bordi, C.; Filloux, A.; Bourne, Y. Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ. Microbiol. 2010, 12, 1775–1786. [Google Scholar] [CrossRef] [PubMed]
- LeRoux, M.; Kirkpatrick, R.L.; Montauti, E.I.; Tran, B.Q.; Peterson, S.B.; Harding, B.N.; Whitney, J.C.; Russell, A.B.; Traxler, B.Y.A.; Goodlett, D.R.; et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 2015, 4, e05701. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.X.; Wheeler, K.M.; Cady, K.C.; Lehoux, S.; Cummings, R.D.; Laub, M.T.; Ribbeck, K. Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Humair, B.; González, N.; Mossialos, D.; Reimmann, C.; Haas, D. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J. 2009, 3, 955–965. [Google Scholar] [CrossRef]
- Busch, A.; Lacal, J.; Martos, A.; Ramos, J.L.; Krell, T. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc. Natl. Acad. Sci. USA 2007, 104, 13774–13779. [Google Scholar] [CrossRef]
- Aguilar, P.S.; Hernandez-Arriaga, A.M.; Cybulski, L.E.; Erazo, A.C.; de Mendoza, D. Molecular basis of thermosensing: A two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001, 20, 1681–1691. [Google Scholar] [CrossRef]
- Purcell, E.B.; Siegal, G.D.; Rawling, D.C.; Fiebig, A.; Crosson, S. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. USA 2007, 104, 18241–18246. [Google Scholar] [CrossRef]
- Bossis, E.; Lemanceau, P.; Latour, X.; Gardan, L. The taxonomy of Pseudomonas fluorescens and Pseudomonas putida: Current status and need for revision. Agron. Sustain. Dev. 2000, 20, 51–63. [Google Scholar] [CrossRef]
- Frimmersdorf, E.; Horatzek, S.; Pelnikevich, A.; Wiehlmann, L.; Schomburg, D. How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Environ. Microbiol. 2010, 12, 1734–1747. [Google Scholar] [CrossRef]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W., II; Kent Lim, C.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Carbon catabolite repression in Pseudomonas: Optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 2010, 34, 658–684. [Google Scholar] [CrossRef]
- Kamilova, F.; Kravchenko, L.V.; Shaposhnikov, A.I.; Makarova, N.; Lugtenberg, B. Effects of the tomato pathogen Fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol. Plant-Microbe Interact. 2006, 19, 1121–1126. [Google Scholar] [CrossRef]
- La Rosa, R.; Johansen, H.K.; Molin, S. Adapting to the airways: Metabolic requirements of Pseudomonas aeruginosa during the infection of cystic fibrosis patients. Metabolites 2019, 9, 234. [Google Scholar] [CrossRef]
- Lemanceau, P.; Corberand, T.; Gardan, L.; Latour, X.; Laguerre, G.; Boeufgras, J.; Alabouvette, C. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 1995, 61, 1004–1012. [Google Scholar] [CrossRef]
- Arai, H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol. 2011, 2, 103. [Google Scholar] [CrossRef]
- Latour, X.; Lemanceau, P. Carbon and energy metabolism of oxidase-positive saprophytic fluorescent Pseudomonas spp. Agronomie 1997, 17, 427–443. [Google Scholar] [CrossRef]
- Valentini, M.; Lapouge, K. Catabolite repression in Pseudomonas aeruginosa PAO1 regulates the uptake of C4-dicarboxylates depending on succinate concentration. Environ. Microbiol. 2013, 15, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, R.; Johansen, H.K.; Molin, S. Convergent metabolic specialization through distinct evolutionary paths in Pseudomonas aeruginosa. mBio 2018, 9, e00269-18. [Google Scholar] [CrossRef] [PubMed]
- Budzikiewicz, H. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol. Rev. 1993, 10, 209–228. [Google Scholar] [CrossRef]
- Dowling, D.N.; O’Gara, F. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 1994, 12, 133–140. [Google Scholar] [CrossRef]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef]
- Wang, M.; Schaefer, A.L.; Dandekar, A.A.; Greenberg, E.P. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl. Acad. Sci. USA 2015, 112, 2187–2191. [Google Scholar] [CrossRef]
- Yan, Q.; Lopes, L.D.; Shaffer, B.T.; Kidarsa, T.E.; Vining, O.; Philmus, B.; Song, C.; Stockwell, V.O.; Raajmakers, J.M.; Mc Phail, K.L.; et al. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio 2018, 9, e01845-17. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C.; Laville, J.; Maurhofer, M.; Oberhänsli, T.; Schnider, U.; Voisard, C.; Wüthrich, B.; Défago, G. Secondary Metabolites of Pseudomonas fluorescens Strain CHA0 Involved in the Suppression of Root Diseases. In Advances in Molecular Genetics of Plant-Microbe Interactions Vol.1, Current Plant Science and Biotechnology in Agriculture; Hennecke, H., Verma, D.P.S., Eds.; Springer: Dordrecht, The Netherlands, 1991; Volume 10, pp. 450–456. [Google Scholar] [CrossRef]
- Heurlier, K.; Williams, F.; Heeb, S.; Dormond, C.; Pessi, G.; Singer, D.; Cámara, M.; Williams, P.; Haas, D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004, 186, 2936–2945. [Google Scholar] [CrossRef]
- Takeuchi, K.; Kiefer, P.; Reimmann, C.; Keel, C.; Dubuis, C.; Rolli, J.; Vorholt, J.A.; Haas, D. Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J. Biol. Chem. 2009, 284, 34976–34985. [Google Scholar] [CrossRef]
- Dubuis, C.; Rolli, J.; Lutz, M.; Défago, G.; Haas, D. Thiamine-auxotrophic mutants of Pseudomonas fluorescens CHA0 are defective in cell-cell signaling and biocontrol factor expression. Appl. Environ. Microbiol. 2006, 72, 2606–2613. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bunik, V.I.; Tylicki, A.; Lukashev, N.V. Thiamin diphosphate-dependent enzymes: From enzymology to metabolic regulation, drug design and disease models. FEBS J. 2013, 280, 6412–6442. [Google Scholar] [CrossRef]
- Park, J.Y.; Kang, B.R.; Ryu, C.M.; Anderson, A.J.; Kim, Y.C. Polyamine is a critical determinant of Pseudomonas chlororaphis o6 for Gacs-dependent bacterial cell growth and biocontrol capacity. Mol. Plant Pathol. 2018, 19, 1257–1266. [Google Scholar] [CrossRef]
- Takeuchi, K. A primary metabolite controlled by the Gac/Rsm regulatory pathway, favors a planktonic over a biofilm lifestyle in Pseudomonas protegens CHA0. Mol. Plant-Microbe Interact. 2018, 31, 274–282. [Google Scholar] [CrossRef]
- Cheng, X.; van der Voort, M.; Raaijmakers, J.M. Gac-mediated changes in pyrroloquinoline quinone biosynthesis enhance the antimicrobial activity of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 2015, 7, 139–147. [Google Scholar] [CrossRef]
- Dubuis, C. Cell-Cell Communication in the Biocontrol Strain Pseudomonas fluorescens CHA0. Ph.D. Thesis, University of Lausanne, Lausanne, Switzerland, 2005. [Google Scholar]
- Dubuis, C.; Haas, D. Cross-species GacA-controlled induction of antibiosis in Pseudomonads. Appl. Environ. Microbiol. 2007, 73, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, M.; Van Gestel, K.; Pirhonen, M.; Taira, S. Soluble plant cell signals induce the expression of the type III secretion system of Pseudomonas syringae and upregulate the production of pilus protein HrpA. Mol. Plant-Microbe Interact. 2009, 22, 282–290. [Google Scholar] [CrossRef]
- Wang, N.; Lu, S.E.; Wang, J.; Chen, Z.J.; Gross, D.C. The expression of genes encoding lipodepsipeptide phytotoxins by Pseudomonas syringae pv. syringae is coordinated in response to plant signal molecules. Mol. Plant-Microbe Interact. 2006, 19, 257–269. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergeau, D.; Mazurier, S.; Barbey, C.; Merieau, A.; Chane, A.; Goux, D.; Bernard, S.; Driouich, A.; Lemanceau, P.; Vicré, M.; et al. Unusual extracellular appendages deployed by the model strain Pseudomonas fluorescens C7R12. PLoS ONE 2019, 14, e0221025. [Google Scholar] [CrossRef]
- Martín, J.F.; Sola, L.A.; Santos, B.F.; Fernández, M.L.T.; Prieto, C.; Rodríguez, G.A. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 2011, 4, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Virolle, M.J.; Millan, O.A.; Esnault, C.; Smirnov, A.; Dulermo, T.; Askora, A. In Streptomyces, the switch between primary and secondary metabolism is underpinned by a transition from glycolytic to oxidative metabolism. New Biotechnol. 2016, 33, S59–S60. [Google Scholar] [CrossRef]
- Entner, N.; Doudoroff, M. Glucose and gluconic oxidation of Pseudomonas saccharophila. J. Biol. Chem. 1952, 196, 853–862. [Google Scholar]
- Daddaoua, A.; Krell, T.; Ramos, J.L. Regulation of glucose metabolism in Pseudomonas: The phosphorylative branch and Entner-Doudoroff enzymes are regulated by a repressor containing a sugar isomerase domain. J. Biol. Chem. 2009, 284, 21360–21368. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B. Quorum-sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Loewen, P.C.; Switala, J.; Fernando, W.G.; de Kievit, T. Genome sequence of Pseudomonas brassicacearum DF41. Genome Announc. 2014, 2, e00390-14. [Google Scholar] [CrossRef]
- Berry, C.; Nandi, M.; Manuel, J.; Brassinga, A.K.C.; Fernando, W.G.D.; Loewen, P.C.; de Kievit, T.R. Characterization of the Pseudomonas sp. DF41 quorum sensing locus and its role in fungal antagonism. Biol. Control 2014, 69, 82–89. [Google Scholar] [CrossRef]
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007, 14, 87–96. [Google Scholar] [CrossRef]
- Diggle, S.P.; Winzer, K.; Chhabra, S.R.; Worall, K.E.; Cámara, M.; Williams, P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 2003, 50, 29–43. [Google Scholar] [CrossRef]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.H. A Cell-Cell Communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Elasri, M.; Delorme, S.; Lemanceau, P.; Stewart, G.; Laue, B.; Glickmann, E.; Oger, P.M.; Dessaux, Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 2001, 67, 1198–1209. [Google Scholar] [CrossRef]
- Kidarsa, T.A.; Shaffer, B.T.; Goebel, N.C.; Roberts, D.P.; Buyer, J.S.; Johnson, A.; Kobayashi, D.Y.; Zabriskie, T.M.; Paulsen, I.; Loper, J.E. Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ. Microbiol. 2013, 15, 716–735. [Google Scholar] [CrossRef] [PubMed]
- Lalaouna, D.; Fochesato, S.; Sanchez, L.; Schmitt, K.P.; Haas, D.; Heulin, T.; Achouak, W. Phenotypic switching in Pseudomonas brassicacearum involves GacS- and GacA-dependent Rsm small RNAs. Appl. Environ. Microbiol. 2012, 78, 1658–1665. [Google Scholar] [CrossRef]
- Latour, X.; Delorme, S.; Mirleau, P.; Lemanceau, P. Identification of Traits Implicated in the Rhizosphere Competence of Fluorescent Pseudomonads: Description of a Strategy Based on Population and Model Strain Studies. In Sustainable Agriculture; Lichtfouse, E., Navarrete, M., Debaeke, P., Véronique, S., Alberola, C., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 285–296. [Google Scholar] [CrossRef]
- Barbey, C.; Chane, A.; Burini, J.F.; Maillot, O.; Merieau, A.; Gallique, M.; Beury, C.A.; Konto, G.Y.; Feuilloley, M.; Gobert, V.; et al. A rhodococcal transcriptional regulatory mechanism detects the common lactone ring of AHL quorum-sensing signals and triggers the quorum-quenching response. Front. Microbiol. 2018, 9, 2800. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Crépin, A.; Barbey, C.; Cirou, A.; Tannières, M.; Orange, N.; Feuilloley, M.; Dessaux, Y.; Burini, J.F.; Faure, D.; Latour, X. Biological control of pathogen communication in the rhizosphere: A novel approach applied to potato soft rot due to Pectobacterium atrosepticum. Plant Soil 2012, 358, 27–37. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N. Metabolism and virulence strategies in Dickeya-host interactions. Prog. Mol. Biol. Transl. Sci. 2016, 142, 93–129. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant. Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Notz, R.; Maurhofer, M.; Schnider, K.U.; Duffy, B.; Haas, D.; Défago, G. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 2001, 91, 873–881. [Google Scholar] [CrossRef][Green Version]
- Koch, B.; Nielsen, T.H.; Sørensen, D.; Andersen, J.B.; Christophersen, C.; Molin, S.; Givskov, M.; Sørensen, J.; Nybroe, O. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 2002, 68, 4509–4516. [Google Scholar] [CrossRef]
- Bais, H.P.; Broeckling, C.D.; Vivanco, J.M. Root Exudates Modulate Plant-Microbe Interactions in the Rhizosphere. In Soil Biology: Secondary Metabolites in Soil Ecology; Karlovsky, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 14, pp. 241–252. [Google Scholar] [CrossRef]
- Planamente, S.; Vigouroux, A.; Mondy, S.; Nicaise, M.; Faure, D.; Moréra, S. A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens. J. Biol. Chem. 2010, 285, 30294–30303. [Google Scholar] [CrossRef]
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Wang, G.; Liu, J.; Zhu, H. Genetic and molecular mechanisms underlying symbiotic specificity in Legume-Rhizobium Interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef]
- Yang, S.; Peng, Q.; San Francisco, M.; Wang, Y.; Zeng, Q.; Yang, C.H. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE 2008, 3, e2973. [Google Scholar] [CrossRef]
- Toyoda, Y.A.; Shimoda, N.; Machida, Y. Genetic analysis of the signal-sensing region of the histidine protein kinase VirA of Agrobacterium tumefaciens. Mol. Gen. Genet. 2000, 263, 939–947. [Google Scholar] [CrossRef]
- Gao, R.; Lynn, D.G. Environmental pH sensing: Resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 2005, 187, 2182–2189. [Google Scholar] [CrossRef]
- Hartmann, A.; Rothballer, M. Role of Quorum Sensing Signals of Rhizobacteria for Plant Growth Promotion. In Rhizotrophs: Plant Growth Promotion to Bioremediation. Microorganisms for Sustainability; Mehnaz, S., Ed.; Springer: Singapore, 2017; Volume 2, pp. 205–217. [Google Scholar] [CrossRef]
- Sall, K.M.; Casabona, M.G.; Bordi, C.; Huber, P.; de Bentzmann, S.; Attrée, I.; Elsen, S. A gacS deletion in Pseudomonas aeruginosa cystic fibrosis isolate CHA shapes its virulence. PLoS ONE 2014, 9, e95936. [Google Scholar] [CrossRef]
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffière, A.; Yan, S.; Dominguez, H.; Thompson, B. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008, 2, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Ausubel, F.M.; Cao, H.; Drenkard, E.; Goumnerov, B.C.; Lau, G.W.; Mahajan, M.S.; Plotnikova, J.; Tan, M.W.; Tsongalis, J.; et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 2000, 97, 8815–8821. [Google Scholar] [CrossRef]
- Gooderham, W.J.; Hancock, R.E.W. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009, 33, 279–294. [Google Scholar] [CrossRef]
- Klockgether, J.; Tümmler, B. Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Res. 2017, 6, 1261. [Google Scholar] [CrossRef]
- Valentini, M.; Gonzalez, D.; Mavridou, D.A.; Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. [Google Scholar] [CrossRef]
- Ertesvåg, H.; Sletta, H.; Senneset, M.; Sun, Y.Q.; Klinkenberg, G.; Konradsen, T.A.; Ellingsen, T.E.; Valla, S. Identification of genes affecting alginate biosynthesis in Pseudomonas fluorescens by screening a transposon insertion library. BMC Genomics 2017, 18, 11. [Google Scholar] [CrossRef]
- Bakkevig, K.; Sletta, H.; Gimmestad, M.; Aune, R.; Ertesvåg, H.; Degnes, K.; Christensen, B.E.; Ellingsen, T.E.; Valla, S. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J. Bacteriol. 2005, 187, 8375–8384. [Google Scholar] [CrossRef]
- Song, C.; van der Voort, M.; van de Mortel, J.; Hassan, K.A.; Elbourne, L.D.; Paulsen, I.T.; Loper, J.E.; Raaijmakers, J.M. The Rsm regulon of plant growth-promoting Pseudomonas fluorescens SS101: Role of small RNAs in regulation of lipopeptide biosynthesis. Microb. Biotechnol. 2015, 8, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Barahona, E.; Navazo, A.; Yousef, C.F.; Aguirre de Cárcer, D.; Martínez, G.F.; Espinosa, U.M.; Martín, M.; Rivilla, R. Efficient rhizosphere colonization by Pseudomonas fluorescens F113 mutants unable to form biofilms on abiotic surfaces. Environ. Microbiol. 2010, 12, 3185–3195. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.M.; Martín, M.; Villacieros, M.; O’Gara, F.; Bonilla, I.; Rivilla, R. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 2002, 184, 1587–1596. [Google Scholar] [CrossRef]
- Nasser, W.; Dorel, C.; Wawrzyniak, J.; Van Gijsegem, F.; Groleau, M.C.; Déziel, E.; Reverchon, S. Vfm a new quorum sensing system controls the virulence of Dickeya dadantii. Environ. Microbiol. 2013, 15, 865–880. [Google Scholar] [CrossRef]
- Valverde, C.; Heeb, S.; Keel, C.; Haas, D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 2003, 50, 1361–1379. [Google Scholar] [CrossRef] [PubMed]
- Maurhofer, M.; Reimmann, C.; Schmidli, S.P.; Heeb, S.; Haas, D.; Défago, G. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 1998, 88, 678–684. [Google Scholar] [CrossRef]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar] [CrossRef]
- Flavier, A.B.; Clough, S.J.; Schell, M.A.; Denny, T.P. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 1997, 26, 251–259. [Google Scholar] [CrossRef]
- Wang, L.H.; He, Y.; Gao, Y.; Wu, J.E.; Dong, Y.H.; He, C.; Wang, S.X.; Weng, L.X.; Xu, J.L.; Tay, L.; et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 2004, 51, 903–912. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Gjersing, E.L.; Herberg, J.L.; Horn, J.; Schaldach, C.M.; Maxwell, R.S. NMR metabolomics of planktonic and biofilm modes of growth in Pseudomonas aeruginosa. Anal. Chem. 2007, 79, 8037–8045. [Google Scholar] [CrossRef]
- Davenport, P.W.; Griffin, J.L.; Welch, M. Quorum sensing is accompanied by global metabolic changes in the opportunistic human pathogen Pseudomonas aeruginosa. J. Bacteriol. 2015, 197, 2072–2082. [Google Scholar] [CrossRef]
- Shahid, I.; Malik, K.A.; Mehnaz, S. A decade of understanding secondary metabolism in Pseudomonas spp. for sustainable agriculture and pharmaceutical applications. Environ. Sustain. 2018, 1, 3–17. [Google Scholar] [CrossRef]
- Dunham, S.J.; Ellis, J.F.; Li, B.; Sweedler, J.V. Mass Spectrometry Imaging of Complex Microbial Communities. Acc. Chem. Res. 2017, 50, 96–104. [Google Scholar] [CrossRef]
- Deveau, A.; Gross, H.; Palin, B.; Mehnaz, S.; Schnepf, M.; Leblond, P.; Dorrestein, P.C.; Aigle, B. Role of secondary metabolites in the interaction between Pseudomonas fluorescens and soil microorganisms under iron-limited conditions. FEMS Microbiol. Ecol. 2016, 92, fiw107. [Google Scholar] [CrossRef]
- Phelan, V.V.; Fang, J.; Dorrestein, P.C. Mass spectrometry analysis of Pseudomonas aeruginosa treated with azithromycin. J. Am. Soc. Mass Spectrom. 2015, 26, 873–877. [Google Scholar] [CrossRef]
- Yang, J.Y.; Phelan, V.V.; Simkovsky, R.; Watrous, J.D.; Trial, R.M.; Fleming, T.C.; Wenter, R.; Moore, B.S.; Golden, S.S.; Pogliano, K.; et al. Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 2012, 194, 6023–6028. [Google Scholar] [CrossRef]
- Huang, W.; Brewer, L.K.; Jones, J.W.; Nguyen, A.T.; Marcu, A.; Wishart, D.S.; Oglesby, S.A.G.; Kane, M.A.; Wilks, A. PAMDB: A comprehensive Pseudomonas aeruginosa metabolome database. Nucleic Acids Res. 2018, 46, D575–D580. [Google Scholar] [CrossRef]
- Takeuchi, K.; Yamada, K.; Haas, D. ppGpp controlled by the Gac/Rsm regulatory pathway sustains biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 2012, 25, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Goo, E.; An, J.H.; Kang, Y.; Hwang, I. Control of bacterial metabolism by quorum sensing. Trends Microbiol. 2015, 23, 567–576. [Google Scholar] [CrossRef]
- Bartell, J.A.; Blazier, A.S.; Yen, P.; Thøgersen, J.C.; Jelsbak, L.; Goldberg, J.B.; Jason, A.; Papin, J.A. Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat. Commun. 2017, 8, 14631. [Google Scholar] [CrossRef]
- Hassan, K.A.; Johnson, A.; Shaffer, B.T.; Ren, Q.; Kidarsa, T.A.; Elbourne, L.D.H.; Hartney, S.; Duboy, R.; Goebel, N.C.; Zabriskie, T.M.; et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far reaching transcriptomic consequences. Environ. Microbiol. 2010, 12, 899–915. [Google Scholar] [CrossRef]
- Wang, D.; Lee, S.H.; Seeve, C.; Yu, J.M.; Pierson, L.S., III; Pierson, E.A. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. MicrobiologyOpen 2013, 2, 505–524. [Google Scholar] [CrossRef]
- Whiteley, M.; Lee, K.M.; Greenberg, E.P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 13904–13909. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef]
- Mellbye, B.; Schuster, M. Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 1155–1164. [Google Scholar] [CrossRef]
- Yan, Q.; Philmus, B.; Chang, J.H.; Loper, J.E. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. eLife 2017, 6, e22835. [Google Scholar] [CrossRef]
- Bowden, S.D.; Eyres, A.; Chung, J.C.S.; Monson, R.E.; Thompson, A.; Salmond, G.P.C.; Spring, D.R.; Welch, M. Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p)ppGpp. Mol. Microbiol. 2013, 90, 457–471. [Google Scholar] [CrossRef]
- Cheung, J.; Wayne, A.; Hendrickson, W.A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265. [Google Scholar] [CrossRef] [PubMed]
- Sevvana, M.; Vijayan, V.; Zweckstetter, M.; Reinelt, S.; Madden, D.R.; Herbst, I.R.; Sheldrick, G.M.; Bott, M.; Griesinger, C.; Becker, S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008, 377, 512–523. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef]
- Hraiech, S.; Hiblot, J.; Lafleur, J.; Lepidi, H.; Papazian, L.; Rolain, J.M.; Raoult, D.; Elias, M.; Silby, M.W.; Bzdrenga, J.; et al. Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat pneumonia. PLoS ONE 2014, 9, e107125. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Bergonzi, C.; Sakkos, J.; Staley, C.; Zhang, Q.; Sadowsky, M.J.; Aksan, A.; Elias, M. Signal disruption leads to changes in bacterial community population. Front Microbiol. 2019, 10, 611. [Google Scholar] [CrossRef]
- Chane, A.; Barbey, C.; Robert, M.; Merieau, A.; Konto, G.Y.; Beury, C.A.; Feuilloley, M.; Pátek, M.; Gobert, V.; Latour, X. Biocontrol of soft rot: Confocal microscopy highlights virulent pectobacterial communication and its jamming by rhodococcal quorum-quenching. Mol. Plant-Microbe Interact. 2019, 32, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Chane, A.; Bourigault, Y.; Bouteiller, M.; Konto, G.Y.; Merieau, A.; Barbey, C.; Latour, X. Close-up on a bacterial informational war in the geocaulosphere. Can. J. Microbiol. 2020, 66, 447–454. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).