Impact of Yeast-Derived β-Glucans on the Porcine Gut Microbiota and Immune System in Early Life
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Study Design
2.3. Experimental Procedures
2.3.1. Dietary Intervention and Oral Vaccination
2.3.2. Blood and Faecal Sampling
2.3.3. Dissection
2.4. Measurements
2.4.1. Microbiota Analysis
2.4.2. Serology
2.4.3. Cell Isolation
2.4.4. Stimulation Assay
2.4.5. Flow Cytometry
2.5. Statistical Analysis
2.5.1. Microbiota Analysis
2.5.2. Immunological Analysis
2.5.3. Correlation Analysis
3. Results
3.1. Microbial Colonization
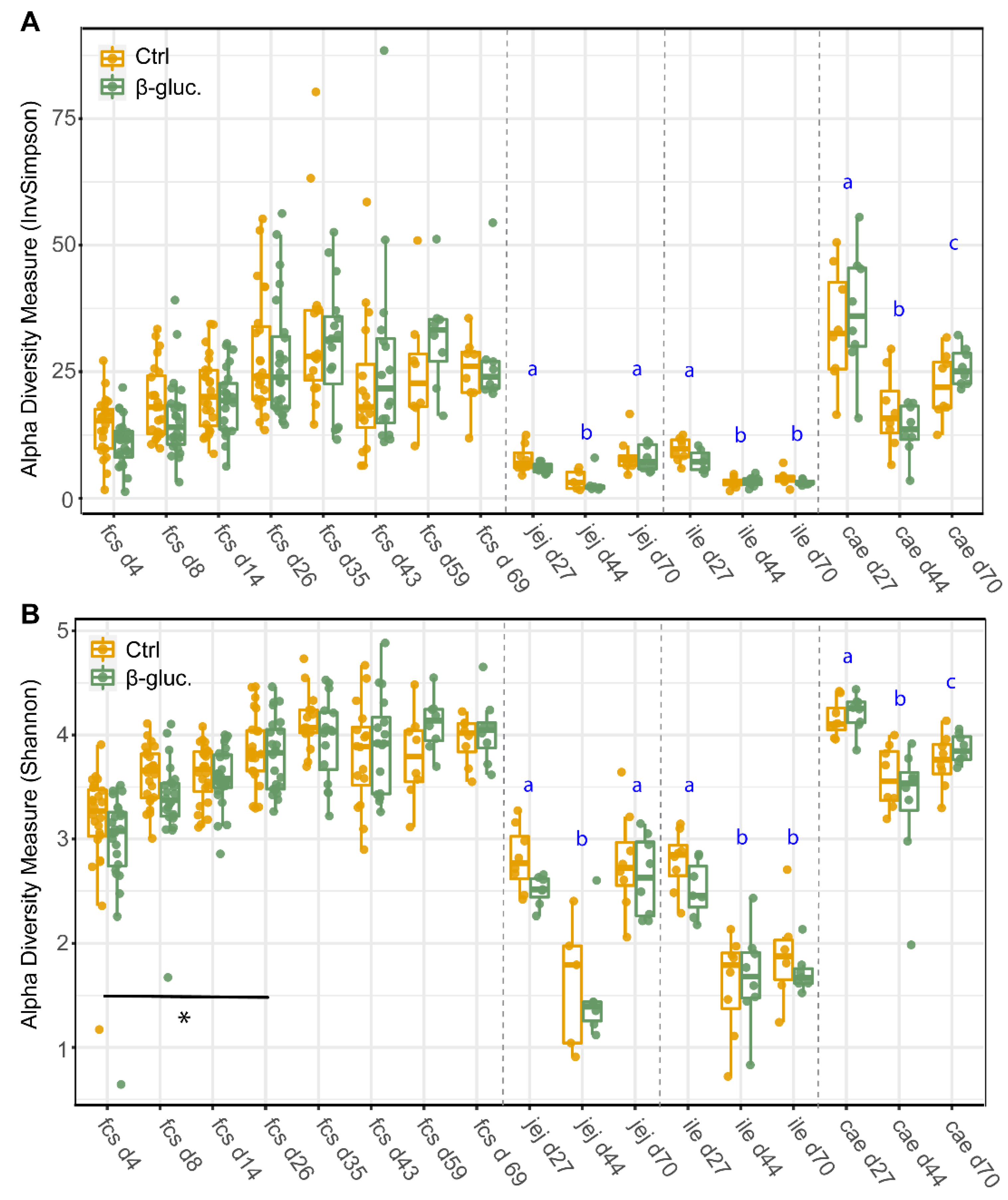
3.1.1. Alpha Diversity
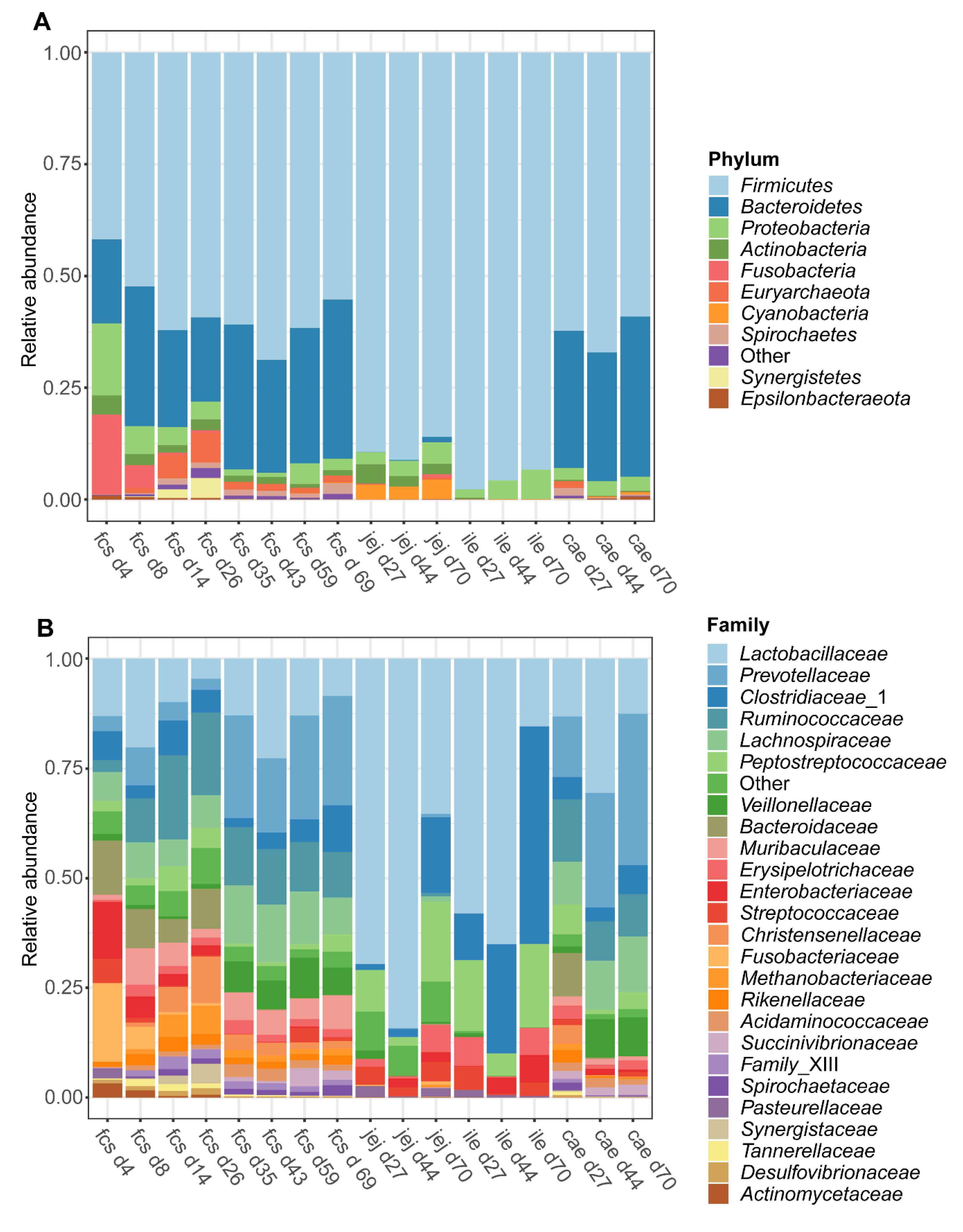
3.1.2. Microbiota Composition over Time
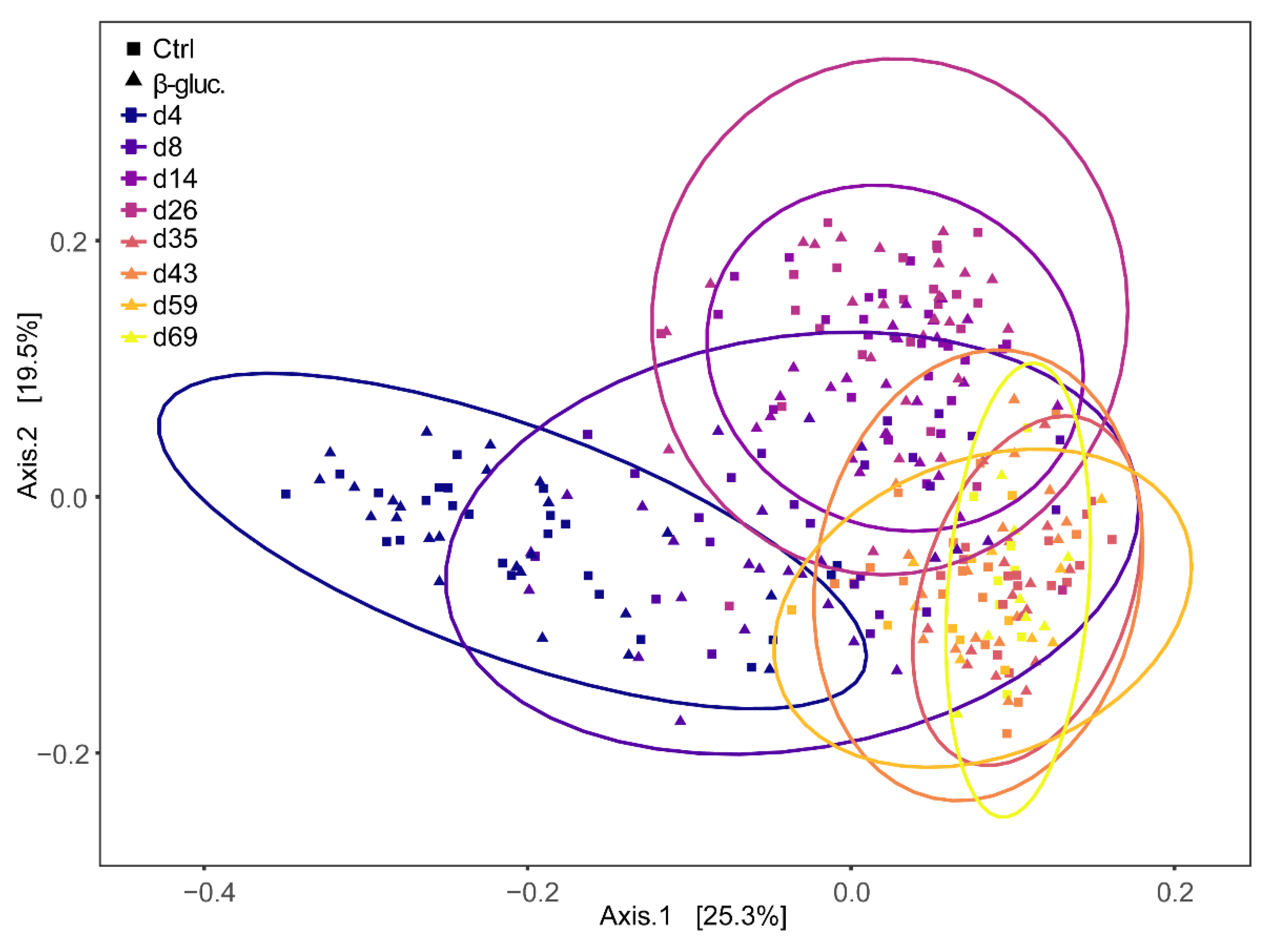
3.1.3. Beta Diversity
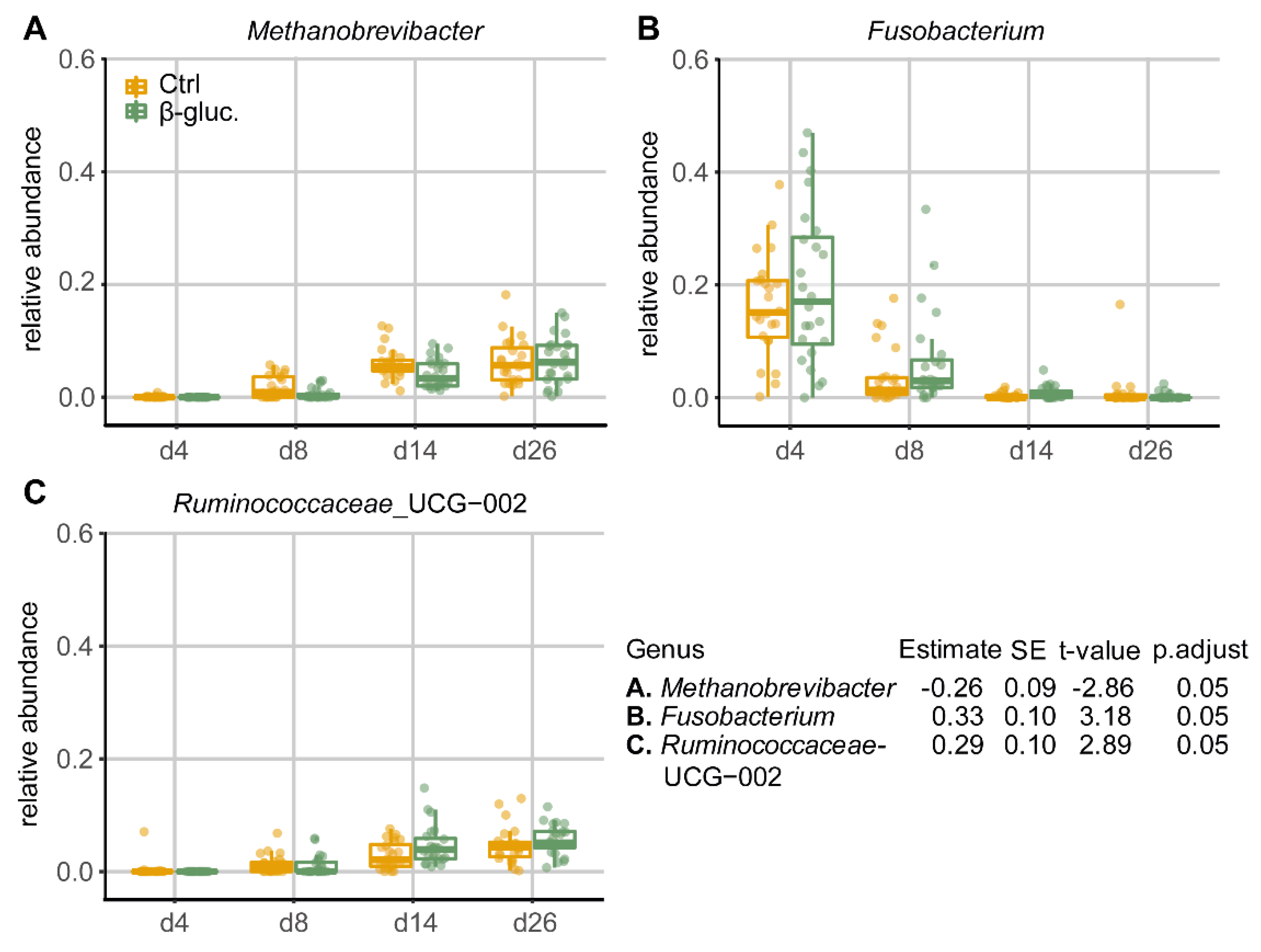
3.1.4. Differentially Abundant Genera between Treatment Groups
3.2. Immunological Analysis
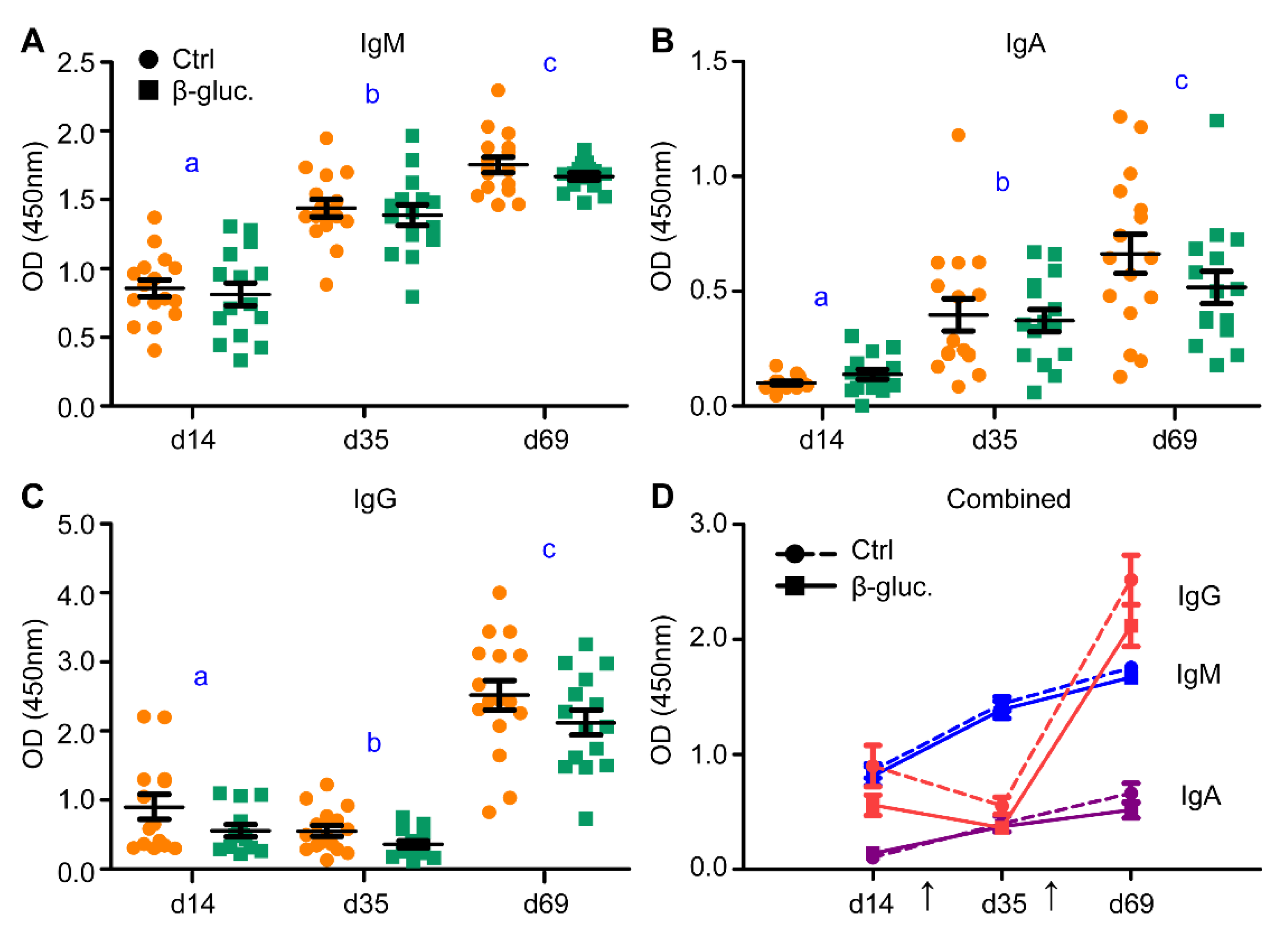
3.2.1. Immunological Response to Oral Vaccination
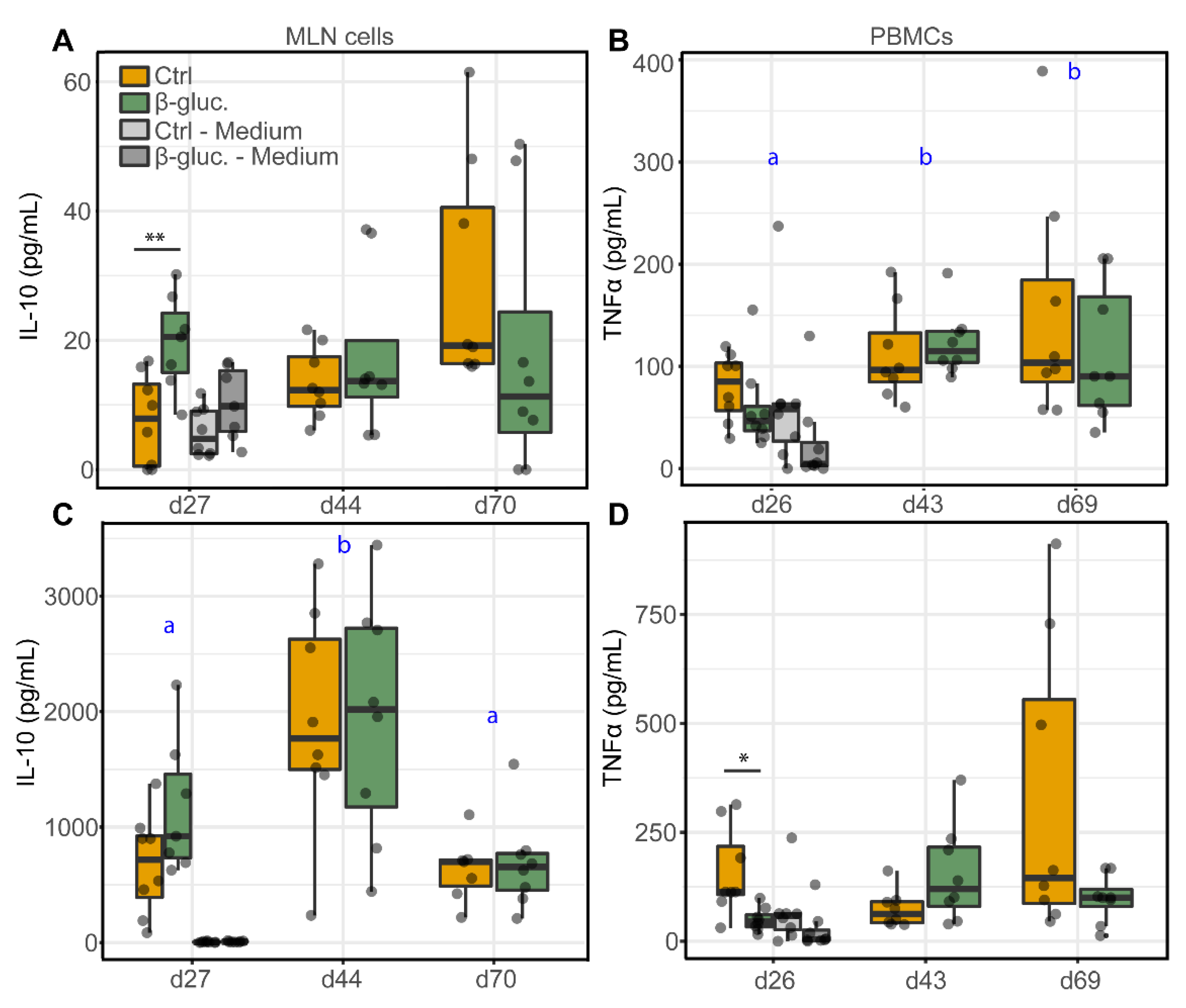
3.2.2. Ex Vivo Stimulation of MLN Cells and PBMCs
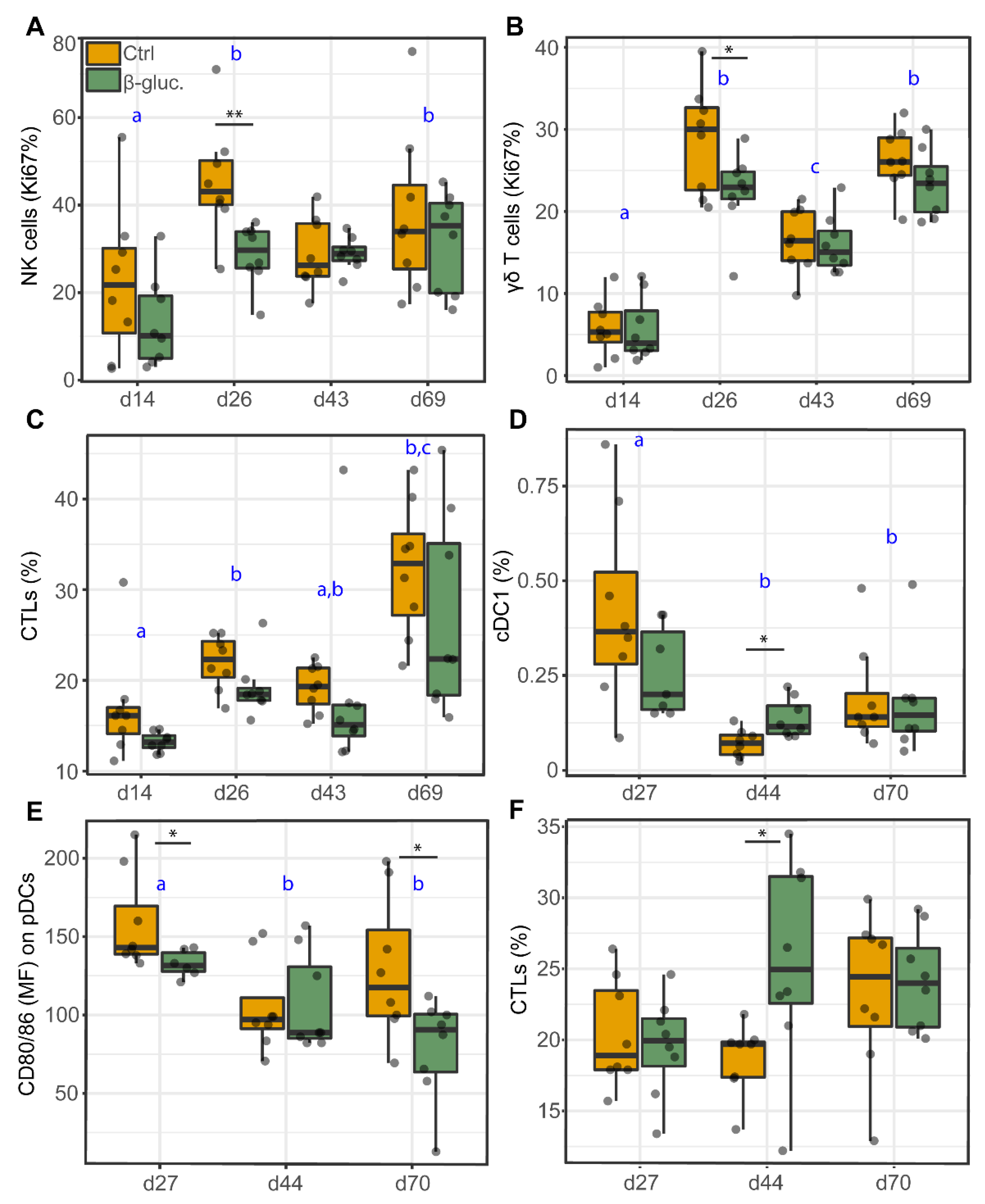
3.2.3. Analysis of Immune Cells in Blood and the MLN
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pluske, J.R.; Turpin, D.L.; Kim, J.-C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2018, 4, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Hornef, M.W.; Torow, N. ‘Layered immunity’ and the ‘neonatal window of opportunity’—Timed succession of non-redundant phases to establish mucosal host–microbial homeostasis after birth. Immunology 2020, 159, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, J.; Ren, E.; Su, Y.; Zhu, W. Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 2018, 49, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Everaert, N.; Van Cruchten, S.; Weström, B.; Bailey, M.; Van Ginneken, C.; Thymann, T.; Pieper, R. A review on early gut maturation and colonization in pigs, including biological and dietary factors affecting gut homeostasis. Anim. Feed Sci. Technol. 2017, 233, 89–103. [Google Scholar] [CrossRef]
- Schmidt, B.; Mulder, I.E.; Musk, C.C.; Aminov, R.I.; Lewis, M.; Stokes, C.R.; Bailey, M.; Prosser, J.I.; Gill, B.P.; Pluske, J.R. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS ONE 2011, 6, e28284. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar]
- Schokker, D.; Zhang, J.; Zhang, L.-L.; Vastenhouw, S.A.; Heilig, H.G.; Smidt, H.; Rebel, J.M.; Smits, M.A. Early-life environmental variation affects intestinal microbiota and immune development in new-born piglets. PLoS ONE 2014, 9, e100040. [Google Scholar]
- Von Mutius, E.; Vercelli, D. Farm living: Effects on childhood asthma and allergy. Nat. Rev. Immunol. 2010, 10, 861–868. [Google Scholar]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef]
- Schokker, D.; Zhang, J.; Vastenhouw, S.A.; Heilig, H.G.; Smidt, H.; Rebel, J.M.; Smits, M.A. Long-lasting effects of early-life antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PLoS ONE 2015, 10, e0116523. [Google Scholar]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [PubMed]
- Goodridge, H.S.; Wolf, A.J.; Underhill, D.M. β-glucan recognition by the innate immune system. Immunol. Rev. 2009, 230, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D. Dectin-1 exerts dual control in the gut. Cell Host Microbe 2015, 18, 139–141. [Google Scholar] [CrossRef]
- Thompson, I.J.; Oyston, P.C.; Williamson, D.E. Potential of the β-glucans to enhance innate resistance to biological agents. Expert Rev. Anti-Infect. Ther. 2010, 8, 339–352. [Google Scholar] [PubMed]
- Volman, J.J.; Ramakers, J.D.; Plat, J. Dietary modulation of immune function by β-glucans. Physiol. Behav. 2008, 94, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Stier, H.; Ebbeskotte, V.; Gruenwald, J. Immune-modulatory effects of dietary Yeast Beta-1, 3/1, 6-D-glucan. Nutr. J. 2014, 13, 38. [Google Scholar] [PubMed]
- Lam, K.-L.; Cheung, P.C.-K. Non-digestible long chain beta-glucans as novel prebiotics. Bioact. Carbohydr. Diet. Fibre 2013, 2, 45–64. [Google Scholar] [CrossRef]
- Atanasov, J.; Schloermann, W.; Trautvetter, U.; Glei, M. The effects of β-glucans on intestinal health. Ernahr. Umsch. 2020, 67, M140–M147. [Google Scholar]
- Elghandour, M.; Tan, Z.; Abu Hafsa, S.; Adegbeye, M.; Greiner, R.; Ugbogu, E.; Cedillo Monroy, J.; Salem, A. Saccharomyces cerevisiae as a probiotic feed additive to non and pseudo-ruminant feeding: A review. J. Appl. Microbiol. 2020, 128, 658–674. [Google Scholar]
- Berto, P.N.; Tse, M.L.; Ramos, D.R.; Saleh, M.A.; Miassi, G.M.; Yamatogi, R.S.; Berto, D.A.; Neto, T.; Messias, A. Dietary supplementation with hydrolyzed yeast and its effect on the performance, intestinal microbiota, and immune response of weaned piglets. An. Acad. Bras. Ciênc. 2020, 92. [Google Scholar] [CrossRef]
- Arfken, A.M.; Frey, J.F.; Summers, K.L. Temporal Dynamics of the Gut Bacteriome and Mycobiome in the Weanling Pig. Microorganisms 2020, 8, 868. [Google Scholar] [CrossRef] [PubMed]
- Geervliet, M.; Lute, L.C.; Jansen, C.A.; Rutten, V.P.; Savelkoul, H.F.; Tijhaar, E. Differential immunomodulation of porcine bone marrow derived dendritic cells by E. coli Nissle 1917 and β-glucans. PLoS ONE 2020, 15, e0233773. [Google Scholar] [CrossRef] [PubMed]
- Sonck, E.; Devriendt, B.; Goddeeris, B.; Cox, E. Varying effects of different β-glucans on the maturation of porcine monocyte-derived dendritic cells. Clin. Vaccine Immunol. 2011, 18, 1441–1446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sonck, E.; Stuyven, E.; Goddeeris, B.; Cox, E. The effect of β-glucans on porcine leukocytes. Vet. Immunol. Immunopathol. 2010, 135, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Sonck, E. Immunomodulation of Porcine Leukocytes and Dendritic Cells by β-(1, 3) Glucans. 2011. Available online: https://biblio.ugent.be/publication/1989184 (accessed on 12 August 2020).
- Ramiro-Garcia, J.; Hermes, G.D.; Giatsis, C.; Sipkema, D.; Zoetendal, E.G.; Schaap, P.J.; Smidt, H. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Poncheewin, W.; Hermes, G.D.; Van Dam, J.C.; Koehorst, J.J.; Smidt, H.; Schaap, P.J. NG-Tax 2.0: A semantic framework for high-throughput amplicon analysis. Front. Genet. 2019, 10, 1366. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar]
- Van Altena, S.; Peen, M.; Van der Linden, F.; Parmentier, H.; Savelkoul, H.; Tijhaar, E. Bovine natural antibodies in antibody-dependent bactericidal activity against Escherichia coli and Salmonella Typhimurium and risk of mastitis. Vet. Immunol. Immunopathol. 2016, 171, 21–27. [Google Scholar] [CrossRef][Green Version]
- Vreman, S.; Auray, G.; Savelkoul, H.F.; Rebel, A.; Summerfield, A.; Stockhofe-Zurwieden, N. Neonatal porcine blood derived dendritic cell subsets show activation after TLR2 or TLR9 stimulation. Dev. Comp. Immunol. 2018, 84, 361–370. [Google Scholar] [CrossRef]
- Gerner, W.; Käser, T.; Saalmüller, A. Porcine T lymphocytes and NK cells–an update. Dev. Comp. Immunol. 2009, 33, 310–320. [Google Scholar] [CrossRef]
- Auray, G.; Keller, I.; Python, S.; Gerber, M.; Bruggmann, R.; Ruggli, N.; Summerfield, A. Characterization and transcriptomic analysis of porcine blood conventional and plasmacytoid dendritic cells reveals striking species-specific differences. J. Immunol. 2016, 197, 1600672. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Team, R.C. Nlme: Linear and nonlinear Mixed Effects Models, R package version 3.1-121; R Foundation for Statistical Computing Vienna: Vienna, Austria, 2015. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Version 2.1.26. Available online: http://microbiome.github.com/microbiome (accessed on 5 June 2020).
- Ho, N.T.; Li, F.; Wang, S.; Kuhn, L. metamicrobiomeR: An R package for analysis of microbiome relative abundance data using zero-inflated beta GAMLSS and meta-analysis across studies using random effects models. BMC Bioinf. 2019, 20, 188. [Google Scholar] [CrossRef] [PubMed]
- Van den Brink, P.J.; Braak, C.J.T. Principal response curves: Analysis of time-dependent multivariate responses of biological community to stress. Environ.Toxicol. Chem. Int. J. 1999, 18, 138–148. [Google Scholar] [CrossRef]
- Shankar, V.; Agans, R.; Paliy, O. Advantages of phylogenetic distance based constrained ordination analyses for the examination of microbial communities. Sci. Rep. 2017, 7, 1–5. [Google Scholar] [CrossRef]
- George, D. SPSS for Windows Step by Step: A Simple Study Guide and Reference, 17.0 Update, 10/e; Pearson Education India: New Delhi, India, 2011. [Google Scholar]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017, 20, 145. [Google Scholar]
- Zhang, L.; Mu, C.; He, X.; Su, Y.; Mao, S.; Zhang, J.; Smidt, H.; Zhu, W. Effects of dietary fibre source on microbiota composition in the large intestine of suckling piglets. FEMS Microbiol. Lett. 2016, 363, fnw138. [Google Scholar]
- Kedia, G.; Vázquez, J.A.; Pandiella, S.S. Enzymatic digestion and in vitro fermentation of oat fractions by human lactobacillus strains. Enzyme Microb. Technol. 2008, 43, 355–361. [Google Scholar] [CrossRef]
- Metzler-Zebeli, B.U.; Zijlstra, R.T.; Mosenthin, R.; Gänzle, M.G. Dietary calcium phosphate content and oat β-glucan influence gastrointestinal microbiota, butyrate-producing bacteria and butyrate fermentation in weaned pigs. FEMS Microbiol. Ecol. 2011, 75, 402–413. [Google Scholar] [CrossRef]
- Van Hees, H.; Davids, M.; Maes, D.; Millet, S.; Possemiers, S.; Den Hartog, L.; Van Kempen, T.; Janssens, G. Dietary fibre enrichment of supplemental feed modulates the development of the intestinal tract in suckling piglets. J. Anim. Sci. Biotechnol. 2019, 10, 83. [Google Scholar]
- Sweeney, T.; Collins, C.; Reilly, P.; Pierce, K.; Ryan, M.; O’doherty, J. Effect of purified β-glucans derived from Laminaria digitata, Laminaria hyperborea and Saccharomyces cerevisiae on piglet performance, selected bacterial populations, volatile fatty acids and pro-inflammatory cytokines in the gastrointestinal tract of pigs. Br. J. Nutr. 2012, 108, 1226–1234. [Google Scholar]
- Chen, L.; Xu, Y.; Chen, X.; Fang, C.; Zhao, L.; Chen, F. The maturing development of gut microbiota in commercial piglets during the weaning transition. Front. Microbiol. 2017, 8, 1688. [Google Scholar] [CrossRef]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gresse, R.; Chaucheyras-Durand, F.; Dunière, L.; Blanquet-Diot, S.; Forano, E. Microbiota composition and functional profiling throughout the gastrointestinal tract of commercial weaning piglets. Microorganisms 2019, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Borewicz, K.; White, B.A.; Singer, R.S.; Sreevatsan, S.; Tu, Z.J.; Isaacson, R.E. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 2011, 153, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016, 6, 27427. [Google Scholar] [CrossRef]
- Mi, J.; Peng, H.; Wu, Y.; Wang, Y.; Liao, X. Diversity and community of methanogens in the large intestine of finishing pigs. BMC Microbiol. 2019, 19, 83. [Google Scholar]
- Taylor, P.R.; Tsoni, S.V.; Willment, J.A.; Dennehy, K.M.; Rosas, M.; Findon, H.; Haynes, K.; Steele, C.; Botto, M.; Gordon, S. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat. Immunol. 2007, 8, 31. [Google Scholar]
- Sonck, E.; Stuyven, E.; Goddeeris, B.; Cox, E. Identification of the porcine C-type lectin dectin-1. Vet. Immunol. Immunopathol. 2009, 130, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Hernanz-Falcon, P.; Arce, I.; Roda-Navarro, P.; Fernandez-Ruiz, E. Cloning of human DECTIN-1, a novel C-type lectin-like receptor gene expressed on dendritic cells. Immunogenetics 2001, 53, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, K.; Shen, G.-L.; Shikano, S.; Xu, S.; Ritter, R.; Kumamoto, T.; Edelbaum, D.; Morita, A.; Bergstresser, P.R.; Takashima, A. Identification of a novel, dendritic cell-associated molecule, dectin-1, by subtractive cDNA cloning. J. Biol. Chem. 2000, 275, 20157–20167. [Google Scholar] [CrossRef] [PubMed]
- Hester, S.N.; Comstock, S.S.; Thorum, S.C.; Monaco, M.H.; Pence, B.D.; Woods, J.A.; Donovan, S.M. Intestinal and systemic immune development and response to vaccination are unaffected by dietary (1, 3/1, 6)-β-D-glucan supplementation in neonatal piglets. Clin. Vaccine Immunol. 2012, 19, 1499–1508. [Google Scholar] [CrossRef]
- Vetvicka, V.; Oliveira, C. β (1-3)(1-6)-D-glucans modulate immune status in pigs: Potential importance for efficiency of commercial farming. Ann. Transl. Med. 2014, 2, 16. [Google Scholar]
- Li, J.; Xing, J.; Li, D.; Wang, X.; Zhao, L.; Lv, S.; Huang, D. Effects of β-glucan extracted from Saccharomyces cerevisiae on humoral and cellular immunity in weaned piglets. Arch. Anim. Nutr. 2005, 59, 303–312. [Google Scholar]
- Dos Santos, J.C.; de Figueiredo, A.M.B.; Silva, M.V.T.; Cirovic, B.; de Bree, L.C.J.; Damen, M.S.; Moorlag, S.J.; Gomes, R.S.; Helsen, M.M.; Oosting, M. β-glucan-induced trained immunity protects against Leishmania braziliensis infection: A crucial role for IL-32. Cell Rep. 2019, 28, 2659–2672.e2656. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar]
- Gerritsen, J. The Genus Romboutsia: Genomic and Functional Characterization of Novel Bacteria Dedicated to Life in the Intestinal Tract. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2015. [Google Scholar]
- Gerritsen, J.; Hornung, B.; Renckens, B.; van Hijum, S.A.; dos Santos, V.A.M.; Rijkers, G.T.; Schaap, P.J.; de Vos, W.M.; Smidt, H. Genomic and functional analysis of Romboutsia ilealis CRIBT reveals adaptation to the small intestine. PeerJ 2017, 5, e3698. [Google Scholar]
- Pence, B.D.; Hester, S.N.; Donovan, S.M.; Woods, J.A. Dietary whole glucan particles do not affect antibody or cell-mediated immune responses to influenza virus vaccination in mice. Immunol. Investig. 2012, 41, 275–289. [Google Scholar] [CrossRef]
- Stuyven, E.; Van Den Broeck, W.; Nauwynck, H.; Goddeeris, B.; Cox, E. Oral administration of beta-1, 3/1, 6-glucan Macrogard® fails to enhance the mucosal immune response following oral F4 fimbrial immunisation in gnotobiotic pigs. Vet. Immunol. Immunopathol. 2010, 137, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Hahn, T.-W.; Lohakare, J.; Lee, S.; Moon, W.; Chae, B. Effects of supplementation of β-glucans on growth performance, nutrient digestibility, and immunity in weanling pigs. J. Anim. Sci. 2006, 84, 1422–1428. [Google Scholar] [PubMed]

| Antibody | Host/Isotype | Clone | Fluorochrome | Company | Dilution |
|---|---|---|---|---|---|
| CD14 | Mouse, IgG2b | MIL-2 | FITC | Bio-Rad | 1:50 |
| CD172a | Mouse, IgG2b | 74-22-15A | PE | BD Biosciences | 1:40 |
| CD4a | Mouse, IgG2b | 74-12-4 | PerCP-Cy5.5 | BD PharmingenTM | 1:320 |
| CADM1 | Chicken, IgY | 3E1 | Biotin | MBL | 1:200 |
| Streptav. | n/a | n/a | BV421 | BD HorizonTM | 1:50 |
| CD152 * | Mouse, IgG2a | n/a | APC | Ancell | 1:320 |
| CD3ε | Mouse, IgG2a | BB23-8E6-8C8 | PE-CyTM7 | BD PharmingenTM | 1:160 |
| CD4a | Mouse, IgG2b | 74-12-4 | PerCP-Cy5.5 | BD PharmingenTM | 1:320 |
| CD8a | Mouse, IgG2a | 76-2-11 | FITC | BD PharmingenTM | 1:10 |
| FoxP3 | Rat, IgG2a | FJK-16s | Alexa Fluor® 700 ** | eBioscienceTM | 1:20 |
| Ki67 | Mouse, IgG1 | B56 | BV421 ** | BD HorizonTM | 1:80 |
| CD25 | Mouse, IgG1 | K231.3B2 | Purified *** | Bio-Rad | 1:200 |
| γδ T cells | Rat, IgG2a | MAC320 | PE | BD PharmingenTM | 1:20 |
| Unpaired T-Test | Two-Way ANOVA | |||||
|---|---|---|---|---|---|---|
| Subset | Phenotype | D14 | D26 | D43 | D69 | |
| p-value [CI] | p-value [CI] | p-value [CI] | p-value [CI] | |||
| pDCs (%) | CD14−CD4+CD172 a+CADM1− | ns | ns | ns | 0.060 ↓ [−0.66 to 34.14] | T (<0.001), I (0.020) |
| pDCs (mat.) | CD14−CD4+CD172 a+CADM1−CTLA4-Ig+ | ns | ns | ns | ns | T (<0.001) |
| cDC1 (%) | CD14−CD4−CD172 alowCADM1+ | ns | ns | ns | ns | T (<0.001) |
| cDC1 (mat.) | CD14−CD4−CD172 alowCADM1+ CTLA4-Ig+ | ns | ns | ns | ns | T (<0.001) |
| cDC2 (%) | CD14−CD4−CD172 a+CADM1+ | ns | ns | ns | ns | T (0.092) |
| cDC2 (mat.) | CD14−CD4−CD172 a+CADM1+ CTLA4-Ig+ | ns | ns | ns | ns | T (<0.001) |
| NK cells (%) | CD3−CD8α+ | ns | 0.060 ↓ [−0.41 to 15.68] | ns | ns | G (0.080), T (0.063) |
| NK cells (act.) | CD3−CD8α+CD25+ | ns | ns | ns | ns | T (<0.001) |
| NK cells (Ki67%) | CD3−CD8α+Ki67+ | ns | 0.009 ↓ [5.15 to 28.53] | ns | ns | G (0.010), T (<0.001) |
| γδ T cells (%) | CD3+TCR-γδ+ | ns | ns | ns | ns | T (0.005) |
| γδ T cells (Ki67%) | CD3+TCR-γδ+Ki67+ | ns | 0.049 ↓ [0.02 to 12.75] | ns | ns | G (0.034), T (<0.001) |
| CTLs (%) | CD3+TCR-γδ−CD8α+ | ns | 0.090 ↓ [−0.53 to 6.13] | ns | ns | G (0.008), T (<0.001) |
| CTLs (Ki67%) | CD3+TCR-γδ−CD8α+Ki67+ | ns | ns | ns | ns | T (<0.001) |
| T helper (%) | CD3+TCR-γδ−CD4+ | ns | ns | ns | ns | T (<0.001) |
| T helper (Ki67%) | CD3+TCR-γδ−CD4+Ki67+ | ns | ns | ns | ns | T (<0.001) |
| Mem./Act. (%) | CD3+TCR-γδ−CD4+CD8α+ | ns | ns | ns | ns | T (<0.001) |
| Mem./Act. (Ki67%) | CD3+TCR-γδ−CD4+CD8α+ki67+ | ns | ns | ns | ns | T (<0.001) |
| Tregs (%) | CD3+TCR-γδ−CD4+CD25 highFoxp3+ | ns | ns | ns | ns | T (0.003) |
| Tregs (Ki67%) | CD3+ TCR-γδ−CD4+CD25 highFoxp3+Ki67+ | ns | ns | ns | ns | T (<0.001) |
| Unpaired T-Test | Two-Way ANOVA | ||||
|---|---|---|---|---|---|
| Subset | Phenotype | D27 | D44 | D70 | |
| p-value [CI] | p-value [CI] | p-value [CI] | |||
| pDCs (%) | CD14−CD4+CD172 a+CADM1− | ns | ns | ns | T (<0.001) |
| pDCs (mat.) | CD14−CD4+CD172 a+CADM1−CTLA4-Ig+ | 0.034 [3.21 to 67.89] ↓ | ns | 0.024 [7.75 to 92.70] ↓ | G (0.007), T (0.001), I (0.08) |
| cDC1 (%) | CD14−CD4−CD172 alowCADM1+ | ns | 0.011 [−0.11 to −0.02] ↑ | ns | T (<0.001), I (0.028) |
| cDC1 (mat.) | CD14−CD4−CD172 alowCADM1+CTLA4-Ig+ | ns | ns | ns | T (<0.001) |
| cDC2 (%) | CD14−CD4−CD172 a+CADM1+ | ns | ns | ns | T (0.001) |
| cDC2 (mat.) | CD14−CD4−CD172 a+CADM1+CTLA4-Ig+ | 0.084 [−62.71 to 4.50] ↑ | ns | ns | ns |
| NK cells (%) | CD3−CD8 a+ | n/a | n/a | n/a | n/a |
| NK cells (act.) | CD3−CD8α+CD25+ | n/a | n/a | n/a | n/a |
| NK cells (Ki67%) | CD3−CD8α+Ki67+ | n/a | n/a | n/a | n/a |
| γδ T cells (%) | CD3+TCR-γδ+ | ns | ns | ns | T (<0.001) |
| γδ T cells (Ki67%) | CD3+TCR-γδ+Ki67+ | 0.089 [−15.45 to 1.26] ↑ | ns | ns | T (<0.001), I (0.086) |
| CTLs (%) | CD3+TCR-γδ−CD8α+ | ns | 0.035 [−13.03 to −0.60] ↑ | ns | T (0.076) |
| CTLs (Ki67%) | CD3+TCR-γδ−CD8α+Ki67+ | ns | ns | ns | T (<0.001) |
| T helper (%) | CD3+TCR-γδ−CD4+ | ns | ns | ns | T (<0.001) |
| T helper (Ki67%) | CD3+TCR-γδ−CD4+Ki67+ | 0.079 [−12.53 to 0.77] ↑ | ns | ns | T (<0.001), I (0.019) |
| Mem./Act. (%) | CD3+CD4+CD8α+ | ns | ns | ns | T (<0.001) |
| CD3+CD4 highCD8αlow | ns | ns | ns | T (0.002) | |
| CD3+CD4 high CD8αlowki67+ | ns | ns | ns | T (<0.001) | |
| CD3+CD4 lowCD8αhigh | ns | ns | ns | ns | |
| CD3+CD4 low CD8αhighki67+ | ns | ns | ns | T (<0.001) | |
| Tregs (%) | CD3+TCR-γδ−CD4+CD25 highFoxp3+ | ns | ns | ns | T (<0.001) |
| Tregs (Ki67%) | CD3+ TCR-γδ−CD4+CD25 highFoxp3+Ki67+ | n/a | n/a | n/a | n/a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Vries, H.; Geervliet, M.; Jansen, C.A.; Rutten, V.P.M.G.; van Hees, H.; Groothuis, N.; Wells, J.M.; Savelkoul, H.F.J.; Tijhaar, E.; Smidt, H. Impact of Yeast-Derived β-Glucans on the Porcine Gut Microbiota and Immune System in Early Life. Microorganisms 2020, 8, 1573. https://doi.org/10.3390/microorganisms8101573
de Vries H, Geervliet M, Jansen CA, Rutten VPMG, van Hees H, Groothuis N, Wells JM, Savelkoul HFJ, Tijhaar E, Smidt H. Impact of Yeast-Derived β-Glucans on the Porcine Gut Microbiota and Immune System in Early Life. Microorganisms. 2020; 8(10):1573. https://doi.org/10.3390/microorganisms8101573
Chicago/Turabian Stylede Vries, Hugo, Mirelle Geervliet, Christine A. Jansen, Victor P. M. G. Rutten, Hubèrt van Hees, Natalie Groothuis, Jerry M. Wells, Huub F. J. Savelkoul, Edwin Tijhaar, and Hauke Smidt. 2020. "Impact of Yeast-Derived β-Glucans on the Porcine Gut Microbiota and Immune System in Early Life" Microorganisms 8, no. 10: 1573. https://doi.org/10.3390/microorganisms8101573
APA Stylede Vries, H., Geervliet, M., Jansen, C. A., Rutten, V. P. M. G., van Hees, H., Groothuis, N., Wells, J. M., Savelkoul, H. F. J., Tijhaar, E., & Smidt, H. (2020). Impact of Yeast-Derived β-Glucans on the Porcine Gut Microbiota and Immune System in Early Life. Microorganisms, 8(10), 1573. https://doi.org/10.3390/microorganisms8101573







