Evidence that Bacteria Packaging by Tetrahymena Is a Widespread Phenomenon
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Co-Cultures
2.3. DAPI Staining
2.4. LIVE/DEAD Staining
2.5. Transmission Electron Microscopy (TEM) Processing
3. Results
3.1. Co-Culture Assays
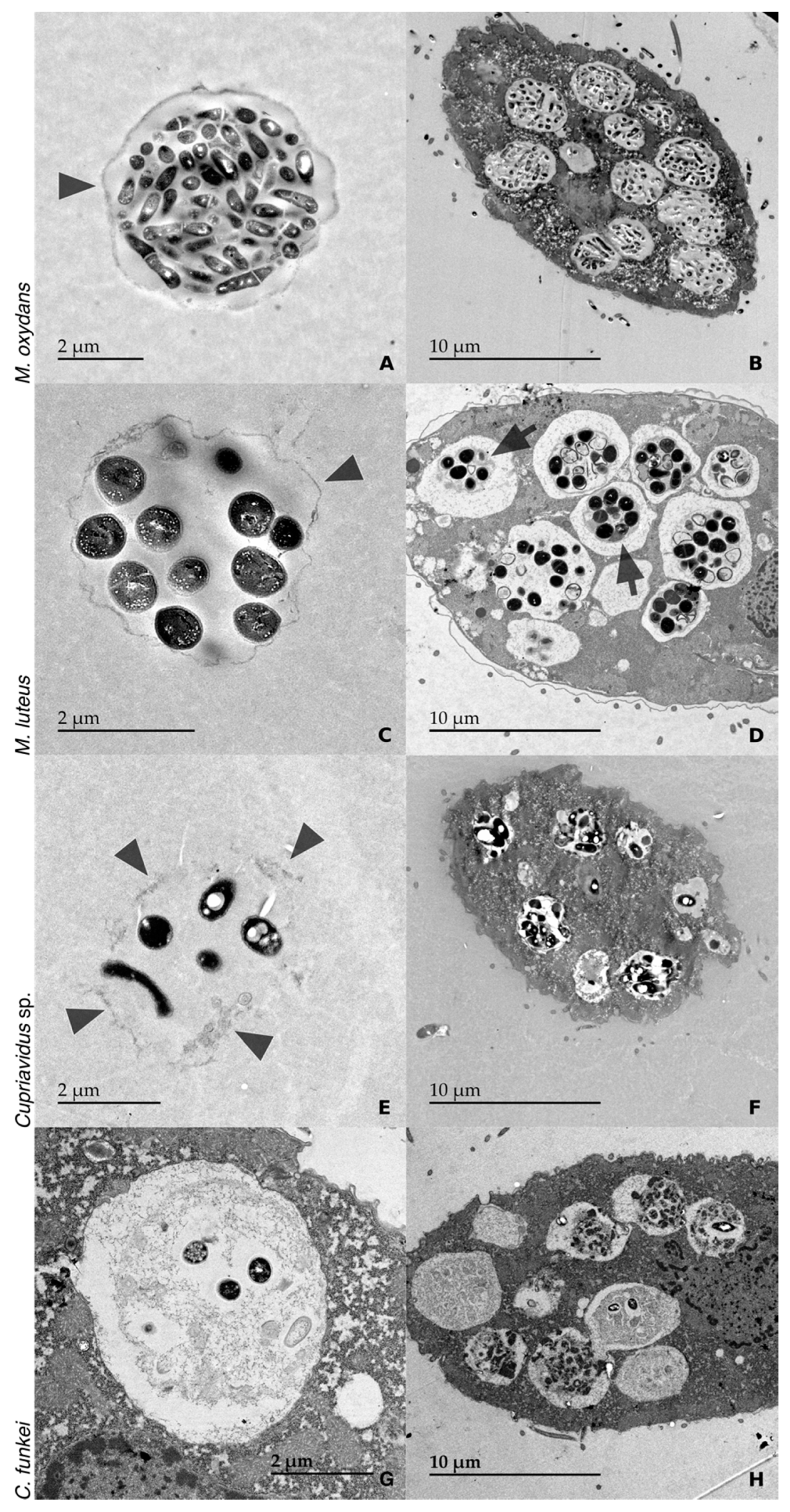
3.2. TEM Observations
3.3. LIVE/DEAD Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berk, S.G.; Faulkner, G.; Garduño, E.; Joy, M.C.; Ortiz-Jimenez, M.A.; Garduño, R.A. Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl. Environ. Microbiol. 2008, 74, 2187–2199. [Google Scholar] [CrossRef] [PubMed]
- Gourabathini, P.; Brandl, M.T.; Redding, K.S.; Gunderson, J.H.; Berk, S.G. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl. Environ. Microbiol. 2008, 74, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Cateau, E.; Delafont, V.; Hechard, Y.; Rodier, M.H. Free-living amoebae: What part do they play in healthcare-associated infections? J. Hosp. Infect. 2014, 87, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Greub, G.; Raoult, D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 2004, 17, 413–433. [Google Scholar] [CrossRef] [PubMed]
- Denoncourt, A.M.; Paquet, V.E.; Charette, S.J. Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Paquet, V.E.; Charette, S.J. Amoeba-resisting bacteria found in multilamellar bodies secreted by Dictyostelium discoideum: Social amoebae can also package bacteria. FEMS Microbiol. Ecol. 2016, 92, fiw025. [Google Scholar] [CrossRef]
- Denoncourt, A.M.; Paquet, V.E.; Charette, S.J. Packaging of Mycobacterium smegmatis bacteria into fecal pellets by the ciliate Tetrahymena pyriformis. FEMS Microbiol. Lett. 2017, 364, fnx237. [Google Scholar] [CrossRef]
- Trigui, H.; Paquet, V.E.; Charette, S.J.; Faucher, S.P. Packaging of Campylobacter jejuni into multilamellar bodies by the ciliate Tetrahymena pyriformis. Appl. Environ. Microbiol. 2016, 82, 2783–2790. [Google Scholar] [CrossRef][Green Version]
- Berk, S.G.; Ting, R.S.; Turner, G.W.; Ashburn, R.J. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 1998, 64, 279–286. [Google Scholar] [CrossRef]
- Brandl, M.T.; Rosenthal, B.M.; Haxo, A.F.; Berk, S.G. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 2005, 71, 1562–1569. [Google Scholar] [CrossRef]
- Espinoza-Vergara, G.; Noorian, P.; Silva-Valenzuela, C.A.; Raymond, B.B.A.; Allen, C.; Hoque, M.M.; Sun, S.; Johnson, M.S.; Pernice, M.; Kjelleberg, S.; et al. Vibrio cholerae residing in food vacuoles expelled by protozoa are more infectious in vivo. Nat. Microbiol. 2019, 4, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Koubar, M.; Rodier, M.H.; Garduño, R.A.; Frère, J. Passage through Tetrahymena tropicalis enhances the resistance to stress and the infectivity of Legionella pneumophila. FEMS Microbiol. Lett. 2011, 325, 10–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Raghu Nadhanan, R.; Thomas, C.J. Colpoda secrete viable Listeria monocytogenes within faecal pellets. Environ. Microbiol. 2014, 16, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Kebbi-Beghdadi, C.; Greub, G. Importance of amoebae as a tool to isolate amoeba-resisting microorganisms and for their ecology and evolution: The Chlamydia paradigm. Environ. Microbiol. Rep. 2014, 6, 309–324. [Google Scholar] [CrossRef]
- Macher, J. Bioaerosols: Assessment and Control; American Conference of Governmental Industrial Hygienists: Cincinatti, OH, USA, 1999; ISBN 978-1-882417-29-1. [Google Scholar]
- Iriberri, J.; Ayo, B.; Santamaria, E.; Barcina, I.; Egea, L. Influence of bacterial density and water temperature on the grazing activity of two freshwater ciliates. Freshw. Biol. 1995, 33, 223–231. [Google Scholar] [CrossRef]
- Fenchel, T. Suspension feeding in ciliated protozoa: Feeding rates and their ecological significance. Microb. Ecol. 1980, 6, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Denoncourt, A.M.; Durocher, A.F.; Paquet, V.E.; Charette, S.J. The fate of multilamellar bodies produced and secreted by Dictyostelium discoideum amoebae. Eur. J. Cell Biol. 2017, 96, 767–773. [Google Scholar] [CrossRef]
- Gorovsky, M.; Yao, M.; Keevert, J.; Pleger, G. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975, 9, 311–327. [Google Scholar]
- Orias, E.; Hamilton, E.P.; Orias, J.D. Tetrahymena as a laboratory organism: Useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 2000, 62, 189–211. [Google Scholar] [CrossRef]
- Paquet, V.E.; Lessire, R.; Domergue, F.; Fouillen, L.; Filion, G.; Sedighi, A.; Charette, S.J. Lipid composition of multilamellar bodies secreted by Dictyostelium discoideum reveals their amoebal origin. Eukaryot. Cell 2013, 12, 1326–1334. [Google Scholar] [CrossRef]
- Berthiaume, C.; Gilbert, Y.; Fournier-Larente, J.; Pluchon, C.; Filion, G.; Jubinville, E.; Sérodes, J.-B.; Rodriguez, M.; Duchaine, C.; Charette, S.J. Identification of dichloroacetic acid degrading Cupriavidus bacteria in a drinking water distribution network model. J. Appl. Microbiol. 2014, 116, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Raghupathi, P.K.; Liu, W.; Sabbe, K.; Houf, K.; Burmølle, M.; Sørensen, S.J. Synergistic interactions within a multispecies biofilm enhance individual species protection against grazing by a pelagic protozoan. Front. Microbiol. 2018, 8, 2649. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.; Berk, S.G.; Garduno, E.; Ortiz-Jimenez, M.A.; Garduno, R.A. Passage through Tetrahymena tropicalis triggers a rapid morphological differentiation in Legionella pneumophila. J. Bacteriol. 2008, 190, 7728–7738. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elliott, A.M.; Clemmons, G.L. An ultrastructural study of ingestion and digestion in Tetrahymena pyriformis. J. Protozool. 1966, 13, 311–323. [Google Scholar] [CrossRef]
- Hausmann, K.; Hülsmann, N.; Machemer, H.; Mulisch, M.; Steinbrück, G.; Corliss, J.O. Chapter entitled “Ingestion, Digestion and Defecation”. In Protozoology, 2nd ed.; Georg Thieme Verlag: Stuttgart, Germany, 1996; ISBN 3131103019. [Google Scholar]
- Thurman, J.; Drinkall, J.; Parry, J. Digestion of bacteria by the freshwater ciliate Tetrahymena pyriformis. Aquat. Microb. Ecol. 2010, 60, 163–174. [Google Scholar] [CrossRef]


| Bacteria | Source | T. thermophila1 | T. pyriformis2 |
|---|---|---|---|
| Cupriavidus basilensis | Isolated from drinking water pipes [22] | + | ++ |
| Cupriavidussp. | Isolated from drinking water pipes [22] | + | ++ |
| Micrococcus luteus | Janet Martha Blatny (NTNU 3, Norway) | ++ | +++ |
| Micrococcus luteus US4 | USDA | + | + |
| Rathayibacter tritici LBUM-572 | Martin Filion (Moncton University, Canada) | + | ++ |
| Rhodococcus erythropolis US1 | USDA | ++ | + |
| Rhodococcus erythropolis US2 | USDA | + | + |
| Rhodococcus fascians US1 | USDA | ++ | ++ |
| Rhodococcus fascians US2 | USDA | + | ++ |
| Cupriavidus necator US1 | USDA | + | +++ |
| Duganella zoogloeoides LBUM-382 | Martin Filion (Moncton University, Canada) | + | + |
| Kocuria kristinae | Janet Martha Blatny (NTNU, Norway) | ++ | +++ |
| Microbacterium oxydans US1 | USDA | ++++ | ++++ |
| Micrococcus luteus | Pierre Cosson (Geneva University, Switzerland) | ++ | + |
| Micrococcus luteus 8_4_14 X2 | Janet Martha Blatny (NTNU, Norway) | + | + |
| Micrococcus luteus D_1_6 X2 | Janet Martha Blatny (NTNU, Norway) | - | ++++ |
| Micrococcus luteus US3 | USDA | + | +++ |
| Rhodococcus erythropolis | Janet Martha Blatny (NTNU, Norway) | + | ++++ |
| Rhodococcus pyridinivorans | Janet Martha Blatny (NTNU, Norway) | + | ++ |
| Cellulosimicrobium funkei | Janet Martha Blatny (NTNU, Norway) | +++ | ++ |
| Bacteria | Gram | Shape | Size 1 (µm) |
|---|---|---|---|
| Microbacterium oxydans US1 | + | Rod | 1 × 0.25 |
| Micrococcus luteus | + | Coccus | 0.5 |
| Cellulosimicrobium funkei | + | Rod | 1 × 0.5 |
| Cupriavidus sp. | - | Rod | 2 × 0.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durocher, A.F.; Denoncourt, A.M.; Paquet, V.E.; Charette, S.J. Evidence that Bacteria Packaging by Tetrahymena Is a Widespread Phenomenon. Microorganisms 2020, 8, 1548. https://doi.org/10.3390/microorganisms8101548
Durocher AF, Denoncourt AM, Paquet VE, Charette SJ. Evidence that Bacteria Packaging by Tetrahymena Is a Widespread Phenomenon. Microorganisms. 2020; 8(10):1548. https://doi.org/10.3390/microorganisms8101548
Chicago/Turabian StyleDurocher, Alicia F., Alix M. Denoncourt, Valérie E. Paquet, and Steve J. Charette. 2020. "Evidence that Bacteria Packaging by Tetrahymena Is a Widespread Phenomenon" Microorganisms 8, no. 10: 1548. https://doi.org/10.3390/microorganisms8101548
APA StyleDurocher, A. F., Denoncourt, A. M., Paquet, V. E., & Charette, S. J. (2020). Evidence that Bacteria Packaging by Tetrahymena Is a Widespread Phenomenon. Microorganisms, 8(10), 1548. https://doi.org/10.3390/microorganisms8101548







