Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. L. salivarius Strains
2.2. PIE Cells
2.3. RT-qPCR
2.4. Purification of Porcine Intestinal Mucins
2.5. Biacore Assay
2.6. Adhesion to PIE Cells
2.7. Scanning Electron Microscopy (SEM) Analysis
2.8. Complete Genome Sequencing
2.9. Bioinformatic Analysis
2.10. Rotavirus Infection
2.11. Statistical Analysis
3. Results and Discussion
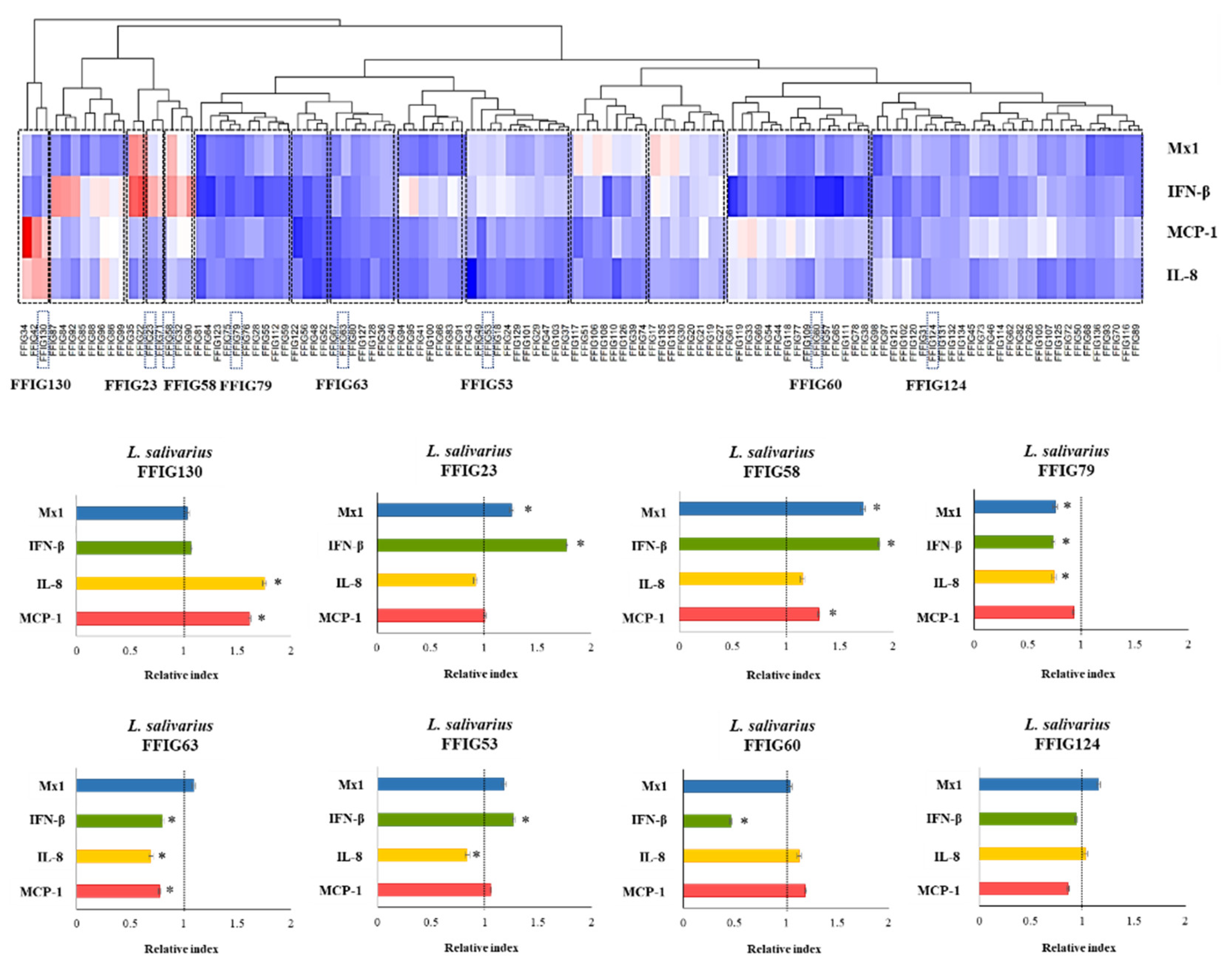
3.1. Characterization of the Ability of L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs to Modulate Innate immune Responses in PIE Cells
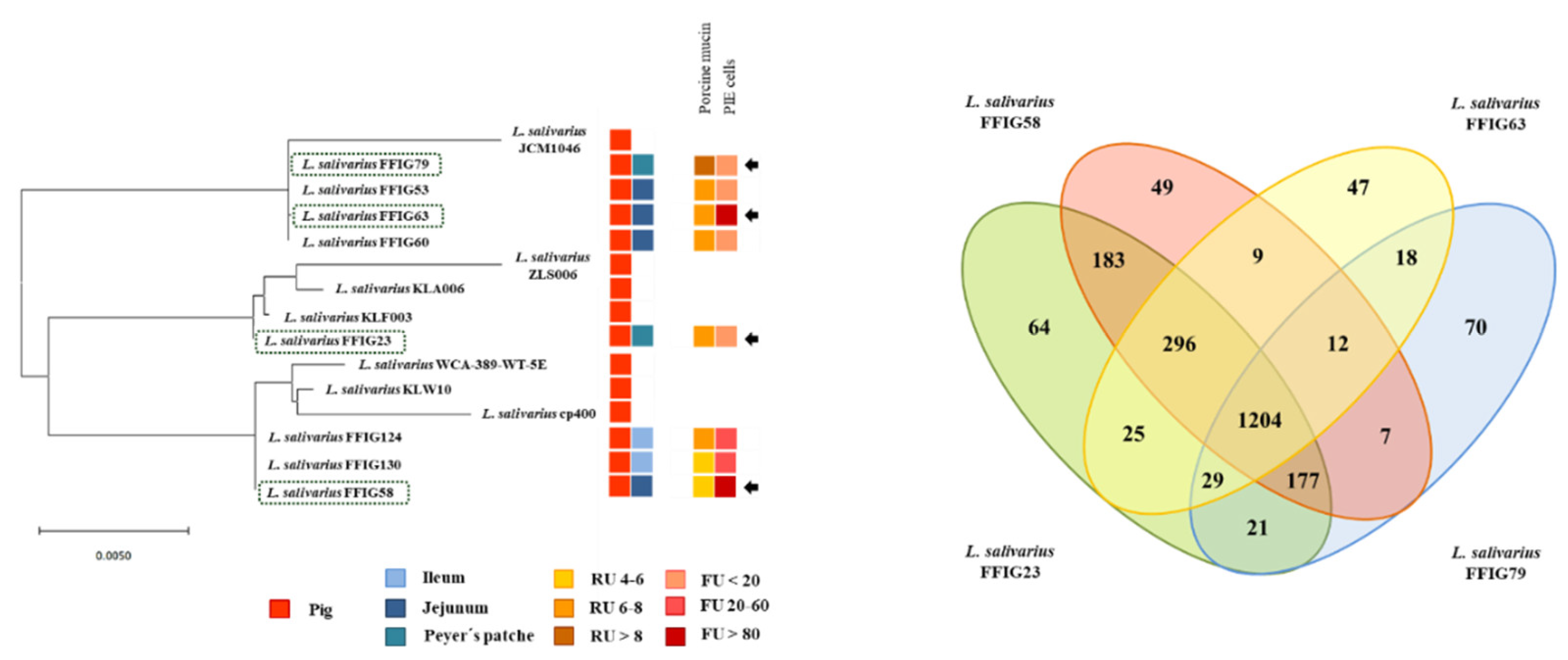
3.2. General Genomic Features of L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs
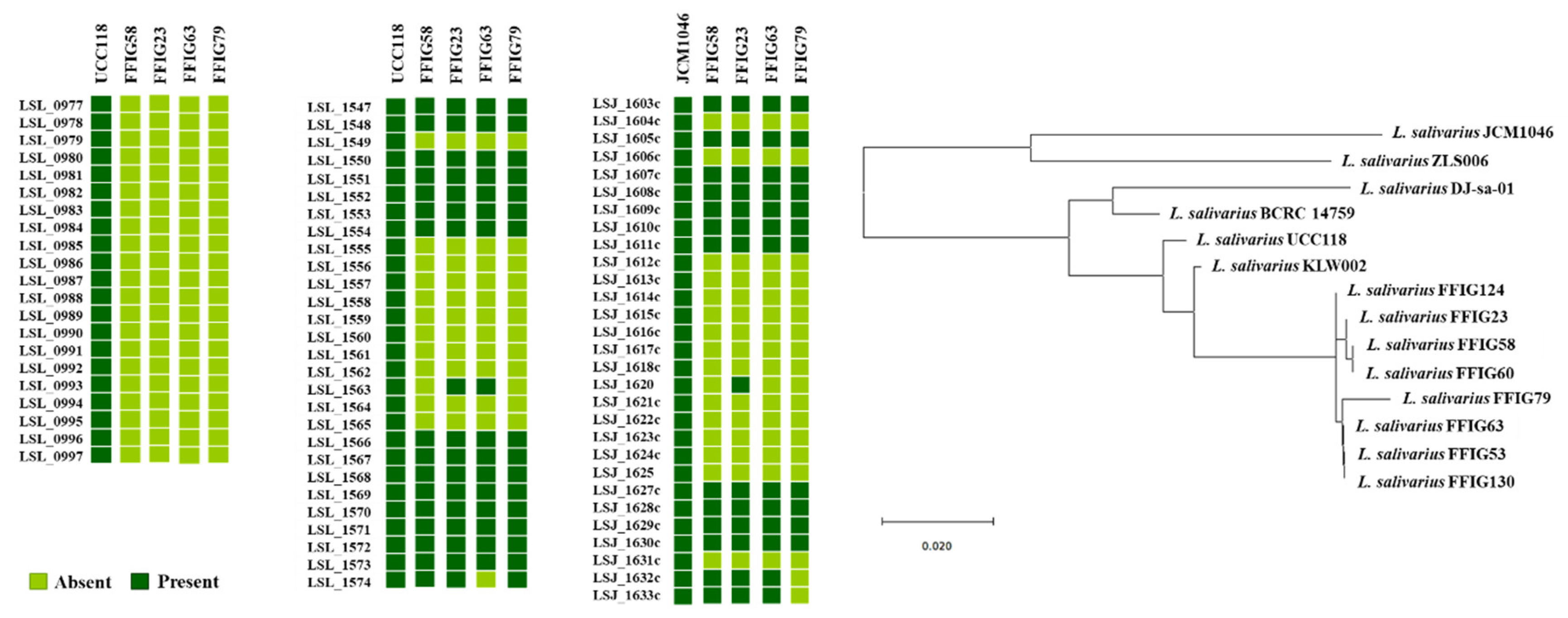
3.3. Comparative Genomic Analysis of L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs Belonging to Different “Immune Phenotypes”
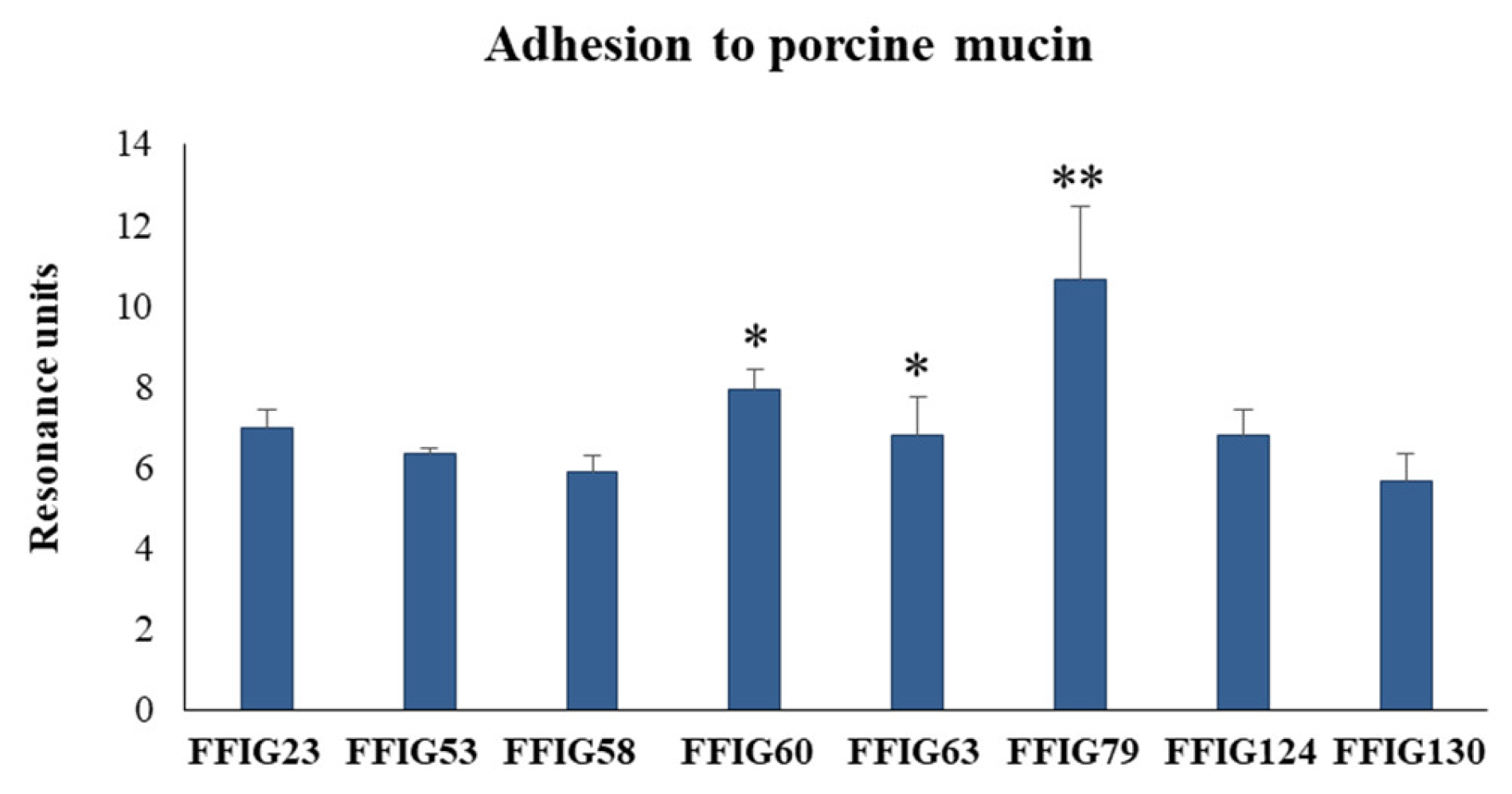
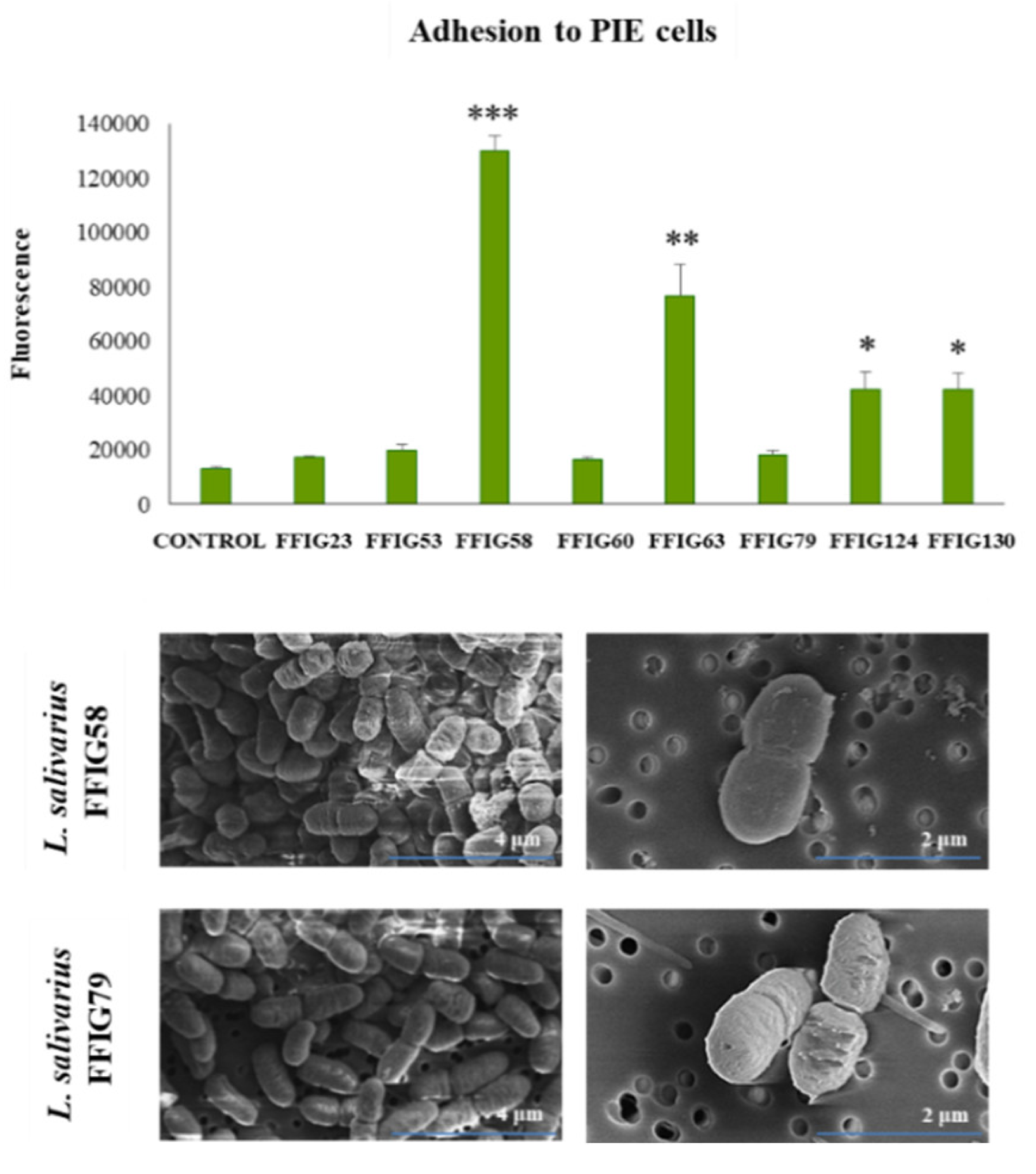
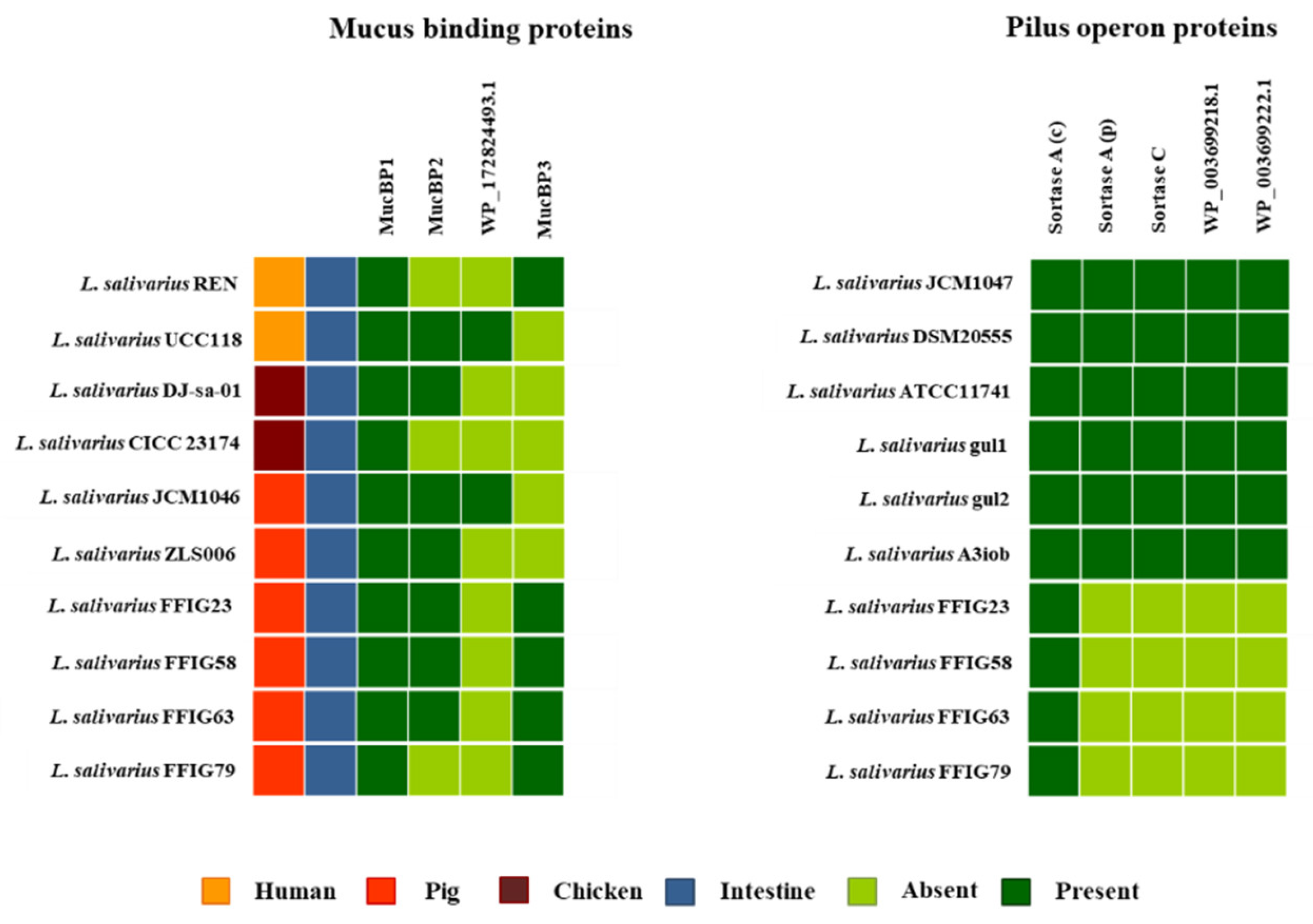
3.4. Characterization of the Ability of L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs to Adhere to Mucins and PIE Cells
3.5. Comparative Genomic Analysis of L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs Belonging to Different “Adhesion Phenotypes”
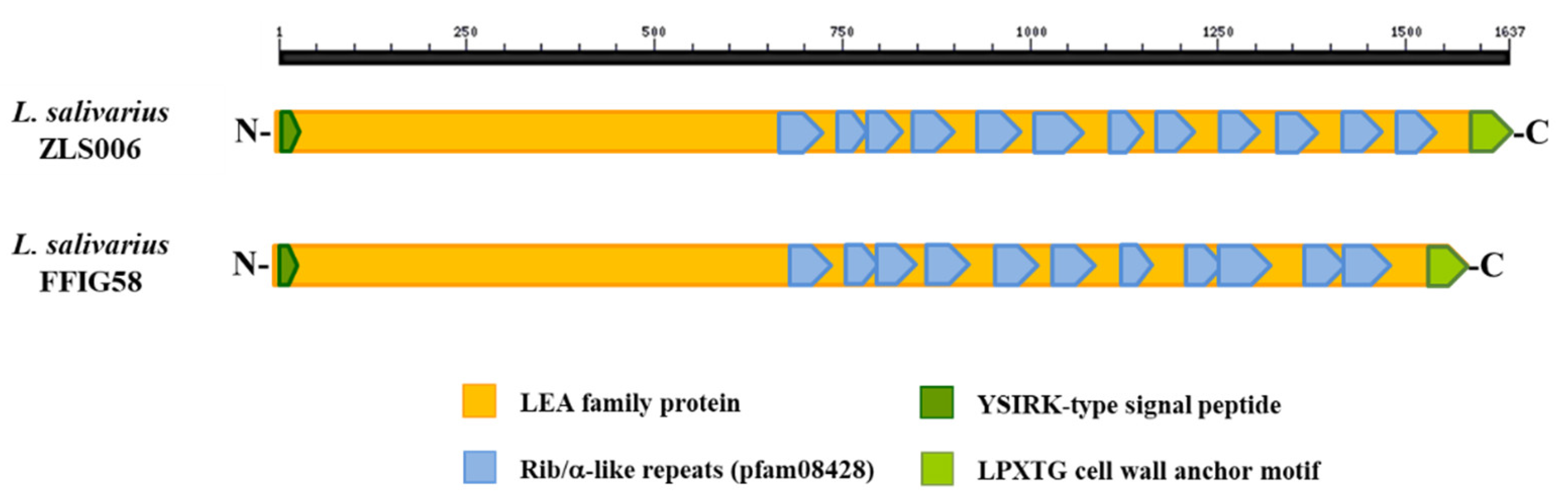
3.6. Analysis of Adhesion Factors in L. salivarius Isolated from the Intestinal Tract of Wakame-Fed Pigs
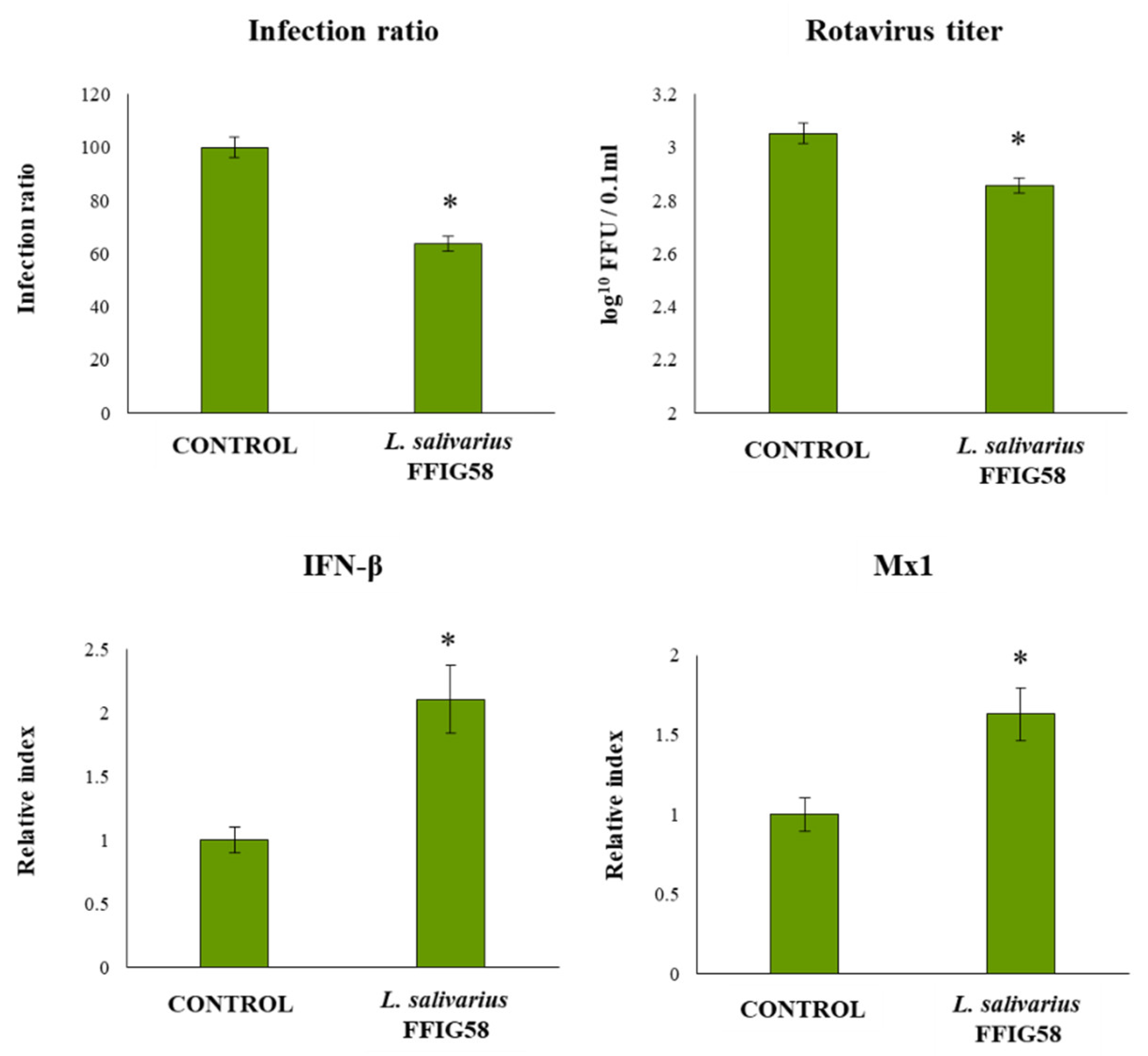
3.7. L. salivarius FFIG58 Increase the Resistance of PIE Cell to Rotavirus Infection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Han, G.G.; Lee, J.Y.; Jin, G.D.; Park, J.; Choi, Y.H.; Kang, S.K.; Chae, B.J.; Kim, E.B.; Choi, Y.J. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Piazuelo, D.; Estellé, J.; Revilla, M.; Criado-Mesas, L.; Ramayo-Caldas, Y.; Óvilo, C.; Fernández, A.I.; Ballester, M.; Folch, J.M. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zeng, X.; Zhang, G.; Hou, C.; Li, N.; Yu, H.; Shang, L.; Zhang, X.; Trevisi, P.; Yang, F.; et al. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the porcine neonatal gut. BMC Biol. 2019, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Suda, Y.; Villena, J.; Takahashi, Y.; Hosoya, S.; Tomosada, Y.; Tsukida, K.; Shimazu, T.; Aso, H.; Tohno, M.; Ishida, M.; et al. Immunobiotic Lactobacillus jensenii as immune-health promoting factor to improve growth performance and productivity in post-weaning pigs. BMC Immunol. 2014, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.F.; Nyachoti, M. Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutr. 2017, 3, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Reyer, H.; Oster, M.; McCormack, U.M.; Muráni, E.; Gardiner, G.E.; Ponsuksili, S.; Lawlor, P.G.; Wimmers, K. Host-microbiota interactions in ileum and caecum of pigs divergent in feed efficiency contribute to nutrient utilization. Microorganisms 2020, 8, 563. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Kitazawa, H. Modulation of intestinal TLR4-inflammatory signaling pathways by probiotic microorganisms: Lessons learned from Lactobacillus jensenii TL2937. Front. Immunol. 2014, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gänzle, M. Toward rational selection criteria for selection of probiotics in pigs. Adv. Appl. Microbiol. 2019, 107, 83–112. [Google Scholar] [CrossRef]
- Aluthge, N.D.; van Sambeek, D.M.; Carney-Hinkle, E.E.; Li, Y.S.; Fernando, S.C.; Burkey, T.E. Board Invited review: The pig microbiota and the potential for harnessing the power of the microbiome to improve growth and health. J. Anim. Sci. 2019, 97, 3741–3757. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Lee, J.Y.; Han, G.G.; Kim, E.B.; Choi, Y.J. Comparative genomics of Lactobacillus salivarius strains focusing on their host adaptation. Microbiol. Res. 2017, 205, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Deng, J.; Wang, Z.; Che, C.; Li, Y.F.; Yang, Q. Modulatory effects of Lactobacillus salivarius on intestinal mucosal immunity of piglets. Curr. Microbiol. 2011, 62, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Lo Verso, L.; Lessard, M.; Talbot, G.; Fernandez, B.; Fliss, I. Isolation and Selection of Potential Probiotic Bacteria from the Pig Gastrointestinal Tract. Probiotics Antimicrob. Proteins 2018, 10, 299–312. [Google Scholar] [CrossRef]
- Yoshinaga, T.; Nishiduka, H.; Nanba, N. Genotype analysis of commercial products of the soft seaweed Undaria pinnatifida (Laminariales, Alariaceae). Coast. Mar. Sci. 2014, 36, 9–15. [Google Scholar] [CrossRef]
- Masumizu, Y.; Zhou, B.; Kober, A.K.M.H.; Islam, M.A.; Iida, H.; Ikeda-Ohtsubo, W.; Suda, Y.; Albarracin, L.; Nochi, T.; Aso, H.; et al. Isolation and immunocharacterization of Lactobacillus salivarius from the intestine of wakame-fed pigs to develop novel “immunosynbiotics”. Microorganisms 2019, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Fukuda, T.; Okamura, T.; Suzuki, E.; Tamura, K.; Shimizu, Y.; Suda, Y.; Suzuki, K. Effect of dietary addition of seaweed and licorice on the immune performance of pigs. Anim. Sci. J. 2011, 82, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Albarracin, L.; Kobayashi, H.; Hebert, E.M.; Saavedra, L.; Komatsu, R.; Gatica, B.; Miyazaki, A.; Ikeda-Ohtsubo, W.; Suda, Y.; et al. Genomic characterization of Lactobacillus delbrueckii TUA4408L and evaluation of the antiviral activities of its extracellular polysaccharides in porcine intestinal epithelial cells. Front. Immunol. 2018, 9, 2178. [Google Scholar] [CrossRef] [PubMed]
- Albarracin, L.; Kobayashi, H.; Iida, H.; Sato, N.; Nochi, T.; Aso, H.; Salva, S.; Alvarez, S.; Kitazawa, H.; Villena, J. Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: Influence of immunobiotic lactobacilli. Front. Immunol. 2017, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Wachi, S.; Kanmani, P.; Tomosada, Y.; Kobayashi, H.; Yuri, T.; Egusa, S.; Shimazu, T.; Suda, Y.; Aso, H.; Sugawara, M.; et al. Lactobacillus delbrueckii TUA4408L and its extracellular polysaccharides attenuate enterotoxigenic Escherichia coli- induced inflammatory response in porcine intestinal epitheliocytes via Toll-like receptor-2 and 4. Mol. Nutr. Food Res. 2014, 58, 2080–2093. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.-N.; Okawara, T.; Watanabe, M.; Kawai, Y.; Kitazawa, H.; Ohnuma, S.; Shibata, C.; Horii, A.; Kimura, K.; Taketomo, N.; et al. New screening methods for probiotics with adhesion properties to sialic acid and sulphate residues in human colonic mucin using the Biacore assay. J. Appl. Microbiol. 2013, 114, 854–860. [Google Scholar] [CrossRef]
- Uchida, H.; Fujitani, K.; Kawai, Y.; Kitazawa, H.; Horii, A.; Shiiba, K.; Saito, K.; Saito, T. A New Assay Using Surface Plasmon Resonance (SPR) to Determine Binding of the Lactobacillus acidophilus Group to Human Colonic Mucin. Biosci. Biotechnol. Biochem. 2004, 68, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Wakahara, N.; Watanabe, M.; Kawasaki, T.; Matsuo, H.; Kawai, Y.; Kitazawa, H.; Ohnuma, S.; Miura, K.; Horii, A.; et al. Cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Lactobacillus plantarum LA 318 recognizes human A and B blood group antigens. Res. Microbiol. 2008, 159, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Albarracin, L.; Masumizu, Y.; Indo, Y.; Islam, M.A.; Garcia-Castillo, V.; Ikeda-Ohtsubo, W.; Suda, Y.; Aso, H.; Villena, J.; et al. Draft Genome Sequence of Ligilactobacillus salivarius FFIG58, Isolated from the Intestinal Tract of Wakame-Fed Pig. Microbiol. Resour. Announc. 2020, 9, e00839-20. [Google Scholar] [CrossRef] [PubMed]
- Audisio, M.C.; Albarracín, L.; Torres, M.J.; Saavedra, L.; Hebert, E.M.; Villena, J. Draft Genome Sequences of Lactobacillus salivarius A3iob and Lactobacillus johnsonii CRL1647, Novel Potential Probiotic Strains for Honeybees (Apis mellifera L.). Microbiol. Resour. Announc. 2018, 7, e00975-18. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Rodriguez-R, L.; Konstantinidis, K. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. Package “gplots”: Various R Programming Tools for Plotting Data, R Packag. version 2.17.0; ScienceOpen: Berlin, Germany, 2015. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Kanmani, P.; Kobayashi, H.; Miyazaki, A.; Soma, J.; Suda, Y.; Aso, H.; Nochi, T.; Iwabuchi, N.; Xiao, J.; et al. Immunobiotic Bifidobacteria Strains Modulate Rotavirus Immune Response in Porcine Intestinal Epitheliocytes via Pattern Recognition Receptor Signaling. PLoS ONE 2016, 11, e0152416. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Villena, J.; Tohno, M.; Fujie, H.; Hosoya, S.; Shimosato, T.; Aso, H.; Suda, Y.; Kawai, Y.; Saito, T.; et al. Immunobiotic Lactobacillus jensenii elicits anti-inflammatory activity in porcine intestinal epithelial cells by modulating negative regulators of the Toll-like receptor signaling pathway. Infect. Immun. 2012, 80, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Albarracin, L.; García-Castillo, V.; Masumizu, Y.; Indo, Y.; Islam, M.A.; Suda, Y.; Garcia, A.; Aso, H.; Takahashi, H.; Kitazawa, H.; et al. Efficient selection of new immunobiotic strains with antiviral effects in local and distal mucosal sites by using porcine intestinal epitheliocytes. Front. Immunol. 2020, 11, 543. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Harris, H.M.B.; Bourin, M.J.B.; Claesson, M.J.; O’Toole, P.W. Phylogenomics and comparative genomics of Lactobacillus salivarius, a mammalian gut commensal. Microb. Genom. 2017, 3. [Google Scholar] [CrossRef]
- Bernard, E.; Rolain, T.; David, B.; André, G.; Dupres, V.; Dufrêne, Y.F.; Hallet, B.; Chapot-Chartier, M.P.; Hols, P. Dual Role for the O-Acetyltransferase OatA in Peptidoglycan Modification and Control of Cell Septation in Lactobacillus plantarum. PLoS ONE 2012, 7, e47893. [Google Scholar] [CrossRef]
- Brott, A.S.; Clarke, A.J. Peptidoglycan O-acetylation as a virulence factor: Its effect on lysozyme in the innate immune system. Antibiotics 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Kolling, Y.; Salva, S.; Villena, J.; Alvarez, S. Are the immunomodulatory properties of Lactobacillus rhamnosus CRL1505 peptidoglycan common for all Lactobacilli during respiratory infection in malnourished mice? PLoS ONE 2018, 13, e0194034. [Google Scholar] [CrossRef] [PubMed]
- Clua, P.; Tomokiyo, M.; Raya Tonetti, F.; Islam, M.A.; García Castillo, V.; Marcial, G.; Salva, S.; Alvarez, S.; Takahashi, H.; Kurata, S.; et al. The Role of Alveolar Macrophages in the Improved Protection against Respiratory Syncytial Virus and Pneumococcal Superinfection Induced by the Peptidoglycan of Lactobacillus rhamnosus CRL1505. Cells 2020, 9, 1653. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Arce, L.; Tomotsune, K.; Albarracin, L.; Funabashi, R.; Vera, D.; Islam, M.A.; Vizoso-Pinto, M.G.; Takahashi, H.; Sasaki, Y.; et al. Lipoteichoic Acid Is Involved in the Ability of the Immunobiotic Strain Lactobacillus plantarum CRL1506 to Modulate the Intestinal Antiviral Innate Immunity Triggered by TLR3 Activation. Front. Immunol. 2020, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Raftis, E.J.; Forde, B.M.; Claesson, M.J.; O’Toole, P.W. Unusual genome complexity in Lactobacillus salivarius JCM1046. BMC Genom. 2014, 15, 771. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.T.; Hsu, I.T.; Chou, C.C.; Lo, P.R.; Yu, R.C. Exopolysaccharide production of Lactobacillus salivarius BCRC 14759 and bifidobacterium bifidum BCRC 14615. World J. Microbiol. Biotechnol. 2009, 25, 883–890. [Google Scholar] [CrossRef]
- Kanmani, P.; Albarracin, L.; Kobayashi, H.; Iida, H.; Komatsu, R.; Humayun Kober, A.K.M.; Ikeda-Ohtsubo, W.; Suda, Y.; Aso, H.; Makino, S.; et al. Exopolysaccharides from Lactobacillus delbrueckii OLL1073R-1 modulate innate antiviral immune response in porcine intestinal epithelial cells. Mol. Immunol. 2018, 93, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Perotti, F.; Masserey, I.; Rouvet, M.; Golliard, M.; Servin, A.; Brassart, D. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 1999, 65, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Živković, M.; Miljković, M.S.; Ruas-Madiedo, P.; Markelić, M.B.; Veljović, K.; Tolinački, M.; Soković, S.; Korać, A.; Golić, N. EPS-SJ exopolisaccharide produced by the strain Lactobacillus paracasei subsp. paracasei BGSJ2-8 is involved in adhesion to epithelial intestinal cells and decrease on E. coli association to Caco-2 cells. Front. Microbiol. 2016, 7, 286. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Cross, B.W.; Ruhl, S. Glycan recognition at the saliva—Oral microbiome interface. Cell. Immunol. 2018, 333, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, B.; Wu, H.; Peng, Z.; Fives-Taylor, P.M. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J. Bacteriol. 2007, 189, 7610–7617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wu, H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 2009, 155, 317–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bensing, B.A.; Seepersaud, R.; Yen, Y.T.; Sullam, P.M. Selective transport by SecA2: An expanding family of customized motor proteins. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1674–1686. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; MacKenzie, D.A.; Peterson, D.A.; Schmaltz, R.; Fangman, T.; Zhou, Y.; Zhang, C.; Benson, A.K.; Cody, L.A.; Mulholland, F.; et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013, 9, e1004057. [Google Scholar] [CrossRef]
- De Boeck, I.; van den Broek, M.F.L.; Allonsius, C.N.; Spacova, I.; Wittouck, S.; Martens, K.; Wuyts, S.; Cauwenberghs, E.; Jokicevic, K.; Vandenheuvel, D.; et al. Lactobacilli have a niche in the human nose. Cell Rep. 2020, 31, 107674. [Google Scholar] [CrossRef] [PubMed]
- Couvigny, B.; Lapaque, N.; Rigottier-Gois, L.; Guillot, A.; Chat, S.; Meylheuc, T.; Kulakauskas, S.; Rohde, M.; Mistou, M.Y.; Renault, P.; et al. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 2017, 19, 3579–3594. [Google Scholar] [CrossRef] [PubMed]
- Couvigny, B.; Kulakauskas, S.; Pons, N.; Quinquis, B.; Abraham, A.L.; Meylheuc, T.; Delorme, C.; Renault, P.; Briandet, R.; Lapaque, N.; et al. Identification of new factors modulating adhesion abilities of the pioneer commensal bacterium Streptococcus salivarius. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Feltcher, M.E.; Braunstein, M. Emerging themes in SecA2-mediated protein export. Nat. Rev. Microbiol. 2012, 10, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, U.; MacKenzie, D.A.; Zheng, J.; Goesmann, A.; Roos, S.; Swarbreck, D.; Walter, J.; Crossman, L.C.; Juge, N. The pan-genome of Lactobacillus reuteri strains originating from the pig gastrointestinal tract. BMC Genom. 2015, 16, 1023. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Benson, A.K.; Tannock, G.W.; Loach, D.M.; Kim, J.; Zhang, M.; Oh, P.L.; Heng, N.C.K.; Patil, P.B.; Juge, N.; et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011, 7, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Wuyts, S.; Wittouck, S.; De Boeck, I.; Allonsius, C.N.; Pasolli, E.; Segata, N.; Lebeer, S. Large-Scale Phylogenomics of the Lactobacillus casei Group Highlights Taxonomic Inconsistencies and Reveals Novel Clade-Associated Features. mSystems 2017, 2, e00061-17. [Google Scholar] [CrossRef] [PubMed]
- Latousakis, D.; Juge, N. How sweet are our gut beneficial bacteria? A focus on protein glycosylation in Lactobacillus. Int. J. Mol. Sci. 2018, 19, 136. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.A.; Tailford, L.E.; Hemmings, A.M.; Juge, N. Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J. Biol. Chem. 2009, 284, 32444–32453. [Google Scholar] [CrossRef] [PubMed]
- Etzold, S.; Kober, O.I.; Mackenzie, D.A.; Tailford, L.E.; Gunning, A.P.; Walshaw, J.; Hemmings, A.M.; Juge, N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 2014, 16, 888–903. [Google Scholar] [CrossRef] [PubMed]
- Etzold, S.; Juge, N. Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 2014, 28, 23–31. [Google Scholar] [CrossRef]
- Gunning, A.P.; Kavanaugh, D.; Thursby, E.; Etzold, S.; Mackenzie, D.A.; Juge, N. Use of atomic force microscopy to study the multi-modular interaction of bacterial adhesins to mucins. Int. J. Mol. Sci. 2016, 17, 1854. [Google Scholar] [CrossRef]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; Von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef]
- Von Ossowski, I. Novel molecular insights about lactobacillar sortase-dependent piliation. Int. J. Mol. Sci. 2017, 18, 1551. [Google Scholar] [CrossRef]
- Edelman, S.M.; Lehti, T.A.; Kainulainen, V.; Antikainen, J.; Kylväjä, R.; Baumann, M.; Westerlund-Wikström, B.; Korhonen, T.K. Identification of a high-molecular-mass Lactobacillus epithelium adhesin (LEA) of Lactobacillus crispatus ST1 that binds to stratified squamous epithelium. Microbiology (United Kingdom) 2012, 158, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Ojala, T.; Kuparinen, V.; Koskinen, J.P.; Alatalo, E.; Holm, L.; Auvinen, P.; Edelman, S.; Westerlund-Wikström, B.; Korhonen, T.K.; Paulin, L.; et al. Genome sequence of Lactobacillus crispatus ST1. J. Bacteriol. 2010, 192, 3547–3548. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, H.; Krogh Andersen, K.; Lin, Y.; Zuo, F.; Zeng, Z.; Larsson, P.G.; Brandsborg, E.; Brønstad, G.; Hammarström, L. Characterization and complete genome sequences of L. rhamnosus DSM 14870 and L. gasseri DSM 14869 contained in the EcoVag® probiotic vaginal capsules. Microbiol. Res. 2017, 205, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Pantoja-Feliciano, I.G.; Clemente, J.C.; Costello, E.K.; Perez, M.E.; Blaser, M.J.; Knight, R.; Dominguez-Bello, M.G. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013, 7, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Kmet, V.; Lucchini, F. Aggregation of sow lactobacilli with diarrhoeagenic Escherichia coli. J. Vet. Med. Ser. B 1999, 46, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Delgado, S.; Maldonado, A.; Jiménez, E.; Olivares, M.; Fernández, L.; Sobrino, O.J.; Rodríguez, J.M. Isolation of lactobacilli from sow milk and evaluation of their probiotic potential. J. Dairy Res. 2009, 76, 418–425. [Google Scholar] [CrossRef] [PubMed]






| Ligilactobacillus salivarius Strain | Host | Sample | Genome Size (bp) | G+C Content (%) | Protein-Coding Genes | GenBank ID | Reference |
|---|---|---|---|---|---|---|---|
| FFIG58 | Sus scrofa | Intestine | 1,984,018 | 32.8 | 1891 | JACBJR000000000.1 | [20] |
| FFIG23 | Sus scrofa | Intestine | 2,041,027 | 32.8 | 1932 | JACBJS000000000.1 | This work |
| FFIG53 | Sus scrofa | Intestine | 1,948,231 | 32.9 | 1863 | JACBJT000000000.1 | This work |
| FFIG60 | Sus scrofa | Intestine | 1,948,639 | 33.0 | 1855 | JACBJU000000000.1 | This work |
| FFIG63 | Sus scrofa | Intestine | 1,777,024 | 33.3 | 1786 | JACBJV000000000.1 | This work |
| FFIG79 | Sus scrofa | Intestine | 1,718,597 | 33.4 | 1728 | JACBJW000000000.1 | This work |
| FFIG124 | Sus scrofa | Intestine | 1,806,583 | 33.2 | 1812 | JACBJX000000000.1 | This work |
| FFIG130 | Sus scrofa | Intestine | 1,862,635 | 33.1 | 1778 | JACBJY000000000.1 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, B.; Albarracin, L.; Indo, Y.; Arce, L.; Masumizu, Y.; Tomokiyo, M.; Islam, M.A.; Garcia-Castillo, V.; Ikeda-Ohtsubo, W.; Nochi, T.; et al. Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies. Microorganisms 2020, 8, 1659. https://doi.org/10.3390/microorganisms8111659
Zhou B, Albarracin L, Indo Y, Arce L, Masumizu Y, Tomokiyo M, Islam MA, Garcia-Castillo V, Ikeda-Ohtsubo W, Nochi T, et al. Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies. Microorganisms. 2020; 8(11):1659. https://doi.org/10.3390/microorganisms8111659
Chicago/Turabian StyleZhou, Binghui, Leonardo Albarracin, Yuhki Indo, Lorena Arce, Yuki Masumizu, Mikado Tomokiyo, Md. Aminul Islam, Valeria Garcia-Castillo, Wakako Ikeda-Ohtsubo, Tomonori Nochi, and et al. 2020. "Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies" Microorganisms 8, no. 11: 1659. https://doi.org/10.3390/microorganisms8111659
APA StyleZhou, B., Albarracin, L., Indo, Y., Arce, L., Masumizu, Y., Tomokiyo, M., Islam, M. A., Garcia-Castillo, V., Ikeda-Ohtsubo, W., Nochi, T., Morita, H., Takahashi, H., Kurata, S., Villena, J., & Kitazawa, H. (2020). Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies. Microorganisms, 8(11), 1659. https://doi.org/10.3390/microorganisms8111659








