Revisiting an Aspergillus flavus Strain Isolated from an Egyptian Sugarcane Field in 1930
Abstract
1. Introduction
2. Materials and Methods
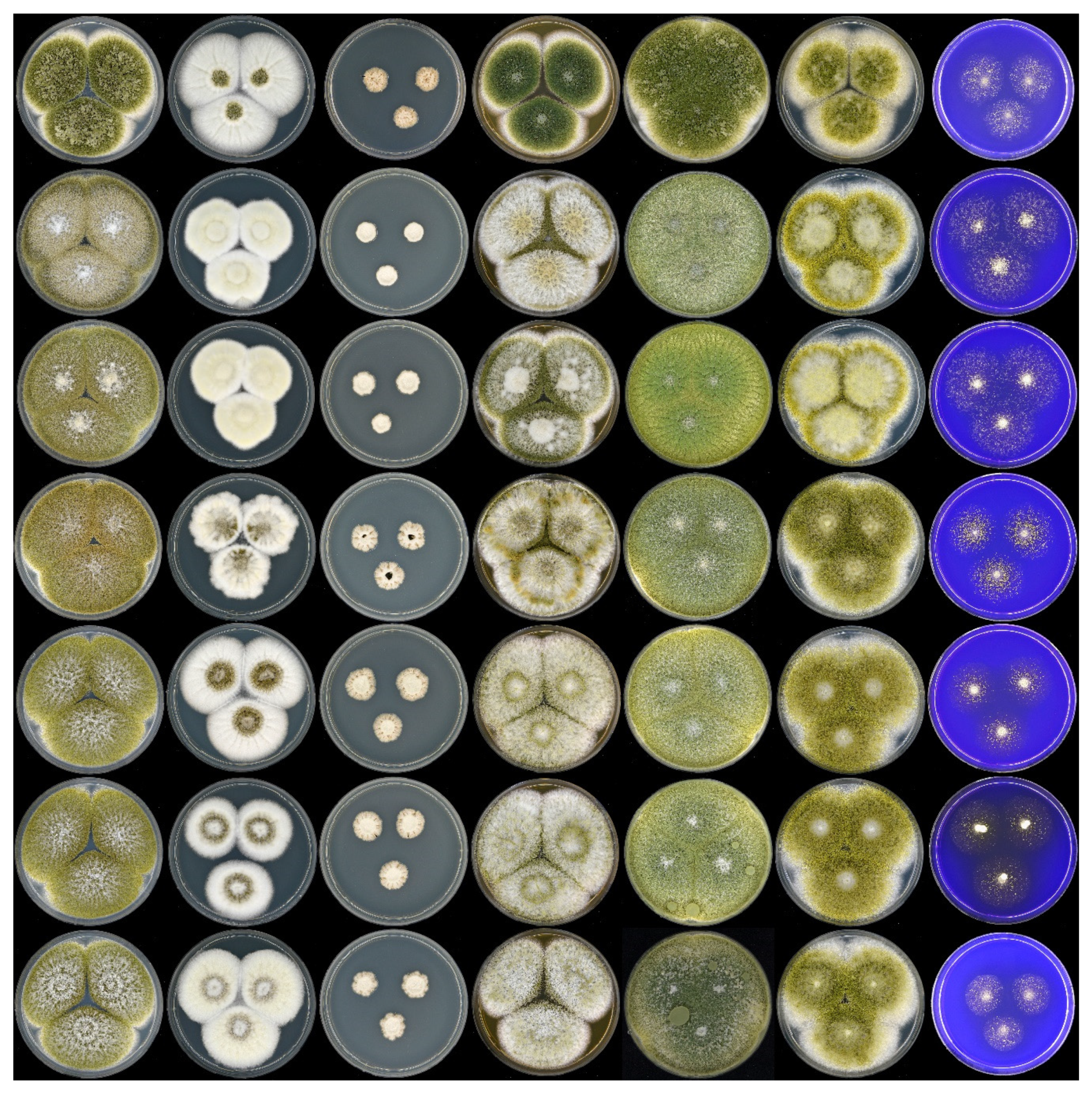
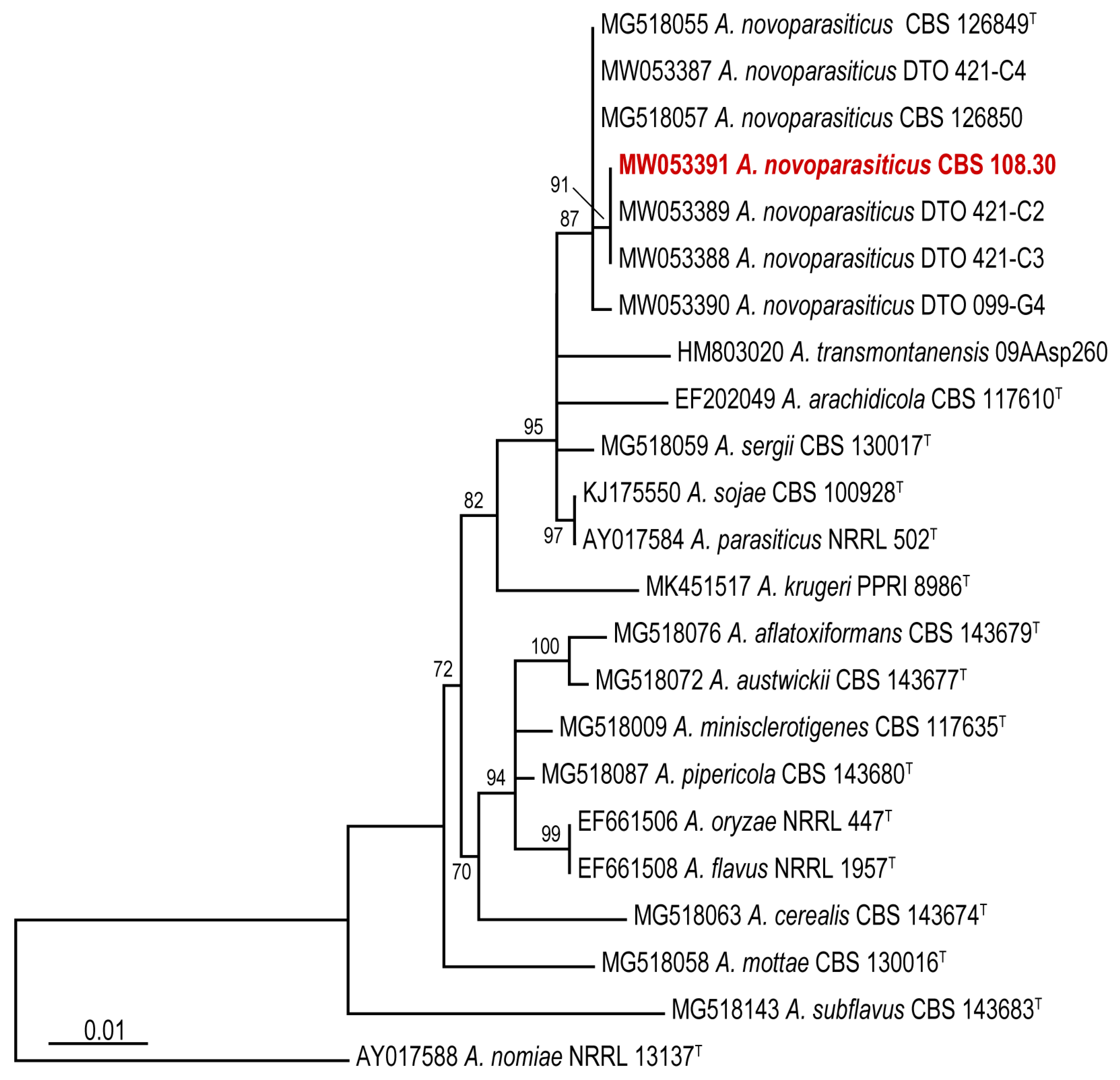
3. Results and Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hall, W.J. The Outbreak of Pseudococcus Sacchari, Ckll., on the Sugar Cane of Egypt; Government Press: Cairo, Egypt, 2011. [Google Scholar]
- Spensley, P.C. Aflatoxin, the active principle in Turkey “X” disease. Endeavour 1963, 22, 75–79. [Google Scholar] [CrossRef]
- Coomes, T.J.; Sanders, J.C. The detection and estimation of aflatoxin in groundnuts and groundnut materials: Part I. Paper-chromatographic procedure. Analyst 1963, 88, 209–213. [Google Scholar] [CrossRef]
- Hartley, R.D.; Nesbitt, B.F.; O’Kelly, J. Toxic metabolites of Aspergillus Flavus. Nature 1963, 198, 1056–1058. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Frisvad, J.C.; Samson, R.A. A reappraisal of fungi producing aflatoxins. World Mycotoxin J. 2009, 2, 263–277. [Google Scholar] [CrossRef]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.F.; Audenaert, K.; Lust, L.; Landschoot, S.; Bekaert, B.; Haesaert, G.; De Boevre, M.; De Saeger, S. Risk characterization and quantification of mycotoxins and their producing fungi in sugarcane juice: A neglected problem in a widely-consumed traditional beverage. Food Control. 2020, 108, 106811. [Google Scholar] [CrossRef]
- Samson, R.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; 2010; Volume 2, ISBN 978-90-70351-82-3. [Google Scholar]
- Klitgaard, A.; Iversen, A.; Andersen, M.R.; Larsen, T.O.; Frisvad, J.C.; Nielsen, K.F. Aggressive dereplication using UHPLC-DAD-QTOF: Screening extracts for up to 3000 fungal secondary metabolites. Anal. Bioanal. Chem. 2014, 406, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, S.S.; Stchigel, A.M.; Cano, J.F.; Godoy-Martinez, P.C.; Colombo, A.L.; Guarro, J. Aspergillus novoparasiticus: A new clinical species of the section Flavi. Med. Mycol. 2012, 50, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Uka, V.; Han, Z.; Buyst, D.; Harris-Coward, P.Y.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Dowd, P.F.; Martens, S.L.; et al. An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet. Biol. 2015, 81, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Adjovi, Y.C.S.; Bailly, S.; Gnonlonfin, B.J.G.; Tadrist, S.; Querin, A.; Sanni, A.; Oswald, I.P.; Puel, O.; Bailly, J.D. Analysis of the contrast between natural occurrence of toxigenic Aspergilliof the Flavi section and aflatoxin B1 in cassava. Food Microbiol. 2014, 38, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Viaro, H.P.; da Silva, J.J.; de Souza Ferranti, L.; Bordini, J.G.; Massi, F.P.; Fungaro, M.H.P. The first report of A. novoparasiticus, A. arachidicola and A. pseudocaelatus in Brazilian corn kernels. Int. J. Food Microbiol. 2017, 243, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.J.; Puel, O.; Lorber, S.; Ferranti, L.S.; Ortiz, L.F.; Taniwaki, M.H.; Iamanaka, B.T.; Fungaro, M.H.P. Occurrence and diversity of Aspergillus in commercial yerba mate elaborated for the Brazilian beverage ‘chimarrão’. Food Res. Int. 2019, 121, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, U.; Wu, H.; Wei, J.; Ou, X.; Qin, P.; Yao, X.; Chen, H.; Chen, A.J.; Liu, B. A polyphasic study of Aspergillus section Flavi isolated from corn in Guangxi, China- a hot spot of aflatoxin contamination. Int. J. Food Microbiol. 2019, 310. [Google Scholar] [CrossRef] [PubMed]
- Iamanaka, B.T.; de Souza Lopes, A.; Martins, L.M.; Frisvad, J.C.; Medina, A.; Magan, N.; Sartori, D.; Massi, F.P.; Fungaro, M.H.P.; Taniwaki, M.H. Aspergillus section Flavi diversity and the role of A. novoparasiticus in aflatoxin contamination in the sugarcane production chain. Int. J. Food Microbiol. 2019, 293, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.F.; Ameye, M.; De Saeger, S.; Audenaert, K.; Haesaert, G. Biological Control of Mycotoxigenic Fungi and Their Toxins: An Update for the Pre-Harvest Approach. In Mycotoxins—Impact and Management Strategies; Njobeh, P.B., Ed.; IntechOpen: London, UK, 2019. [Google Scholar]
- Kjærbølling, I.; Vesth, T.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kildgaard, S.; Petersen, T.I.; Kuo, A.; Sato, A.; Lyhne, E.K.; et al. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Ferrara, M.; Medina, A.; Pascale, M.; Magan, N. Toxigenic Fungi and Mycotoxins in a Climate Change Scenario: Ecology, Genomics, Distribution, Prediction and Prevention of the Risk. Microorganisms 2020, 8, 1496. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Pascale, M.; Logrieco, A.F. Mycotoxin risks under a climate change scenario in Europe. Trends Food Sci. Technol. 2019, 84, 38–40. [Google Scholar] [CrossRef]

| Metabolites | Chemical Formula | Exact Mass (Da) | Adduct (M+H+) | MS/MS Fragments (Da) | Retention Time (min) |
|---|---|---|---|---|---|
| AFB1 | C17H12O6 | 312.06339 | 313.07066 | 298.0483, 285.0764, 241.0504, 214.0632 | 4.88 |
| AFG1 | C17H12O7 | 328.0583 | 329.06558 | 311.0562, 296.0342, 283.0612, 243.0661 | 4.58 |
| Kojic acid | C6H6O4 | 142.02661 | 143.03389 | 125.0227, 97.0281, 69.0334 | 1.10 |
| Aspergillic acid | C12H20N2O2 | 224.15248 | 225.15975 | 207.1507, 165.1024, 139.0491 | 4.35 |
| Neoaspergillic acid | C12H20N2O2 | 224.15248 | 225.15975 | 207.1507, 165.1024, 123.0548 | 4.25 |
| Hydroxyaspergillic acid | C12H20N2O3 | 240.14739 | 241.15467 | 312.1580, 181.1324, 153.0650, 100.0757 | 4.15 |
| Neohydroxyaspergillic acid | C12H20N2O3 | 240.14739 | 241.15467 | 312.1580, 181.1324, 153.0650, 100.0757 | 3.99 |
| Leporin B | C22H25NO3 | 351.18344 | 352.19072 | 278.1151, 216.0679,199.0608, 171.0654 | 8.08 |
| Leporin C | C22H25NO2 | 335.18853 | 336.19581 | 254.1179, 214.0865, 200.0709, 188.0712 | 7.78 |
| Flavacol | C12H20N2O | 208.15756 | 209.16484 | 191.1529, 167.1185, 137.0704, 109.0743 | 5.40 |
| O-methylsterigmatocystin | C19H14O6 | 338.07904 | 339.08631 | 324.0634, 306.0534, 295.0609, 295.0609 | 5.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdallah, M.F.; Audenaert, K.; Saeger, S.D.; Houbraken, J. Revisiting an Aspergillus flavus Strain Isolated from an Egyptian Sugarcane Field in 1930. Microorganisms 2020, 8, 1633. https://doi.org/10.3390/microorganisms8111633
Abdallah MF, Audenaert K, Saeger SD, Houbraken J. Revisiting an Aspergillus flavus Strain Isolated from an Egyptian Sugarcane Field in 1930. Microorganisms. 2020; 8(11):1633. https://doi.org/10.3390/microorganisms8111633
Chicago/Turabian StyleAbdallah, Mohamed F., Kris Audenaert, Sarah De Saeger, and Jos Houbraken. 2020. "Revisiting an Aspergillus flavus Strain Isolated from an Egyptian Sugarcane Field in 1930" Microorganisms 8, no. 11: 1633. https://doi.org/10.3390/microorganisms8111633
APA StyleAbdallah, M. F., Audenaert, K., Saeger, S. D., & Houbraken, J. (2020). Revisiting an Aspergillus flavus Strain Isolated from an Egyptian Sugarcane Field in 1930. Microorganisms, 8(11), 1633. https://doi.org/10.3390/microorganisms8111633








