Weak Influence of Paleoenvironmental Conditions on the Subsurface Biosphere of Lake Ohrid over the Last 515 ka
Abstract
1. Introduction
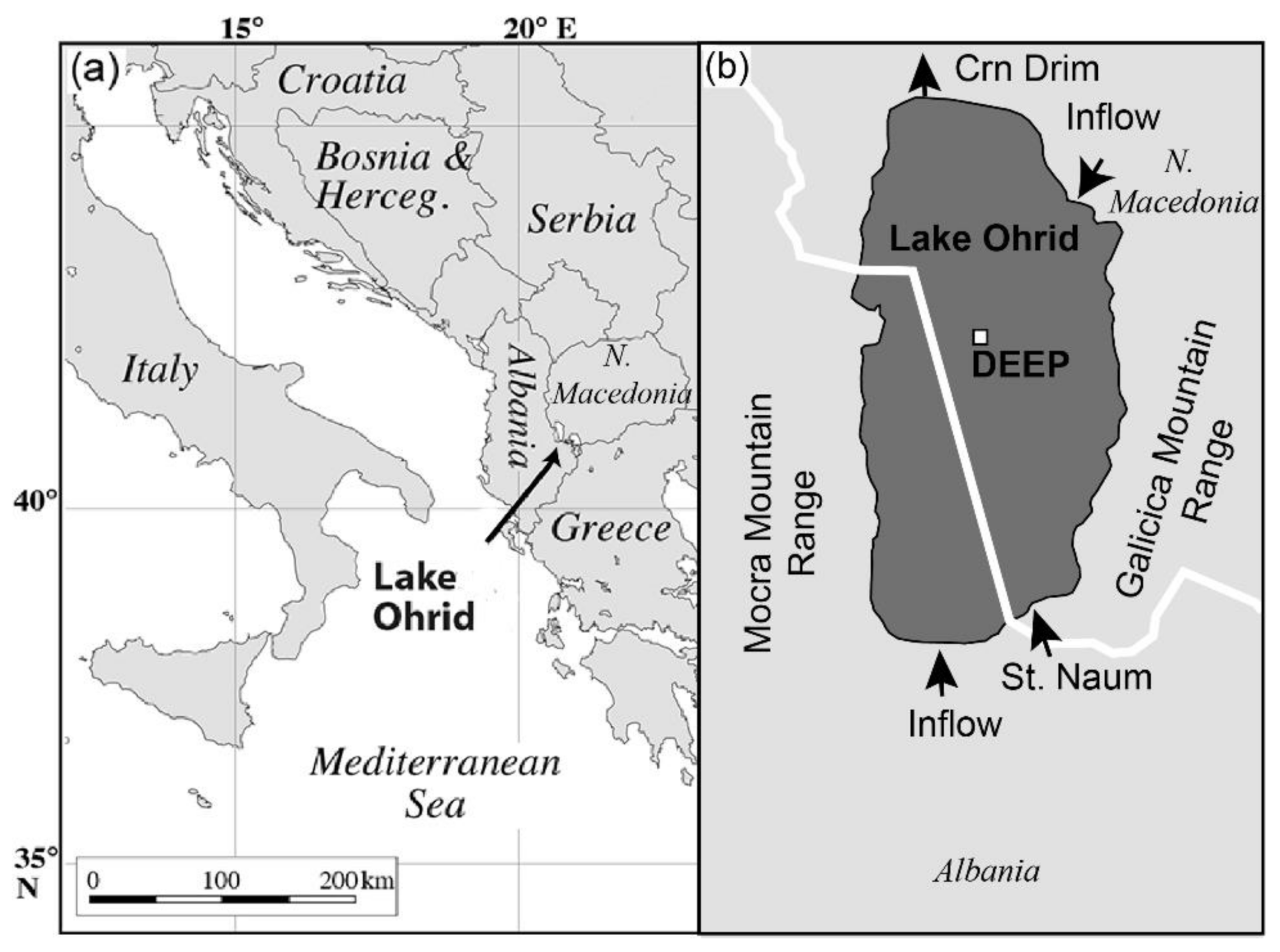
2. Geological and Limnological Settings
3. Materials and Methods
3.1. Sampling Material
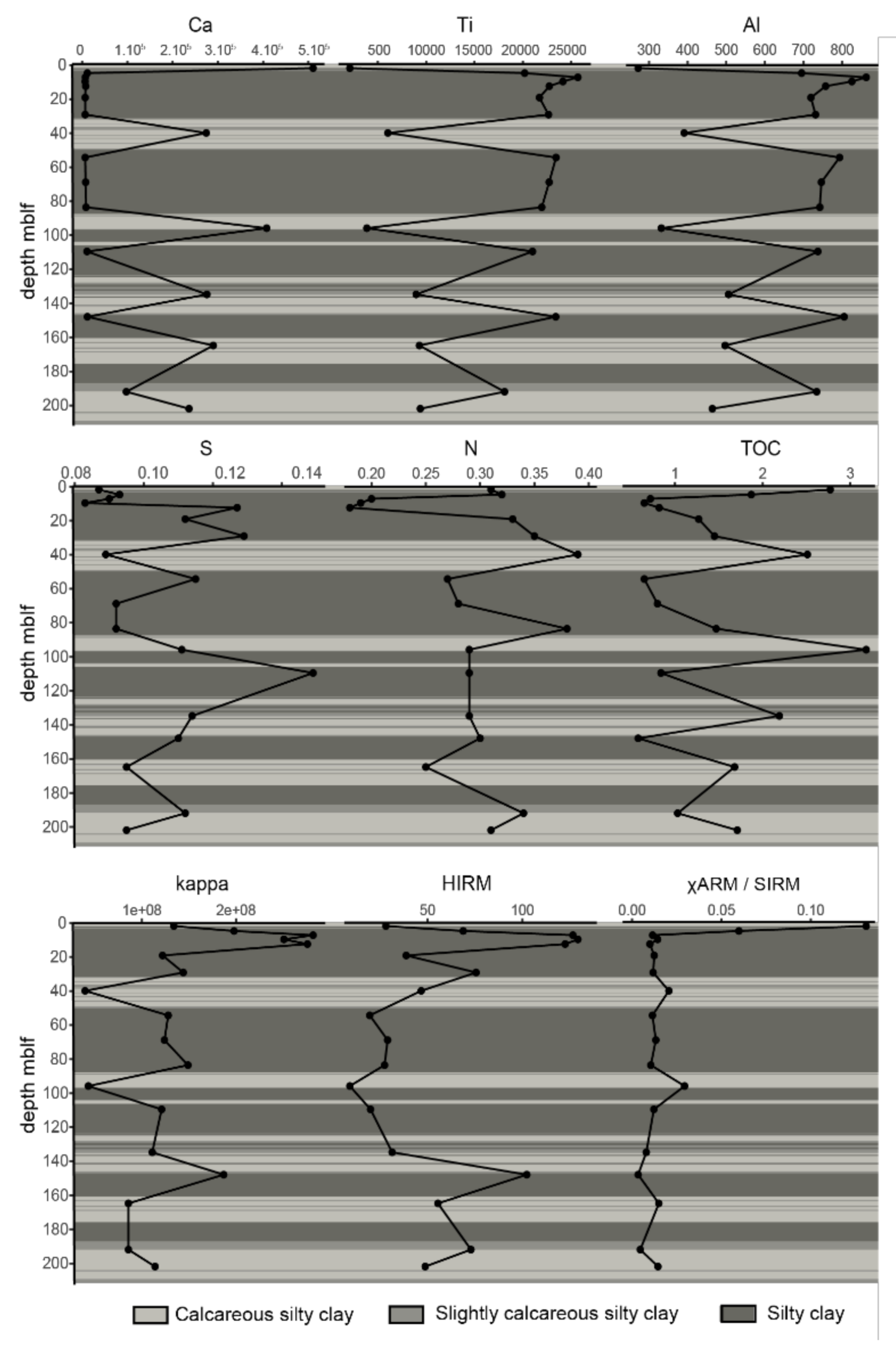
3.2. Sediment Chemistry
3.3. DNA Extraction and Sequencing
3.4. DNA Sequence Processing
3.5. Data Analysis
4. Results
4.1. Lake and Sediment Characteristics
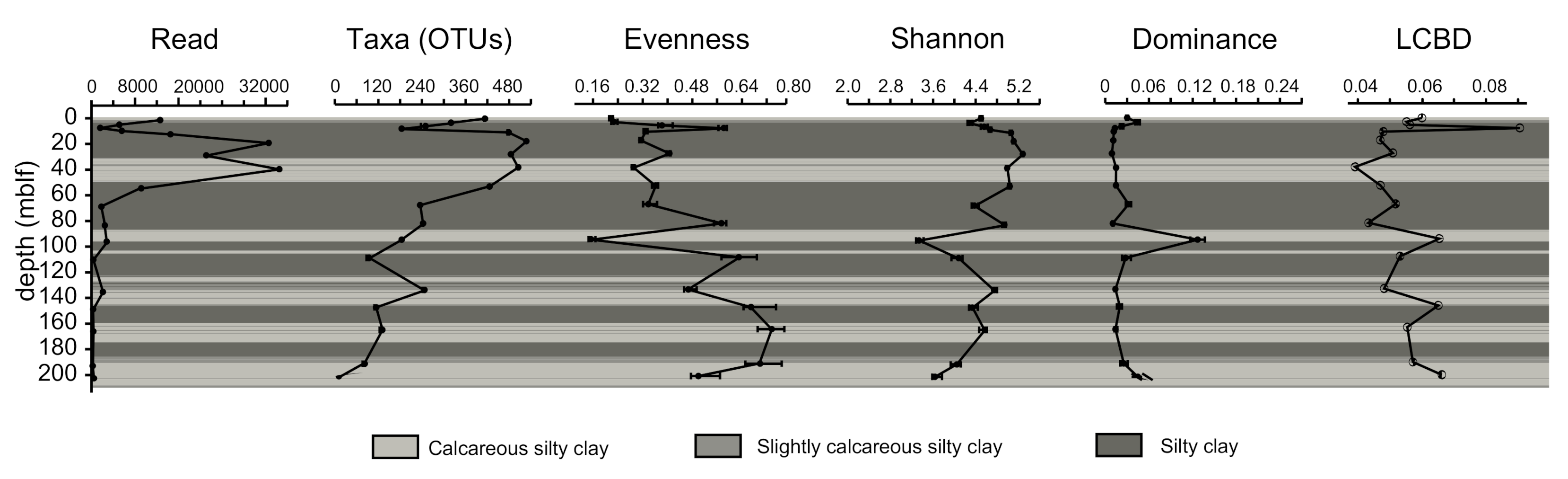
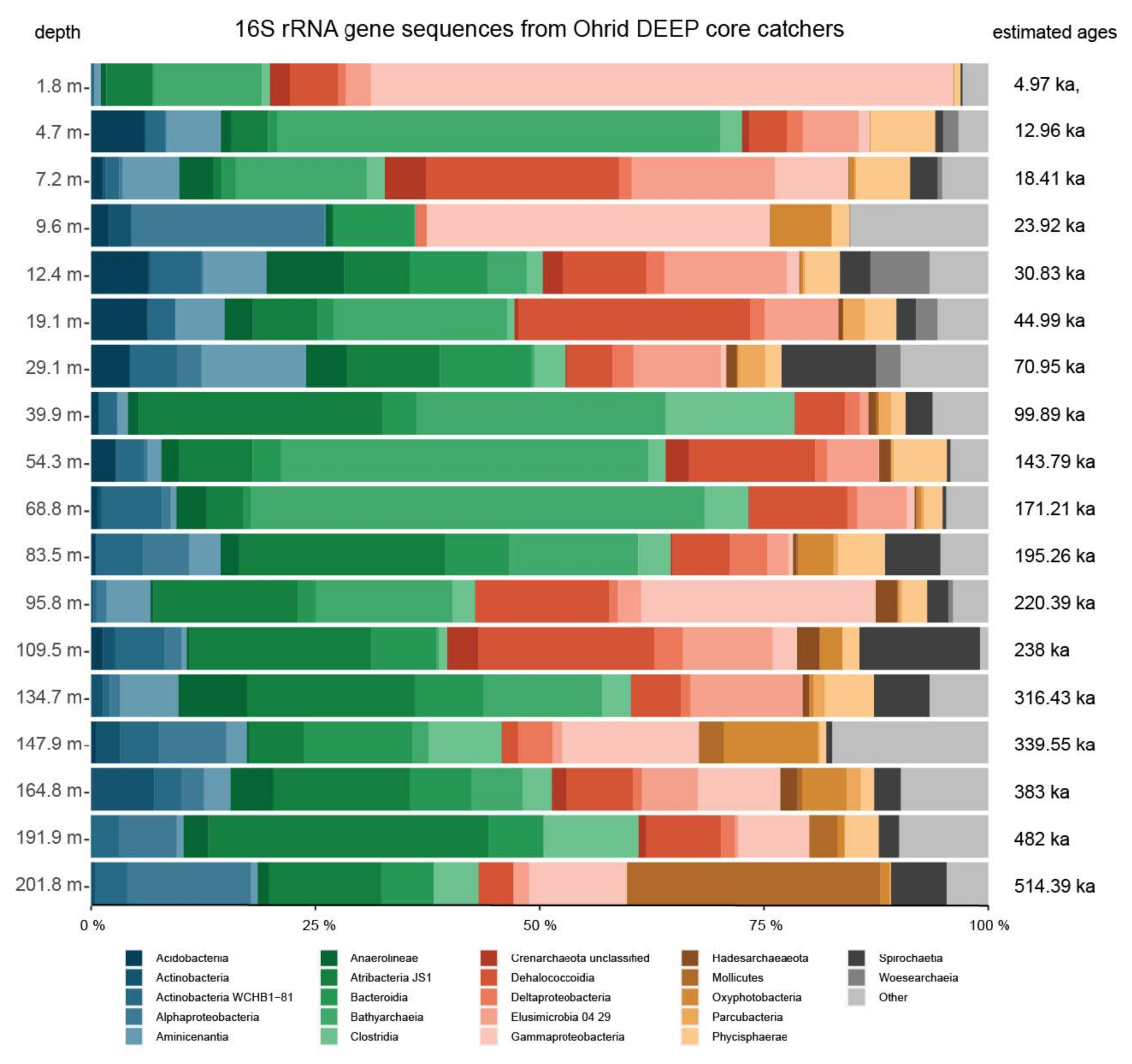
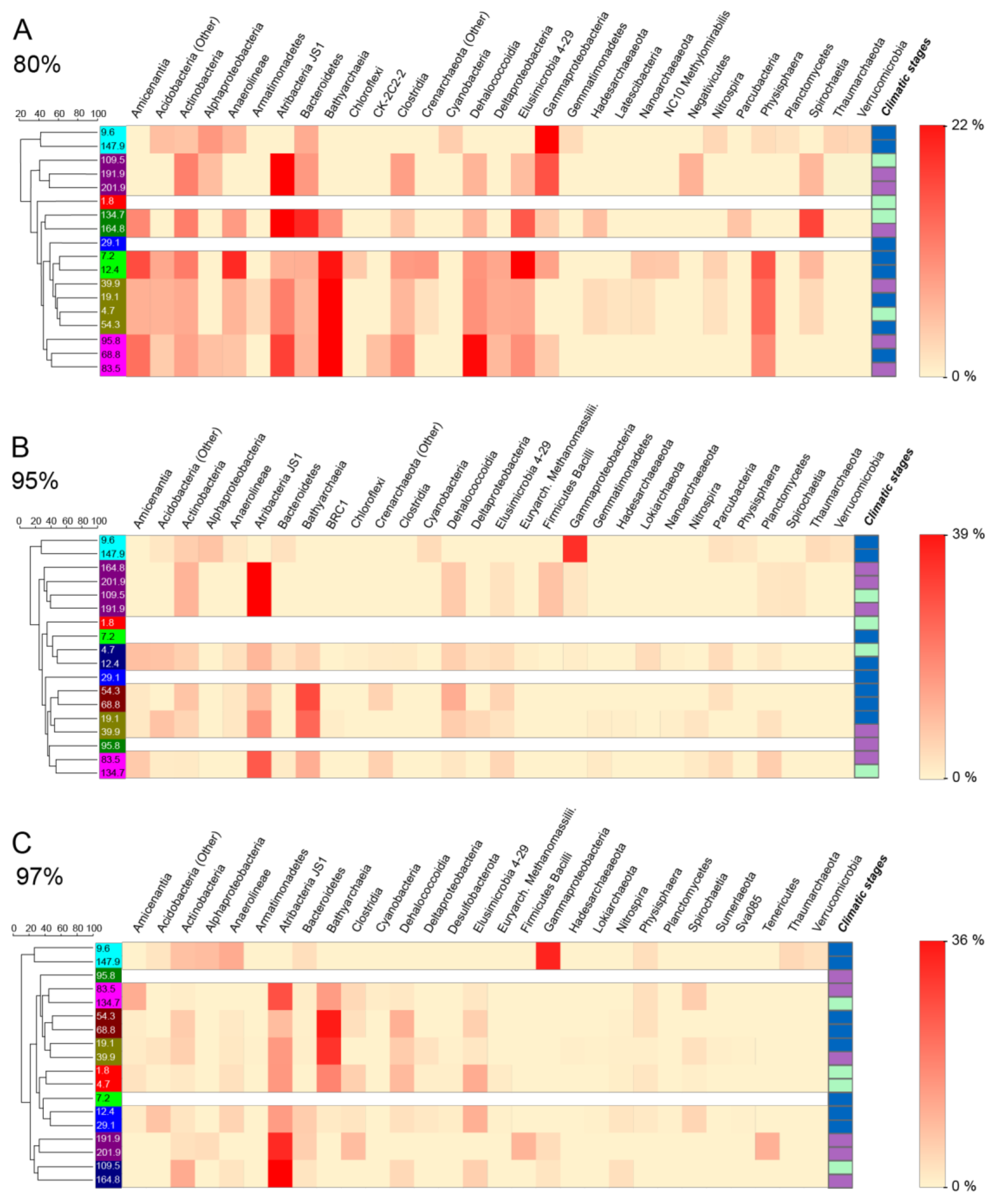
4.2. Microbial Community Composition and Variation
5. Discussion
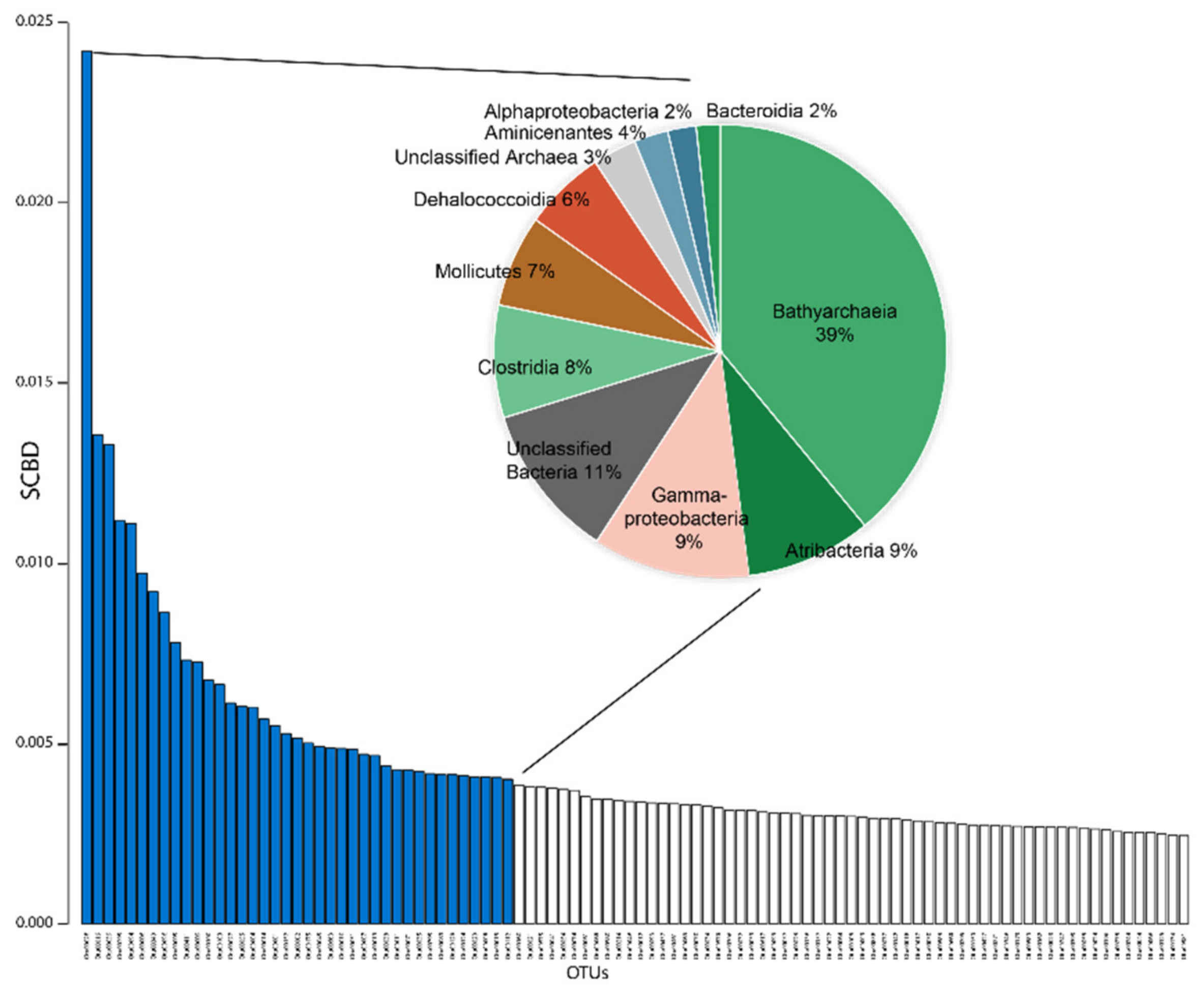
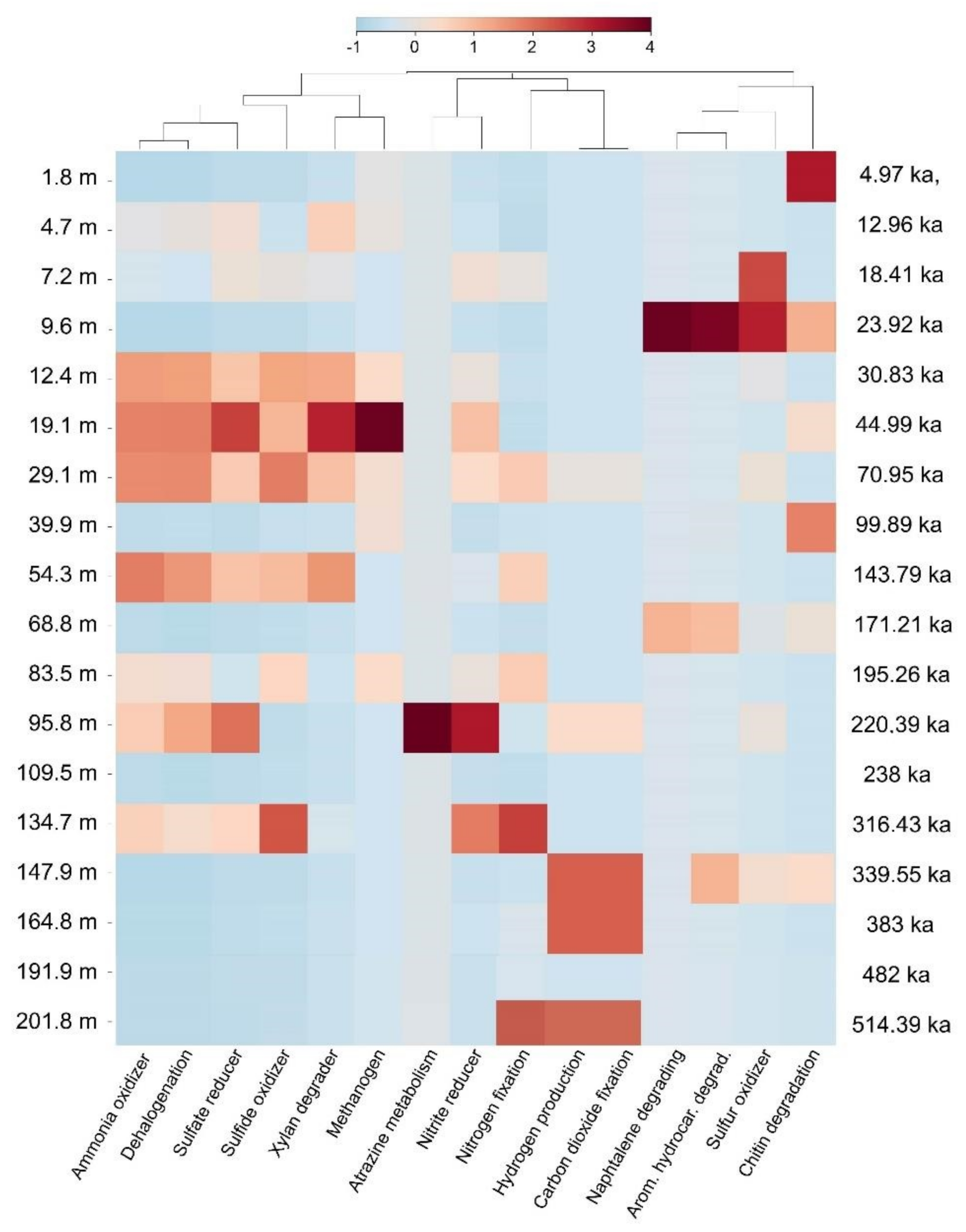
5.1. Dominant Taxa and Associated Metabolisms in the Ohrid Sediment
5.2. Diversity Changes by Depth
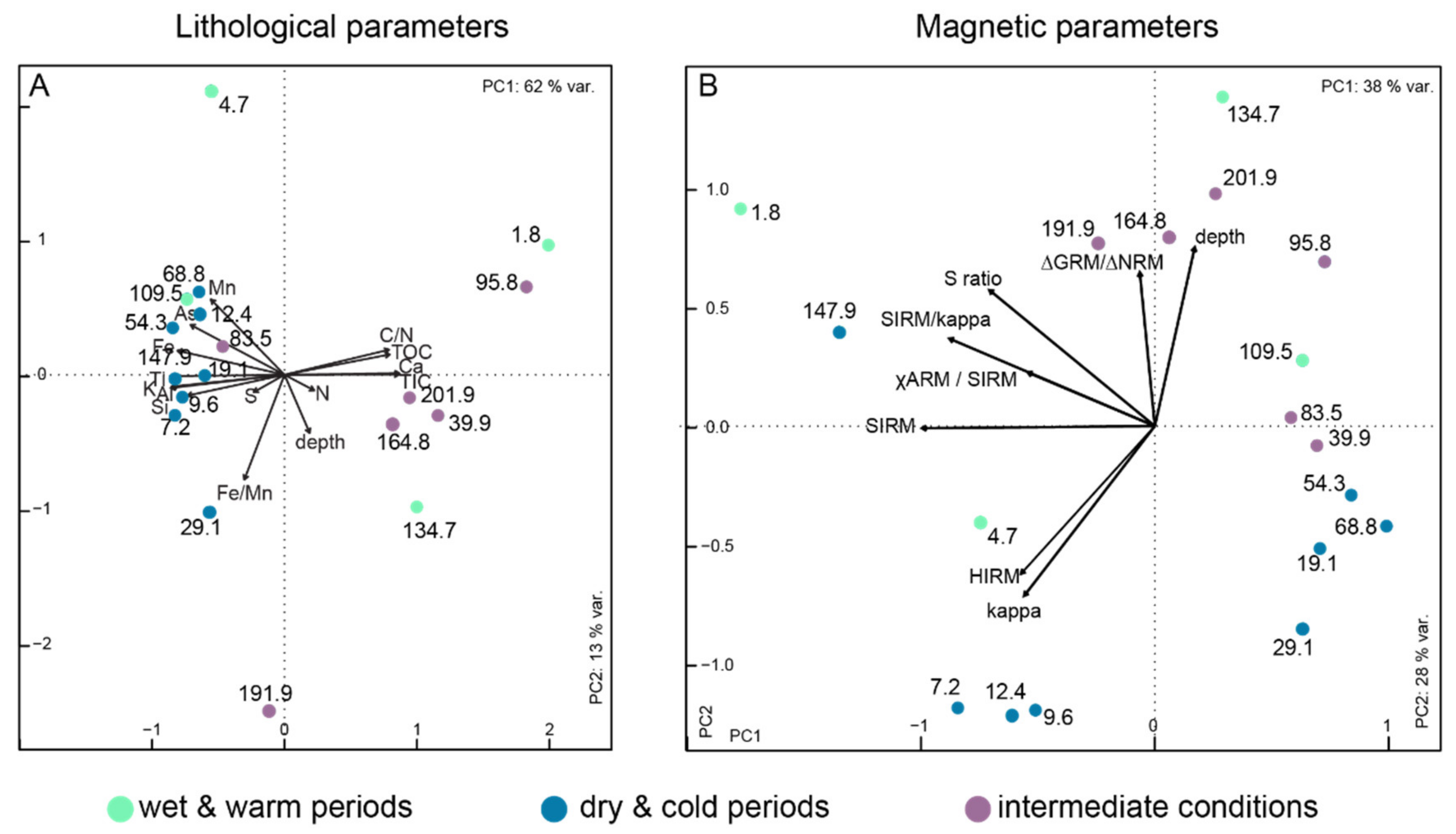
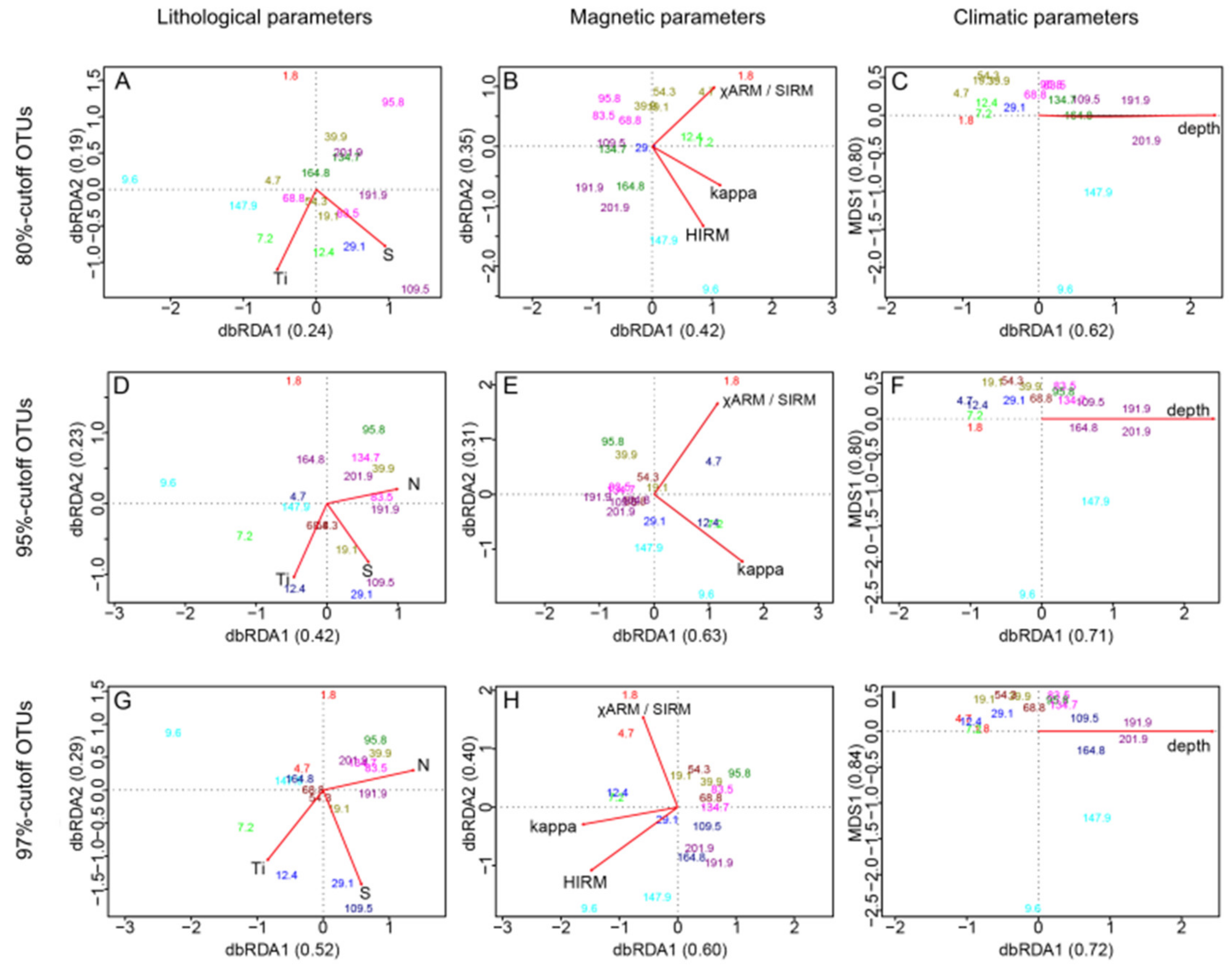
5.3. Impact of Environmental Parameters on Current Communities
5.4. Lake Ohrid Specificity
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wagner, B.; Vogel, H.; Francke, A.; Friedrich, T.; Donders, T.; Lacey, J.H.; Leng, M.J.; Regattieri, E.; Sadori, L.; Wilke, T.; et al. Mediterranean winter rainfall in phase with African monsoons during the past 1.36 million years. Nature 2019. [Google Scholar] [CrossRef] [PubMed]
- Francke, A.; Wagner, B.; Just, J.; Leicher, N.; Gromig, R.; Baumgarten, H.; Vogel, H.; Lacey, J.H.; Sadori, L.; Wonik, T.; et al. Sedimentological processes and environmental variability at Lake Ohrid (Macedonia, Albania) between 637 ka and the present. Biogeosciences 2016, 13, 1179–1196. [Google Scholar] [CrossRef]
- Lacey, J.H.; Leng, M.J.; Francke, A.; Sloane, H.H.; Milodowski, A.; Vogel, H.; Baumgarten, H.; Zanchetta, G.; Wagner, B. Northern Mediterranean climate since the Middle Pleistocene: A 637 ka stable isotope record from Lake Ohrid (Albania/Macedonia). Biogeosciences 2016, 13, 1801–1820. [Google Scholar] [CrossRef]
- Reed, J.M.; Cvetkoska, A.; Levkov, Z.; Vogel, H.; Wagner, B. The last glacial-interglacial cycle in Lake Ohrid (Macedonia/Albania): Testing diatom response to climate. Biogeosciences 2010, 7, 3083–3094. [Google Scholar] [CrossRef]
- Vogel, H.; Wagner, B.; Zanchetta, G.; Sulpizio, R.; Rosén, P. A paleoclimate record with tephrochronological age control for the last glacial-interglacial cycle from Lake Ohrid, Albania and Macedonia. J. Paleolimnol. 2010, 44, 295–310. [Google Scholar] [CrossRef]
- Wagner, B.; Vogel, H.; Zanchetta, G.; Sulpizio, R. Environmental change within the Balkan region during the past ca. 50 ka recorded in the sediments from lakes Prespa and Ohrid. Biogeosciences 2010, 7, 3187–3198. [Google Scholar] [CrossRef]
- Wagner, B.; Lotter, A.F.; Nowaczyk, N.; Reed, J.M.; Schwalb, A.; Sulpizio, R.; Valsecchi, V.; Wessels, M.; Zanchetta, G. A 40,000-year record of environmental change from ancient Lake Ohrid (Albania and Macedonia). J. Paleolimnol. 2009, 41, 407–430. [Google Scholar] [CrossRef]
- Zanchetta, G.; Baneschi, I.; Francke, A.; Boschi, C.; Regattieri, E.; Wagner, B.; Lacey, J.H.; Leng, M.J.; Vogel, H.; Sadori, L. Evidence for carbon cycling in a large freshwater lake in the Balkans over the last 0.5 million years using the isotopic composition of bulk organic matter. Quat. Sci. Rev. 2018, 202, 154–165. [Google Scholar] [CrossRef]
- Holtvoeth, J.; Vogel, H.; Valsecchi, V.; Lindhorst, K.; Schouten, S.; Wagner, B.; Wolff, G.A. Linear and non-linear responses of vegetation and soils to glacial-interglacial climate change in a Mediterranean refuge. Sci. Rep. 2017, 7, 8121. [Google Scholar] [CrossRef] [PubMed]
- Holtvoeth, J.; Vogel, H.; Wagner, B.; Wolff, G.A. Lipid biomarkers in Holocene and glacial sediments from ancient Lake Ohrid (Macedonia, Albania). Biogeosciences 2010, 7, 3473–3489. [Google Scholar] [CrossRef]
- Just, J.; Nowaczyk, N.R.; Sagnotti, L.; Francke, A.; Vogel, H.; Lacey, J.H.; Wagner, B. Environmental control on the occurrence of high-coercivity magnetic minerals and formation of iron sulfides in a 640 ka sediment sequence from Lake Ohrid (Balkans). Biogeosciences 2016, 13, 2093–2109. [Google Scholar] [CrossRef]
- Lacey, J.H.; Francke, A.; Leng, M.J.; Vane, C.H.; Wagner, B. A high-resolution Late Glacial to Holocene record of environmental change in the Mediterranean from Lake Ohrid (Macedonia/Albania). Int. J. Earth Sci. 2015, 104, 1623–1638. [Google Scholar] [CrossRef]
- Leng, M.J.; Baneschi, I.; Zanchetta, G.; Jex, C.N.; Wagner, B.; Vogel, H. Late Quaternary palaeoenvironmental reconstruction from Lakes Ohrid and Prespa (Macedonia/Albania border) using stable isotopes. Biogeosci. Discuss. 2010, 7, 3815–3853. [Google Scholar] [CrossRef]
- Wagner, B.; Wilke, T.; Francke, A.; Albrecht, C.; Baumgarten, H.; Bertini, A.; Combourieu-Nebout, N.; Cvetkoska, A.; Addabbo, M.; Donders, T.H.; et al. The environmental and evolutionary history of Lake Ohrid (FYROM/Albania): Interim results from the SCOPSCO deep drilling project. Biogeosciences 2017, 14, 2033–2054. [Google Scholar] [CrossRef]
- Jovanovska, E.; Cvetkoska, A.; Hauffe, T.; Levkov, Z.; Wagner, B.; Sulpizio, R.; Francke, A.; Albrecht, C.; Wilke, T. Differential resilience of ancient sister lakes Ohrid and Prespa to environmental disturbances during the Late Pleistocene. Biogeosciences 2016, 13, 1149–1161. [Google Scholar] [CrossRef]
- Wilke, T.; Hauffe, T.; Jovanovska, E.; Cvetkoska, A.; Donders, T.; Ekschmitt, K.; Francke, A.; Lacey, J.H.; Levkov, Z.; Marshall, C.R.; et al. Deep drilling reveals massive shifts in evolutionary dynamics after formation of ancient ecosystem. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Föller, K.; Stelbrink, B.; Hauffe, T.; Albrecht, C.; Wilke, T. Constant diversification rates of endemic gastropods in ancient Lake Ohrid: Ecosystem resilience likely buffers environmental fluctuations. Biogeosciences 2015, 12, 7209–7222. [Google Scholar] [CrossRef]
- Baulch, H.M.; Schindler, D.W.; Turner, M.A.; Findlay, D.L.; Paterson, M.J.; Vinebrooke, R.D. Effects of warming on benthic communities in a boreal lake: Implications of climate change. Limnol. Oceanogr. 2005, 50, 1377–1392. [Google Scholar] [CrossRef]
- De Senerpont Domis, L.N.; Elser, J.J.; Gsell, A.S.; Huszar, V.L.M.; Ibelings, B.W.; Jeppesen, E.; Kosten, S.; Mooij, W.M.; Roland, F.; Sommer, U.; et al. Plankton dynamics under different climatic conditions in space and time. Freshw. Biol. 2013, 58, 463–482. [Google Scholar] [CrossRef]
- Zwirglmaier, K.; Keiz, K.; Engel, M.; Geist, J.; Raeder, U. Seasonal and spatial patterns of microbial diversity along a trophic gradient in the interconnected lakes of the Osterseen Lake District, Bavaria. Front. Microbiol. 2015, 6, 1168. [Google Scholar] [CrossRef]
- Do Nam, Y.; Sung, Y.; Chang, H.W.; Roh, S.W.; Kim, K.H.; Rhee, S.K.; Kim, J.C.; Kim, J.Y.; Yoon, J.H.; Bae, J.W. Characterization of the depth-related changes in the microbial communities in Lake Hovsgol sediment by 16S rRNA gene-based approaches. J. Microbiol. 2008, 46, 125–136. [Google Scholar] [CrossRef]
- Dong, H.; Jiang, H.; Yu, B.; Liu, X.; Zhang, C.; Chan, M.A. Impacts of environmental change and human activity on microbial ecosystems on the Tibetan Plateau, NW China. GSA Today 2010, 20, 4–10. [Google Scholar] [CrossRef][Green Version]
- Ariztegui, D.; Thomas, C.; Vuillemin, A. Present and future of subsurface biosphere studies in lacustrine sediments through scientific drilling. Int. J. Earth Sci. 2015, 104, 1655–1665. [Google Scholar] [CrossRef]
- Wilke, T.; Wagner, B.; Van Bocxlaer, B.; Albrecht, C.; Ariztegui, D.; Delicado, D.; Francke, A.; Harzhauser, M.; Hauffe, T.; Holtvoeth, J.; et al. Scientific drilling projects in ancient lakes: Integrating geological and biological histories. Glob. Planet. Chang. 2016, 143, 118–151. [Google Scholar] [CrossRef]
- Vuillemin, A.; Ariztegui, D.; Leavitt, P.R.; Bunting, L. Recording of climate and diagenesis through fossil pigments and sedimentary DNA at Laguna Potrok Aike, Argentina. Biogeosci. Discuss. 2015, 12, 18345–18388. [Google Scholar] [CrossRef]
- Vuillemin, A.; Ariztegui, D.; Horn, F.; Kallmeyer, J.; Orsi, W.D. Microbial community composition along a 50 000-year lacustrine sediment sequence. FEMS Microbiol. Ecol. 2018. [Google Scholar] [CrossRef]
- Glombitza, C.; Stockhecke, M.; Schubert, C.J.; Vetter, A.; Kallmeyer, J. Sulfate reduction controlled by organic matter availability in deep sediment cores from the saline, alkaline Lake Van (Eastern Anatolia, Turkey). Front. Microbiol. 2013, 4, 209. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Ionescu, D.; Ariztegui, D. Impact of paleoclimate on the distribution of microbial communities in the subsurface sediment of the Dead Sea. Geobiology 2015, 13, 546–561. [Google Scholar] [CrossRef]
- Thomas, C.; Grossi, V.; Antheaume, I.; Ariztegui, D. Recycling of Archaeal Biomass as a New Strategy for Extreme Life in the Dead Sea Deep Sediment. Geology 2019, 47, 479–482. [Google Scholar] [CrossRef]
- Thomas, C.; Ebert, Y.; Kiro, Y.; Stein, M.; Ariztegui, D. Microbial sedimentary imprint on the deep Dead Sea sediment. Depos. Rec. 2016, 2, 118–138. [Google Scholar] [CrossRef]
- Vuillemin, A.; Ariztegui, D.; Coninck, A.; Lücke, A.; Mayr, C.; Schubert, C. Origin and significance of diagenetic concretions in sediments of Laguna Potrok Aike, southern Argentina. J. Paleolimnol. 2013, 50, 275–291. [Google Scholar] [CrossRef]
- Wurzbacher, C.; Fuchs, A.; Attermeyer, K.; Frindte, K.; Grossart, H.P.; Hupfer, M.; Casper, P.; Monaghan, M.T.; Li, H.; Ettema, T.; et al. Shifts among Eukaryota, Bacteria, and Archaea define the vertical organization of a lake sediment. Microbiome 2017, 5, 41. [Google Scholar] [CrossRef]
- Orsi, W.D. Ecology and evolution of seafloor and subseafloor microbial communities. Nat. Rev. Microbiol. 2018, 16, 671–683. [Google Scholar] [CrossRef]
- Lindhorst, K.; Krastel, S.; Reicherter, K.; Stipp, M.; Wagner, B.; Schwenk, T. Sedimentary and tectonic evolution of Lake Ohrid (Macedonia/Albania). Basin Res. 2015, 27, 84–101. [Google Scholar] [CrossRef]
- Matzinger, A.; Schmid, M.; Veljanoska-Sarafiloska, E.; Patceva, S.; Guseska, D.; Wagner, B.; Müller, B.; Sturm, M.; Wüest, A. Eutrophication of ancient Lake Ohrid: Global warming amplifies detrimental effects of increased nutrient inputs. Limnol. Oceanogr. 2007, 52, 338–353. [Google Scholar] [CrossRef]
- Wagner, B.; Wilke, T.; Krastel, S.; Zanchetta, G.; Sulpizio, R.; Reicherter, K.; Leng, M.J.; Grazhdani, A.; Trajanovski, S.; Francke, A.; et al. The SCOPSCO drilling project recovers more than 1.2 million years of history from Lake Ohrid. Sci. Drill. 2014, 17, 19–29. [Google Scholar] [CrossRef]
- Leicher, N.; Zanchetta, G.; Sulpizio, R.; Giaccio, B.; Wagner, B.; Nomade, S.; Francke, A.; Del Carlo, P. First tephrostratigraphic results of the DEEP site record from Lake Ohrid (Macedonia and Albania). Biogeosciences 2016, 13, 2151–2178. [Google Scholar] [CrossRef]
- Meyers, P.A.; Ishiwatari, R. Lacustrine organic geochemistry-an overview of indicators of organic matter sources and diagenesis in lake sediments. Org. Geochem. 1993, 20, 867–900. [Google Scholar] [CrossRef]
- Holtvoeth, J.; Vogel, H.; Wagner, T.; Wolff, G.A. Improved end-member characterisation of modern organic matter pools in the Ohrid Basin (Albania, Macedonia) and evaluation of new palaeoenvironmental proxies. Biogeosciences 2016, 13, 795–816. [Google Scholar] [CrossRef]
- Vogel, H.; Wessels, M.; Albrecht, C.; Stich, H.B.; Wagner, B. Spatial variability of recent sedimentation in Lake Ohrid (Albania/Macedonia). Biogeosciences 2010, 7, 3333–3342. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Aronesty, E. ea-utils: Command-Line Tools for Processing Biological Sequencing Data; Exprience Analysis: Durham, NC, USA, 2011. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Sheik, C.S.; Reese, B.K.; Twing, K.I.; Sylvan, J.B.; Grim, S.L.; Schrenk, M.O.; Sogin, M.L.; Colwell, F. Identification and removal of contaminant sequences from ribosomal gene databases: Lessons from the Census of Deep Life. Front. Microbiol. 2018, 9, 840. [Google Scholar] [CrossRef]
- Legendre, P.; De Cáceres, M. Beta diversity as the variance of community data: Dissimilarity coefficients and partitioning. Ecol. Lett. 2013, 16, 951–963. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005, 71, 1501–1506. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.; Ryan, P. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbø, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, W88–W95. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.E.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Nobu, M.K.; Dodsworth, J.A.; Murugapiran, S.K.; Rinke, C.; Gies, E.A.; Webster, G.; Schwientek, P.; Kille, P.; Parkes, R.J.; Sass, H.; et al. Phylogeny and physiology of candidate phylum “Atribacteria” (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 2016, 10, 273–286. [Google Scholar] [CrossRef]
- Lazar, C.S.; Baker, B.J.; Seitz, K.; Hyde, A.S.; Dick, G.J.; Hinrichs, K.U.; Teske, A.P. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 2016, 18, 1200–1211. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Schreiber, L.; Petersen, D.G.; Kjeldsen, K.U.; Lever, M.A.; Steen, A.D.; Stepanauskas, R.; Richter, M.; Kleindienst, S.; Lenk, S.; et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 2013, 496, 215–218. [Google Scholar] [CrossRef]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef]
- Kawai, M.; Futagami, T.; Toyoda, A.; Takaki, Y.; Nishi, S.; Hori, S.; Arai, W.; Tsubouchi, T.; Morono, Y.; Uchiyama, I.; et al. High frequency of phylogenetically diverse reductive dehalogenase-homologous genes in deep subseafloor sedimentary metagenomes. Front. Microbiol. 2014, 5, 80. [Google Scholar] [CrossRef]
- Sewell, H.L.; Kaster, A.K.; Spormann, A.M. Homoacetogenesis in Deep-Sea Chloroflexi, as inferred by Single-Cell Genomics, Provides a link to Reductive Dehalogenation in Terrestrial Dehalococcoidetes. MBio 2017, 8, e02022-17. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, A.; Horn, F.; Alawi, M.; Henny, C.; Wagner, D.; Crowe, S.A.; Kallmeyer, J. Preservation and Significance of Extracellular DNA in Ferruginous Sediments from Lake Towuti, Indonesia. Front. Microbiol. 2017, 8, 1440. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.Y.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Kirkpatrick, J.B.; Walsh, E.A.; D’Hondt, S. Microbial Selection and Survival in Subseafloor Sediment. Front. Microbiol. 2019, 10, 956. [Google Scholar] [CrossRef]
- Bird, J.T.; Tague, E.D.; Zinke, L.; Schmidt, J.M.; Steen, A.D.; Reese, B.; Marshall, I.P.G.; Webster, G.; Weightman, A.; Castro, H.F.; et al. Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in baltic sea sediments. MBio 2019, 10, e02376-18. [Google Scholar] [CrossRef] [PubMed]
- Francke, A.; Dosseto, A.; Panagiotopoulos, K.; Leicher, N.; Lacey, J.H.; Kyrikou, S.; Wagner, B.; Zanchetta, G.; Kouli, K.; Leng, M.J. Sediment residence time reveals Holocene shift from climatic to vegetation control on catchment erosion in the Balkans. Glob. Planet. Chang. 2019, 177, 186–200. [Google Scholar] [CrossRef]
- Reynolds, C.S. Cyanobacterial Water-Blooms. Adv. Bot. Res. 1987, 13, 67–143. [Google Scholar]
- Levich, A.P. The role of nitrogen-phosphorus ratio in selecting for dominance of phytoplankton by cyanobacteria or green algae and its application to reservoir management. J. Aquat. Ecosyst. Health 1996, 5, 55–61. [Google Scholar] [CrossRef]
- Teramoto, T.; Yoshimura, M.; Azai, C.; Terauchi, K.; Ohta, T. Determination of carbon-to-nitrogen ratio in the filamentous and heterocystous cyanobacterium Anabaena sp. PCC 7120 with single-cell soft X-ray imaging. J. Phys. Conf. Ser. 2017, 849, 3–7. [Google Scholar] [CrossRef]
- Scanlan, D.J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W.R.; Post, A.F.; Hagemann, M.; Paulsen, I.; Partensky, F. Ecological Genomics of Marine Picocyanobacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 249–299. [Google Scholar] [CrossRef]
- Vuillemin, A.; Ariztegui, D.; Nobbe, G.; Schubert, C.J. Influence of methanogenic populations in Holocene lacustrine sediments revealed by clone libraries and fatty acid biogeochemistry. Geomicrobiol. J. 2013, 31, 285–298. [Google Scholar] [CrossRef]
- Amend, J.P.; McCollom, T.M.; Hentscher, M.; Bach, W. Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim. Cosmochim. Acta 2011, 75, 5736–5748. [Google Scholar] [CrossRef]
- Lever, M.A.; Alperin, M.J.; Teske, A.; Heuer, V.B.; Schmidt, F.; Hinrichs, K.U.; Morono, Y.; Masui, N.; Inagaki, F. Acetogenesis in Deep Subseafloor Sediments of The Juan de Fuca Ridge Flank: A Synthesis of Geochemical, Thermodynamic, and Gene-based Evidence. Geomicrobiol. J. 2010, 27, 183–211. [Google Scholar] [CrossRef]
- Vuillemin, A.; Vargas, S.; Coskun, Ö.; Pockalny, R.; Murray, R.; Smith, D.; D’Hondt, S.; Orsi, W. Atribacteria reproducing over millions of years in the Atlantic abyssal subseafloor. MBio 2020. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, C.; Francke, A.; Vogel, H.; Wagner, B.; Ariztegui, D. Weak Influence of Paleoenvironmental Conditions on the Subsurface Biosphere of Lake Ohrid over the Last 515 ka. Microorganisms 2020, 8, 1736. https://doi.org/10.3390/microorganisms8111736
Thomas C, Francke A, Vogel H, Wagner B, Ariztegui D. Weak Influence of Paleoenvironmental Conditions on the Subsurface Biosphere of Lake Ohrid over the Last 515 ka. Microorganisms. 2020; 8(11):1736. https://doi.org/10.3390/microorganisms8111736
Chicago/Turabian StyleThomas, Camille, Alexander Francke, Hendrik Vogel, Bernd Wagner, and Daniel Ariztegui. 2020. "Weak Influence of Paleoenvironmental Conditions on the Subsurface Biosphere of Lake Ohrid over the Last 515 ka" Microorganisms 8, no. 11: 1736. https://doi.org/10.3390/microorganisms8111736
APA StyleThomas, C., Francke, A., Vogel, H., Wagner, B., & Ariztegui, D. (2020). Weak Influence of Paleoenvironmental Conditions on the Subsurface Biosphere of Lake Ohrid over the Last 515 ka. Microorganisms, 8(11), 1736. https://doi.org/10.3390/microorganisms8111736




