Biofilm Produced In Vitro by Piscirickettsia salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
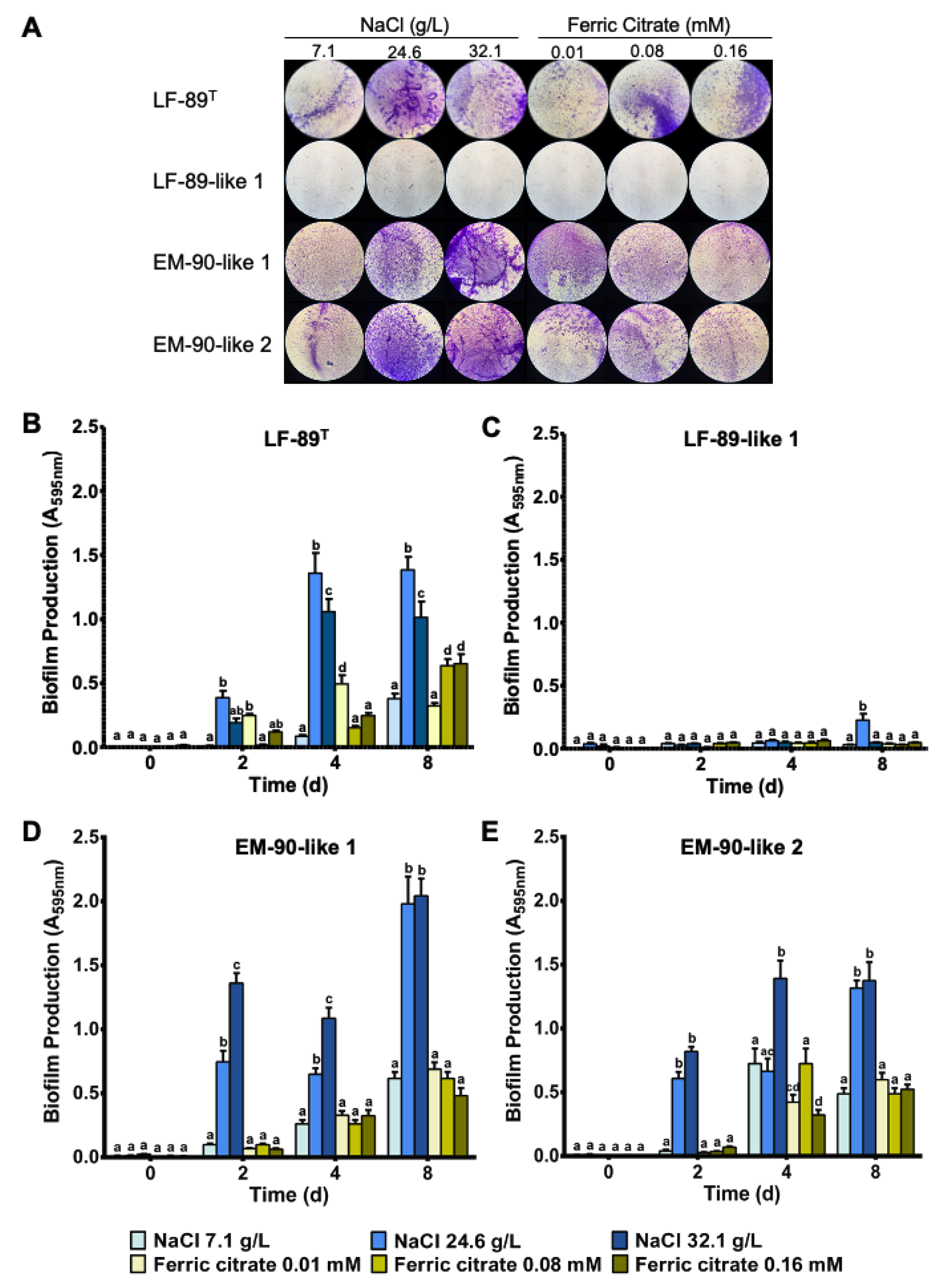
2.2. Quantitative Assessment of P. salmonis Biofilm Formation in Restrictive Liquid Media
2.3. Planktonic and Sessile P. salmonis Infection in SHK-1 Cells
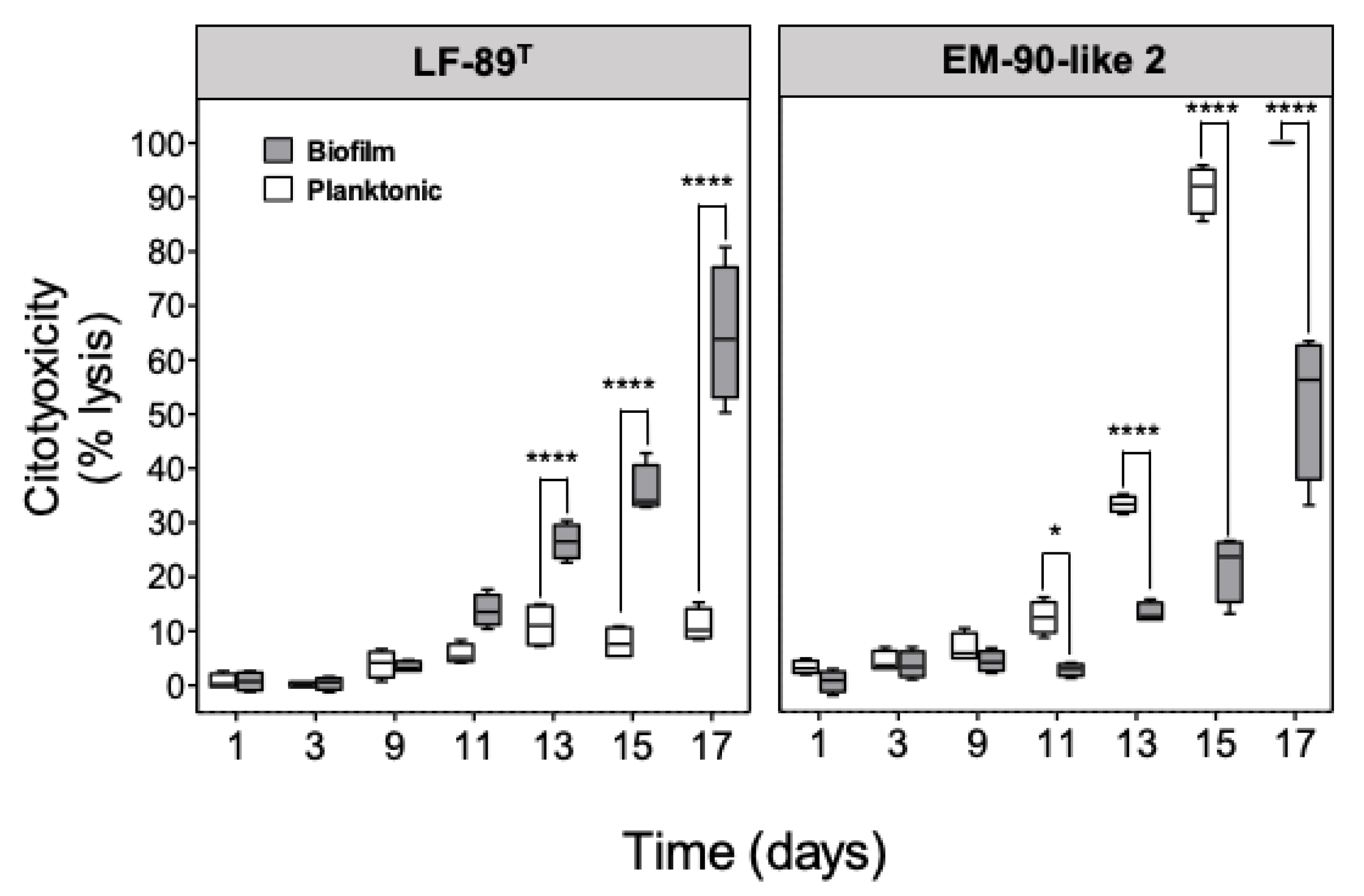
2.4. Determination of Cytotoxicity by Lactate Dehydrogenase Assay
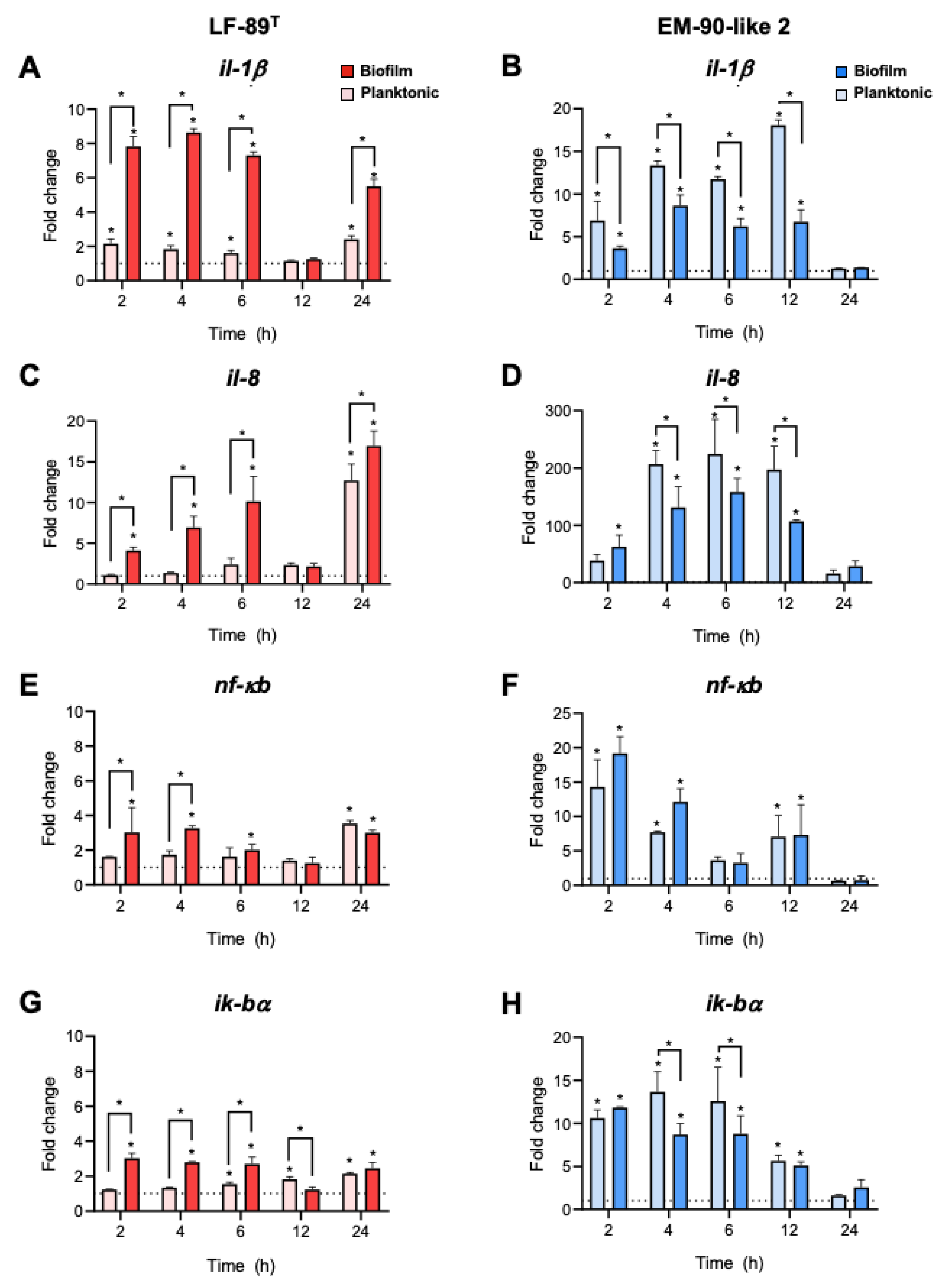
2.5. RNA Purification and Gene Expression Analysis
2.6. Statistical Analyses
3. Results
3.1. P. salmonis Biofilm Formation
3.2. Planktonic and Sessile P. salmonis Infection in SHK-1 Cells
3.3. Immune Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Figueroa, J.; Cárcamo, J.; Yañez, A.; Olavarria, V.; Ruiz, P.; Manríquez, R.; Muñoz, C.; Romero, A.; Avendaño-Herrera, R. Addressing viral and bacterial threats to salmon farming in Chile: Historical contexts and perspectives for management and control. Rev. Aquac. 2019, 11, 299–324. [Google Scholar] [CrossRef]
- Bravo, C.; Martinez, V. Whole-genome comparative analysis of the pathogen Piscirickettsia salmonis. Vet. Microbiol. 2016, 196, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Nourdin-Galindo, G.; Sanchez, P.; Molina, C.F.; Espinoza-Rojas, D.A.; Oliver, C.; Ruiz, P.; Vargas-Chacoff, L.; Carcamo, J.G.; Figueroa, J.E.; Mancilla, M.; et al. Comparative Pan-Genome Analysis of Piscirickettsia salmonis Reveals Genomic Divergences within Genogroups. Front. Cell Infect. Microbiol. 2017, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Cortés, M.; Sánchez, P.; Ruiz, P.; Haro, R.; Sáez, J.; Sánchez, F.; Hernández, M.; Oliver, C.; Yáñez, A.J. In vitro expression of Sec-dependent pathway and type 4B secretion system in Piscirickettsia salmonis. Microb. Pathog. 2017, 110, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Gomez, F.A.; Tobar, J.A.; Henriquez, V.; Sola, M.; Altamirano, C.; Marshall, S.H. Evidence of the Presence of a Functional Dot/Icm Type IV-B Secretion System in the Fish Bacterial Pathogen Piscirickettsia salmonis. PLoS ONE 2013, 8, e54934. [Google Scholar] [CrossRef]
- Mancilla, M.; Saavedra, J.; Grandon, M.; Tapia, E.; Navas, E.; Grothusen, H.; Bustos, P. The mutagenesis of a type IV secretion system locus of Piscirickettsia salmonis leads to the attenuation of the pathogen in Atlantic salmon, Salmo salar. J. Fish Dis. 2018, 41, 625–634. [Google Scholar] [CrossRef]
- Oliver, C.; Valenzuela, K.; Hernandez, M.; Sandoval, R.; Haro, R.E.; Avendano-Herrera, R.; Carcamo, J.G.; Villar, M.T.; Artigues, A.; Garduno, R.; et al. Characterization and pathogenic role of outer membrane vesicles produced by the fish pathogen Piscirickettsia salmonis under in vitro conditions. Vet. Microbiol. 2016, 184, 94–101. [Google Scholar] [CrossRef]
- Oliver, C.; Hernandez, M.A.; Tandberg, J.I.; Valenzuela, K.N.; Lagos, L.X.; Haro, R.E.; Sanchez, P.; Ruiz, P.A.; Sanhueza-Oyarzun, C.; Cortes, M.A.; et al. The Proteome of Biologically Active Membrane Vesicles from Piscirickettsia salmonis LF-89 Type Strain Identifies Plasmid-Encoded Putative Toxins. Front. Cell Infect. Microbiol. 2017, 7, 420. [Google Scholar] [CrossRef]
- Chen, T.; Dong, G.; Zhang, S.; Zhang, X.; Zhao, Y.; Cao, J.; Zhou, T.; Wu, Q. Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess. BMC Microbiol. 2020, 20, 36. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Hostacka, A.; Ciznar, I.; Stefkovicova, M. Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. (Prague) 2010, 55, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Epstein, A.K.; Pokroy, B.; Seminara, A.; Aizenberg, J. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc. Natl. Acad. Sci. USA 2011, 108, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Tote, K.; Horemans, T.; Vanden Berghe, D.; Maes, L.; Cos, P. Inhibitory effect of biocides on the viable masses and matrices of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2010, 76, 3135–3142. [Google Scholar] [CrossRef]
- Isiaku, A.I.; Sabri, M.Y.; Ina-Salwany, M.Y.; Hassan, M.D.; Tanko, P.N.; Bello, M.B. Biofilm is associated with chronic streptococcal meningoencephalitis in fish. Microb. Pathog. 2017, 102, 59–68. [Google Scholar] [CrossRef]
- Thuptimdang, P.; Limpiyakorn, T.; McEvoy, J.; Pruss, B.M.; Khan, E. Effect of silver nanoparticles on Pseudomonas putida biofilms at different stages of maturity. J. Hazard. Mater. 2015, 290, 127–133. [Google Scholar] [CrossRef]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Nguyen, U.T.; Burrows, L.L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 2014, 187, 26–32. [Google Scholar] [CrossRef]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef]
- Marshall, S.H.; Gomez, F.A.; Ramirez, R.; Nilo, L.; Henriquez, V. Biofilm generation by Piscirickettsia salmonis under growth stress conditions: A putative in vivo survival/persistence strategy in marine environments. Res. Microbiol. 2012, 163, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Contreras, A.; Magarinos, B.; Godoy, M.; Irgang, R.; Toranzo, A.E.; Avendano-Herrera, R. Surface properties of Streptococcus phocae strains isolated from diseased Atlantic salmon, Salmo salar L. J. Fish Dis. 2011, 34, 203–215. [Google Scholar] [CrossRef]
- Ruiz, P.; Poblete, M.; Yanez, A.J.; Irgang, R.; Toranzo, A.E.; Avendano-Herrera, R. Cell-surface properties of Vibrio ordalii strains isolated from Atlantic salmon Salmo salar in Chilean farms. Dis. Aquat. Organ. 2015, 113, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; De La Fuente, L.; Arias, C.R. Biofilm formation by the fish pathogen Flavobacterium columnare: Development and parameters affecting surface attachment. Appl. Environ. Microbiol. 2013, 79, 5633–5642. [Google Scholar] [CrossRef] [PubMed]
- Sundell, K.; Wiklund, T. Effect of biofilm formation on antimicrobial tolerance of Flavobacterium psychrophilum. J. Fish. Dis. 2011, 34, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Arias, C.R. Biofilm Formation on Aquaculture Substrates by Selected Bacterial Fish Pathogens. J. Aquat. Anim. Health 2017, 29, 95–104. [Google Scholar] [CrossRef]
- Coquet, L.; Cosette, P.; Junter, G.-A.; Beucher, E.; Saiter, J.-M.; Jouenne, T. Adhesion of Yersinia ruckeri to fish farm materials: Influence of cell and material surface properties. Colloid Surf. B 2002, 26, 373–378. [Google Scholar] [CrossRef]
- King, R. The Presence of Bacterial Pathogens in Recirculating Aquaculture System Biofilms and their Response to Various Sanitizers. Ph.D. Thesis, Faculty of the Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2001. [Google Scholar]
- Levipan, H.A.; Irgang, R.; Yáñez, A.; Avendaño-Herrera, R. Improved understanding of biofilm development by Piscirickettsia salmonis reveals potential risks for the persistence and dissemination of piscirickettsiosis. Sci. Rep. 2020, 10, 12224. [Google Scholar] [CrossRef]
- Karatas, S.; Mikalsen, J.; Steinum, T.M.; Taksdal, T.; Bordevik, M.; Colquhoun, D.J. Real time PCR detection of Piscirickettsia salmonis from formalin-fixed paraffin-embedded tissues. J. Fish Dis. 2008, 31, 747–753. [Google Scholar] [CrossRef]
- Vera, T.; Isla, A.; Cuevas, A.; Figueroa, J. A new liquid medium for the pathogen Piscirickettsia salmonis. Arch. Med. Vet. 2012, 44, 273–277. [Google Scholar] [CrossRef][Green Version]
- Contreras-Lynch, S.; Olmos, P.; Vargas, A.; Figueroa, J.; Gonzalez-Stegmaier, R.; Enriquez, R.; Romero, A. Identification and genetic characterization of Piscirickettsia salmonis in native fish from southern Chile. Dis. Aquat. Organ. 2015, 115, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Albornoz, R.; Valenzuela, K.; Pontigo, J.P.; Sanchez, P.; Ruiz, P.; Avendano-Herrera, R.; Romero, A.; Oliver, C.; Yanez, A. Identification of chemotaxis operon cheYZA and cheA gene expression under stressful conditions in Piscirickettsia salmonis. Microb. Pathog. 2017, 107, 436–441. [Google Scholar] [CrossRef] [PubMed]
- McBain, A.J.; Bartolo, R.G.; Catrenich, C.E.; Charbonneau, D.; Ledder, R.G.; Rickard, A.H.; Symmons, S.A.; Gilbert, P. Microbial Characterization of Biofilms in Domestic Drains and the Establishment of Stable Biofilm Microcosms. Appl. Environ. Microbiol. 2003, 69, 177–185. [Google Scholar] [CrossRef]
- Oliver, C.; Valenzuela, K.; Silva, H.; Haro, R.E.; Cortes, M.; Sandoval, R.; Pontigo, J.P.; Alvarez, C.; Figueroa, J.E.; Avendano-Herrera, R.; et al. Effectiveness of egg yolk immunoglobulin against the intracellular salmonid pathogen Piscirickettsia salmonis. J. Appl. Microbiol. 2015, 119, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Manríquez, R.; Alvarez, C.; Gajardo, C.; Vásquez, J.; Kausel, G.; Monrás, M.; Olavarría, V.H.; Yáñez, A.; Enríquez, R.; et al. Prolactin-releasing peptide is a potent mediator of the innate immune response in leukocytes from Salmo salar. Vet. Immunol. Immunopathol. 2012, 147, 170–179. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Makrinos, D.L.; Bowden, T.J. Growth characteristics of the intracellular pathogen, Piscirickettsia salmonis, in tissue culture and cell-free media. J. Fish Dis. 2017, 40, 1115–1127. [Google Scholar] [CrossRef]
- Yanez, A.J.; Valenzuela, K.; Silva, H.; Retamales, J.; Romero, A.; Enriquez, R.; Figueroa, J.; Claude, A.; Gonzalez, J.; Avendano-Herrera, R.; et al. Broth medium for the successful culture of the fish pathogen Piscirickettsia salmonis. Dis. Aquat. Organ. 2012, 97, 197–205. [Google Scholar] [CrossRef]
- Calquin, P.; Ruiz, P.; Oliver, C.; Sanchez, P.; Haro, R.; Oliva, H.; Vargas-Chacoff, L.; Avendano-Herrera, R.; Yanez, A.J. Physiological evidence that Piscirickettsia salmonis produces siderophores and uses iron from different sources. J. Fish Dis. 2018, 41, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Bury, N.; Grosell, M. Iron acquisition by teleost fish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2003, 135, 97–105. [Google Scholar] [CrossRef]
- Lannan, C.N.; Fryer, J.L. Extracellular Survival of Piscirickettsia salmonis. J. Fish Dis. 1994, 17, 545–548. [Google Scholar] [CrossRef]
- Olivares, J.; Marshall, S.H. Determination of minimal concentration of Piscirickettsia salmonis in water columns to establish a fallowing period in salmon farms. J. Fish Dis. 2010, 33, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G. Microbial Colonization in Marine Environments: Overview of Current Knowledge and Emerging Research Topics. J. Mar. Sci. Eng. 2020, 8, 78. [Google Scholar] [CrossRef]
- Chiu, J.M.Y.; Thiyagarajan, V.; Tsoi, M.M.Y.; Qian, P.Y. Qualitative and quantitative changes in marine biofilms as a function of temperature and salinity in summer and winter. Biofilms 2005, 2, 183–195. [Google Scholar] [CrossRef]
- Gu, L.; Chen, Q.; Guo, A.; Liu, W.; Ruan, Y.; Zhang, X.; Nou, X. Differential Effects of Growth Medium Salinity on Biofilm Formation of Two Salmonella enterica Strains. J. Food Prot. 2020, 83, 196–203. [Google Scholar] [CrossRef]
- Marsden, A.E.; Grudzinski, K.; Ondrey, J.M.; DeLoney-Marino, C.R.; Visick, K.L. Impact of Salt and Nutrient Content on Biofilm Formation by Vibrio fischeri. PLoS ONE 2017, 12, e0169521. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Ashrafudoulla, M.; Sadekuzzaman, M.; Kang, I.; Ha, S.-D. Effects of NaCl, glucose, and their combinations on biofilm formation on black tiger shrimp (Penaeus monodon) surfaces by Vibrio parahaemolyticus. Food Control. 2018, 89, 203–209. [Google Scholar] [CrossRef]
- Smith, P.A.; Diaz, F.E.; Rojas, M.E.; Diaz, S.; Galleguillos, M.; Carbonero, A. Effect of Piscirickettsia salmonis inoculation on the ASK continuous cell line. J. Fish Dis. 2015, 38, 321–324. [Google Scholar] [CrossRef]
- Alvarez, C.A.; Gomez, F.A.; Mercado, L.; Ramirez, R.; Marshall, S.H. Piscirickettsia salmonis Imbalances the Innate Immune Response to Succeed in a Productive Infection in a Salmonid Cell Line Model. PLoS ONE 2016, 11, e0163943. [Google Scholar] [CrossRef] [PubMed]
- Dannevig, B.H.; Brudeseth, B.E.; Gjøen, T.; Rode, M.; Wergeland, H.I.; Evensen, Ø.; Press, C.M. Characterisation of a long-term cell line (SHK-1) developed from the head kidney of Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 1997, 7, 213–226. [Google Scholar] [CrossRef]
- Dannevig, B.H.; Falk, K.; Namork, E. Isolation of the causal virus of infectious salmon anaemia (ISA) in a long-term cell line from Atlantic salmon head kidney. J. Gen. Virol. 1995, 76, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Levicán-Asenjo, J.; Soto-Rifo, R.; Aguayo, F.; Gaggero, A.; Leon, O. Salmon cells SHK-1 internalize infectious pancreatic necrosis virus by macropinocytosis. J. Fish Dis. 2019, 42, 1035–1046. [Google Scholar] [CrossRef]
- Olavarría, V.H.; Gallardo, L.; Figueroa, J.E.; Mulero, V. Lipopolysaccharide primes the respiratory burst of Atlantic salmon SHK-1 cells through protein kinase C-mediated phosphorylation of p47phox. Dev. Comp. Immunol. 2010, 34, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.J.; Romero, A.; Gonzalez-Stegmaier, R.; Dantagnan, P. The effects of supplemented diets with a phytopharmaceutical preparation from herbal and macroalgal origin on disease resistance in rainbow trout against Piscirickettsia salmonis. Aquaculture 2016, 454, 109–117. [Google Scholar] [CrossRef]
- Oliver, C.; Sánchez, P.; Valenzuela, K.; Hernández, M.; Pontigo, J.P.; Rauch, M.C.; Garduño, R.A.; Avendaño-Herrera, R.; Yáñez, A.J. Subcellular Location of Piscirickettsia salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis. Microorganisms 2020, 8, 117. [Google Scholar] [CrossRef]
- Zuniga, A.; Aravena, P.; Pulgar, R.; Travisany, D.; Ortiz-Severin, J.; Chavez, F.P.; Maass, A.; Gonzalez, M.; Cambiazo, V. Transcriptomic Changes of Piscirickettsia salmonis During Intracellular Growth in a Salmon Macrophage-Like Cell Line. Front. Cell Infect. Microbiol. 2019, 9, 426. [Google Scholar] [CrossRef]
- Abu Khweek, A.; Fernández Dávila, N.; Caution, K.; Akhter, A.; Abdulrahman, B.; Tazi, M.; Hassan, H.; Novotny, L.; Bakaletz, L.; Amer, A. Biofilm-derived Legionella pneumophila evades the innate immune response in macrophages. Front. Cell Infect. Microbiol. 2013, 3, 18. [Google Scholar] [CrossRef]
- McCarthy, U.M.; Bron, J.E.; Brown, L.; Pourahmad, F.; Bricknell, I.R.; Thompson, K.D.; Adams, A.; Ellis, A.E. Survival and replication of Piscirickettsia salmonis in rainbow trout head kidney macrophages. Fish Shellfish Immunol. 2008, 25, 477–484. [Google Scholar] [CrossRef]
- Perez-Stuardo, D.; Morales-Reyes, J.; Tapia, S.; Ahumada, D.E.; Espinoza, A.; Soto-Herrera, V.; Brianson, B.; Ibaceta, V.; Sandino, A.M.; Spencer, E.; et al. Non-lysosomal Activation in Macrophages of Atlantic Salmon (Salmo salar) After Infection With Piscirickettsia salmonis. Front. Immunol. 2019, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Baldassarri, L.; Bertuccini, L.; Creti, R.; Filippini, P.; Ammendolia, M.G.; Koch, S.; Huebner, J.; Orefici, G. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J. Infect. Dis. 2005, 191, 1253–1262. [Google Scholar] [CrossRef]
- Daw, K.; Baghdayan, A.S.; Awasthi, S.; Shankar, N. Biofilm and planktonic Enterococcus faecalis elicit different responses from host phagocytes in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Pena, A.; Maldonado, L. Transcriptomic profiles of post-smolt Atlantic salmon challenged with Piscirickettsia salmonis reveal a strategy to evade the adaptive immune response and modify cell-autonomous immunity. Dev. Comp. Immunol. 2018, 81, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Van der Aa, L.M.; Chadzinska, M.; Tijhaar, E.; Boudinot, P.; Verburg-van Kemenade, B.M.L. CXCL8 chemokines in teleost fish: Two lineages with distinct expression profiles during early phases of inflammation. PLoS ONE 2010, 5, e12384. [Google Scholar] [CrossRef]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef]
- Stevens, N.T.; Sadovskaya, I.; Jabbouri, S.; Sattar, T.; O’Gara, J.P.; Humphreys, H.; Greene, C.M. Staphylococcus epidermidis polysaccharide intercellular adhesin induces IL-8 expression in human astrocytes via a mechanism involving TLR2. Cell. Microbiol. 2009, 11, 421–432. [Google Scholar] [CrossRef]
- Watters, C.; Fleming, D.; Bishop, D.; Rumbaugh, K.P. Host Responses to Biofilm. Prog. Mol. Biol. Transl. Sci. 2016, 142, 193–239. [Google Scholar]
- Mittal, R.; Sharma, S.; Chhibber, S.; Harjai, K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp. Immunol. Microbiol. Infect. Dis. 2006, 29, 12–26. [Google Scholar] [CrossRef]
- Arima, S.; Ochi, H.; Mitsuhashi, M.; Kibe, R.; Takahashi, K.; Kataoka, Y. Staphylococcus pseudintermedius biofilms secrete factors that induce inflammatory reactions in vitro. Lett. Appl. Microbiol. 2018, 67, 214–219. [Google Scholar] [CrossRef]
- Rathore, S.S.; Cheepurupalli, L.; Gangwar, J.; Raman, T.; Ramakrishnan, J. Biofilm of Klebsiella pneumoniae minimize phagocytosis and cytokine expression by macrophage cell line. bioRxiv 2019, 623546. [Google Scholar] [CrossRef]
- Guilhen, C.; Miquel, S.; Charbonnel, N.; Joseph, L.; Carrier, G.; Forestier, C.; Balestrino, D. Colonization and immune modulation properties of Klebsiella pneumoniae biofilm-dispersed cells. NPJ Biofilms Microbiomes 2019, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Alboslemy, T.; Yu, B.; Rogers, T.; Kim, M.H. Staphylococcus aureus Biofilm-Conditioned Medium Impairs Macrophage-Mediated Antibiofilm Immune Response by Upregulating KLF2 Expression. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Schommer, N.N.; Christner, M.; Hentschke, M.; Ruckdeschel, K.; Aepfelbacher, M.; Rohde, H. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect. Immun. 2011, 79, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Rise, M.L.; Jones, S.R.M.; Brown, G.D.; von Schalburg, K.R.; Davidson, W.S.; Koop, B.F. Microarray analyses identify molecular biomarkers of Atlantic salmon macrophage and hematopoietic kidney response to Piscirickettsia salmonis infection. Physiol. Genom. 2004, 20, 21–35. [Google Scholar] [CrossRef]
- Rozas-Serri, M.; Peña, A.; Arriagada, G.; Enríquez, R.; Maldonado, L. Comparison of gene expression in post-smolt Atlantic salmon challenged by LF-89-like and EM-90-like Piscirickettsia salmonis isolates reveals differences in the immune response associated with pathogenicity. J. Fish Dis. 2018, 41, 539–552. [Google Scholar] [CrossRef]
- Bottero, V.; Imbert, V.; Frelin, C.; Formento, J.L.; Peyron, J.F. Monitoring NF-kappa B transactivation potential via real-time PCR quantification of I kappa B-alpha gene expression. Mol. Diagn. 2003, 7, 187–194. [Google Scholar]
- Ferreiro, D.U.; Komives, E.A. Molecular mechanisms of system control of NF-kappaB signaling by IkappaBalpha. Biochemistry 2010, 49, 1560–1567. [Google Scholar] [CrossRef]
- Parrino, B.; Carbone, D.; Cirrincione, G.; Diana, P.; Cascioferro, S. Inhibitors of antibiotic resistance mechanisms: Clinical applications and future perspectives. Future Med. Chem. 2020, 12, 357–359. [Google Scholar] [CrossRef]
- Azad, I.S.; Shankar, K.M.; Mohan, C.V.; Kalita, B. Uptake and processing of biofilm and free-cell vaccines of Aeromonas hydrophila in Indian major carps and common carp following oral vaccination-antigen localization by a monoclonal antibody. Dis. Aquat. Organ. 2000, 43, 103–108. [Google Scholar] [CrossRef]
- Kahieshesfandiari, M.; Sabri, M.Y.; Ina-salwany, M.Y.; Hassan, M.D.; Noraini, O.; Ajadi, A.A.; Isiaku, A.I. Streptococcosis in Oreochromis sp.: Is feed-based biofilm vaccine of Streptococcus agalactiae effective? Aquacult. Int. 2019, 27, 817–832. [Google Scholar] [CrossRef]
| Gene | Sequence (5′- > 3′) | GenBank Accession No. |
|---|---|---|
| ef-1α | F-CCCCTCCAGGACGTTTACAAA R-CACACGGCCCACAGGTACA | NM_001123629.1 |
| il-1β | F-CAAGCTGCCTCAGGGTCT R-CGGCACCCTTTAACCTCTCC | NM_001123582.1 |
| il-8 | F-GCAACAGCGGTCAGGAGATT R-TGGAATGATTCCCCTTCTTCA | HM162835.1 |
| nf-κb | F-ACCTGGCCATCATTCACCAG R-TGGTTGAGCTTGTCGAGGAC | NM_001173583.1 |
| Iκb-α | F-GGAGAGTGAGGAGGAGTGCAT R-CTGCTTCAATTCTGCCCAAATGTAA | XM_014204687.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santibañez, N.; Vega, M.; Pérez, T.; Yáñez, A.; González-Stegmaier, R.; Figueroa, J.; Enríquez, R.; Oliver, C.; Romero, A. Biofilm Produced In Vitro by Piscirickettsia salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1. Microorganisms 2020, 8, 1609. https://doi.org/10.3390/microorganisms8101609
Santibañez N, Vega M, Pérez T, Yáñez A, González-Stegmaier R, Figueroa J, Enríquez R, Oliver C, Romero A. Biofilm Produced In Vitro by Piscirickettsia salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1. Microorganisms. 2020; 8(10):1609. https://doi.org/10.3390/microorganisms8101609
Chicago/Turabian StyleSantibañez, Natacha, Matías Vega, Tatiana Pérez, Alejandro Yáñez, Roxana González-Stegmaier, Jaime Figueroa, Ricardo Enríquez, Cristian Oliver, and Alex Romero. 2020. "Biofilm Produced In Vitro by Piscirickettsia salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1" Microorganisms 8, no. 10: 1609. https://doi.org/10.3390/microorganisms8101609
APA StyleSantibañez, N., Vega, M., Pérez, T., Yáñez, A., González-Stegmaier, R., Figueroa, J., Enríquez, R., Oliver, C., & Romero, A. (2020). Biofilm Produced In Vitro by Piscirickettsia salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1. Microorganisms, 8(10), 1609. https://doi.org/10.3390/microorganisms8101609






