Culturomics Discloses Anti-Tubercular Enterococci Exclusive of Pulmonary Tuberculosis: A Preliminary Report
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Population Information
2.2. Culturomics
2.3. Real-Time Quantitative PCR
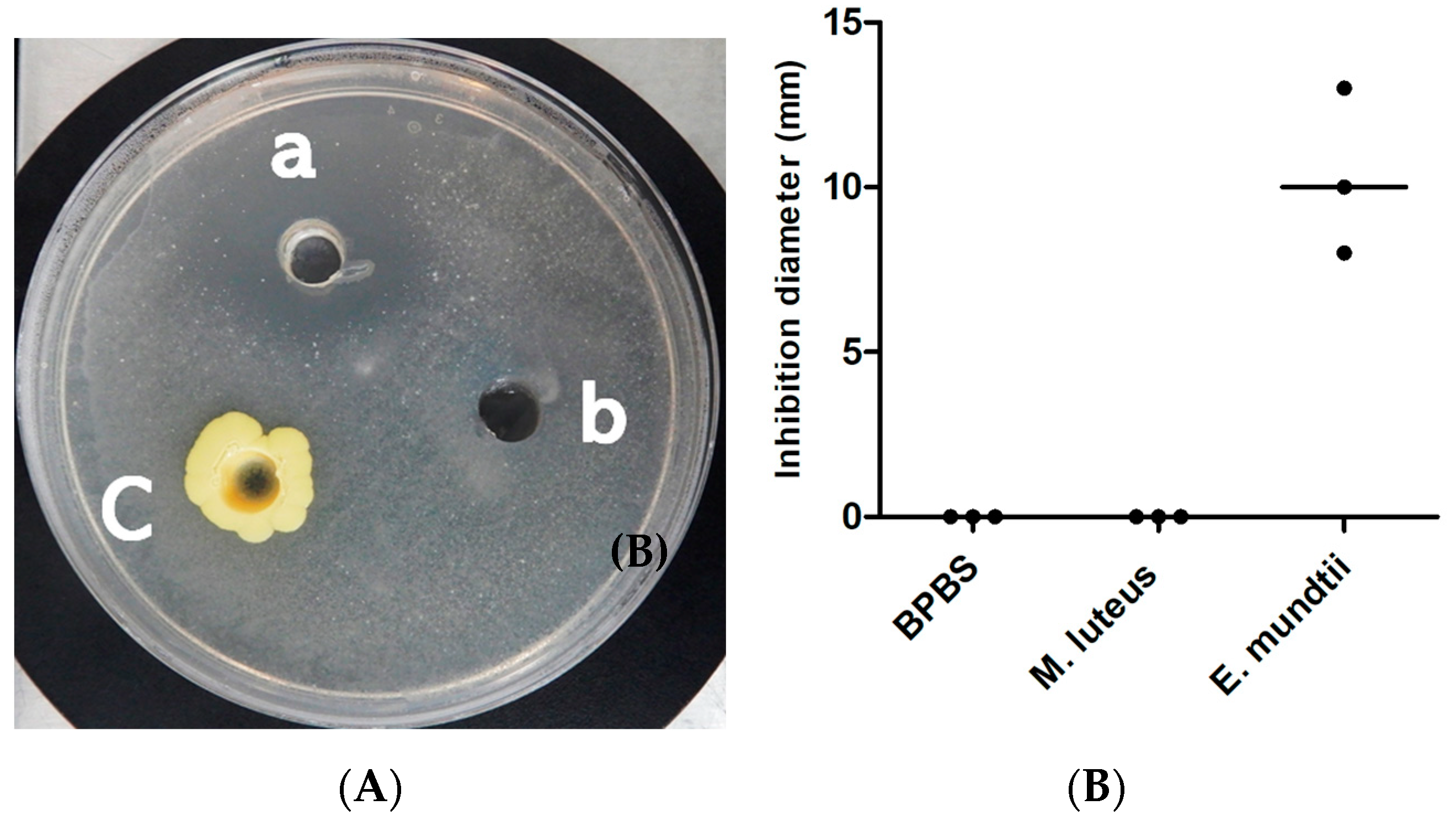
2.4. Anti-Tuberculosis Activity Assays
2.5. Statistical Analyses
3. Results
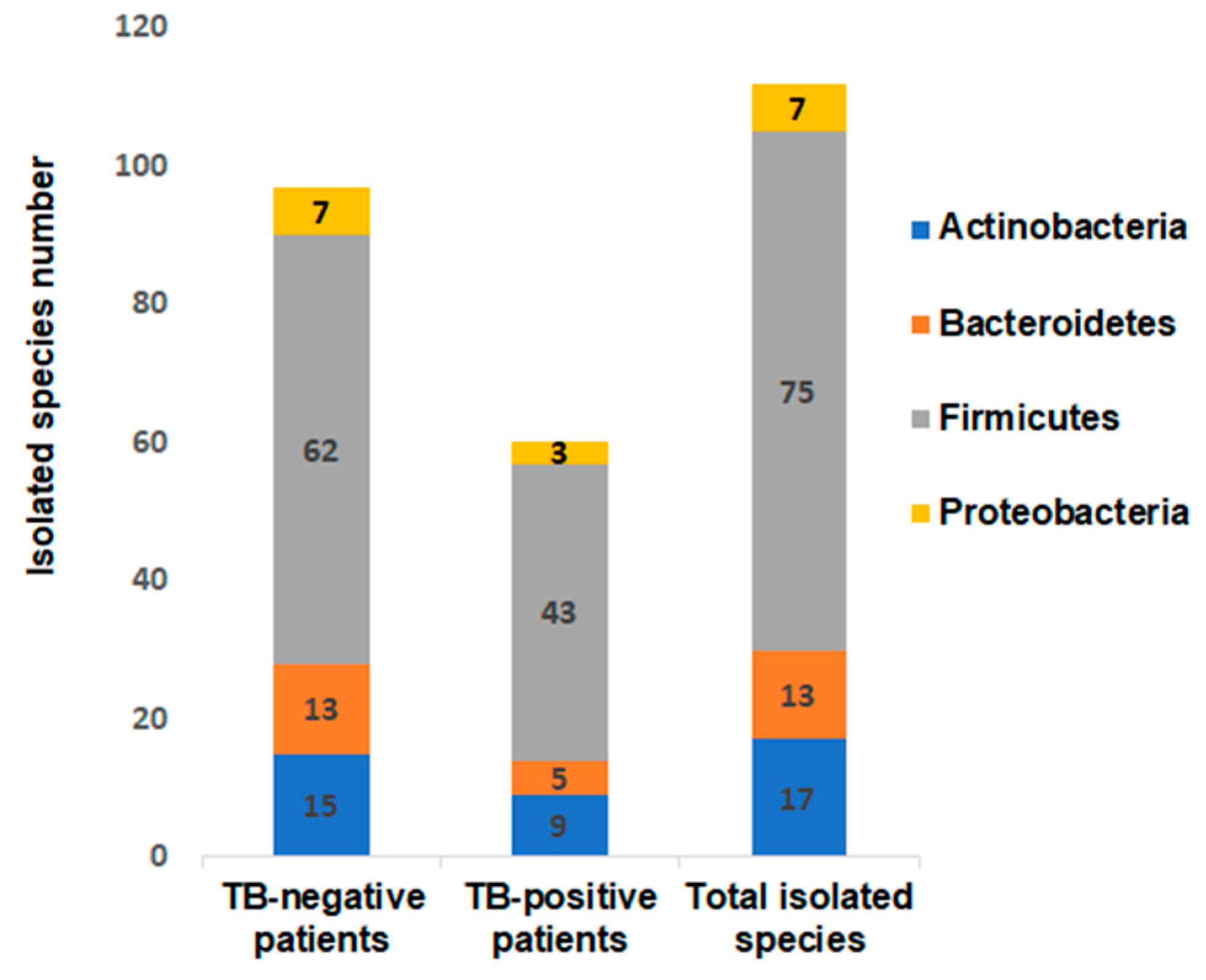
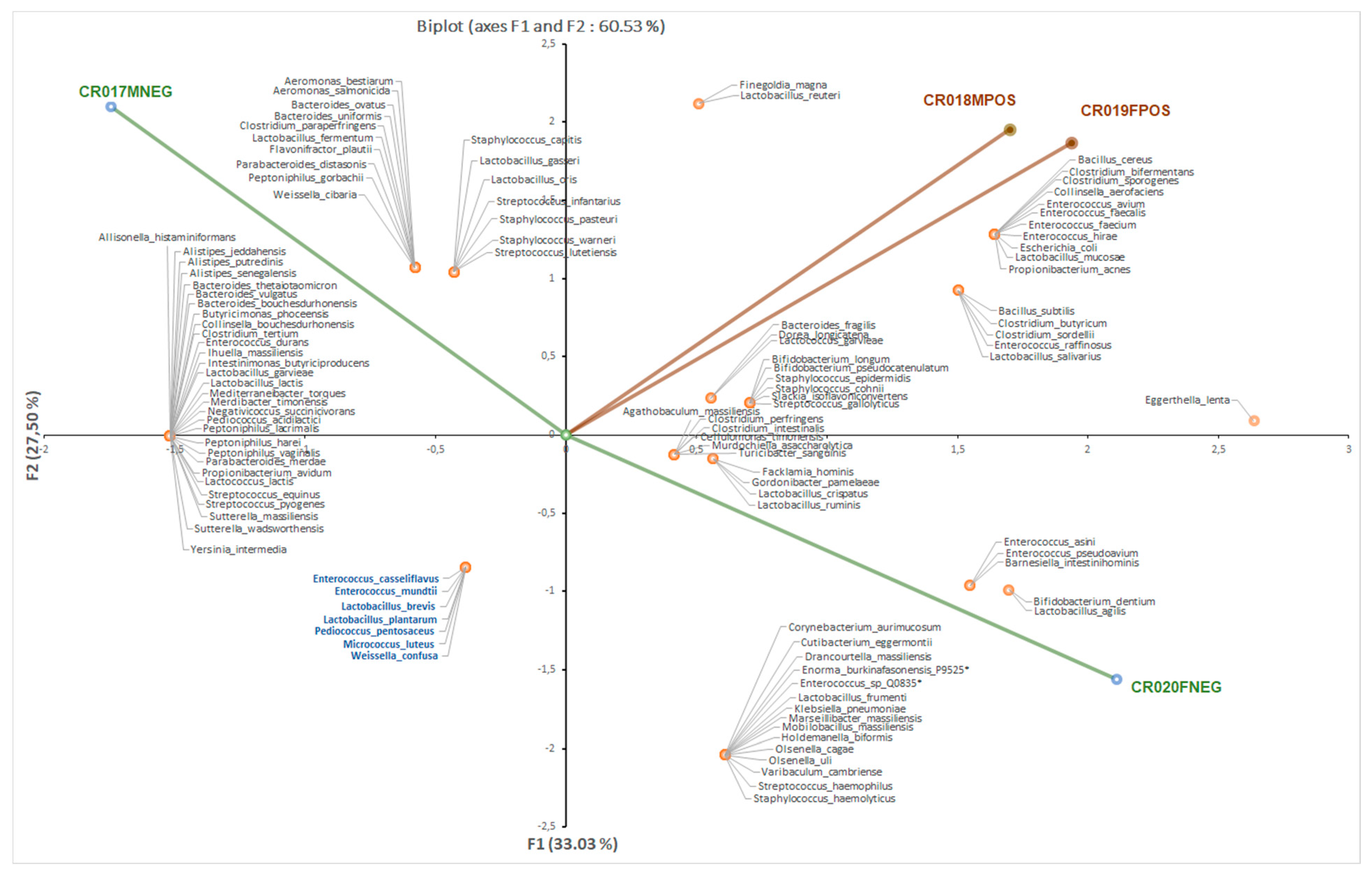
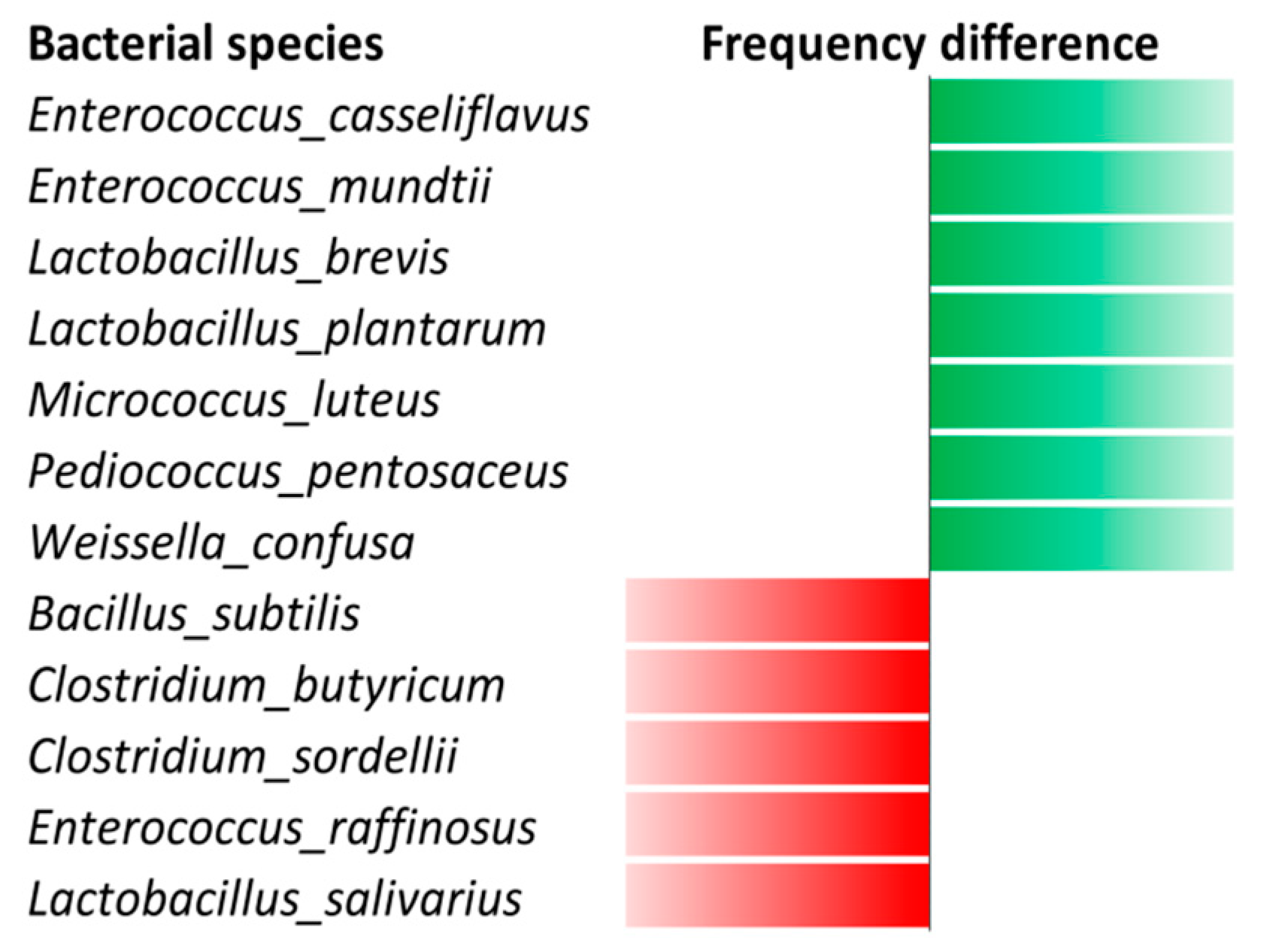
3.1. Bacterial Culturomics
3.2. Real-Time PCR Assays
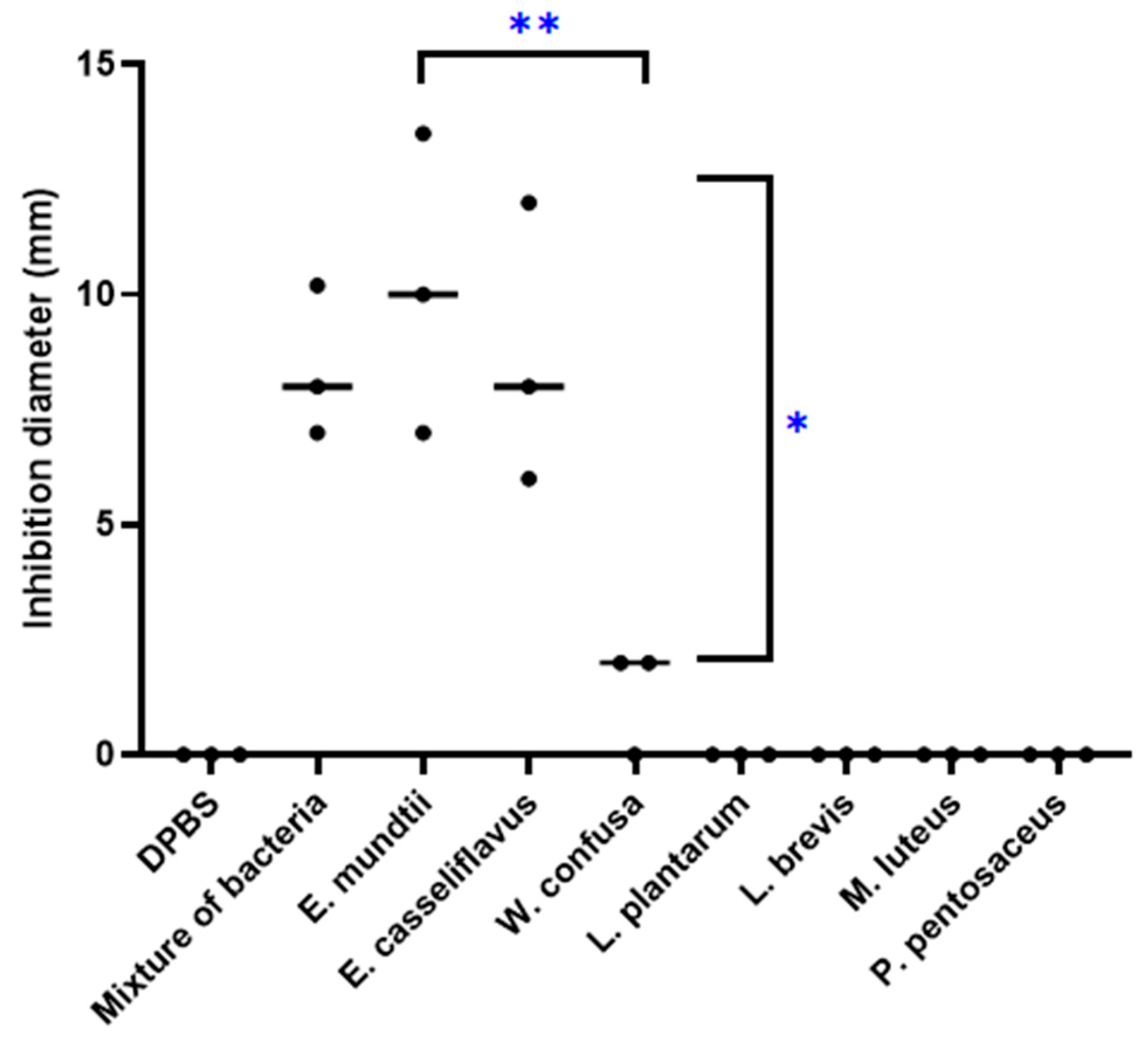
3.3. Antitubercular Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aboubaker, O.D.; Bouzid, F.; Canaan, S.; Drancourt, M. Smooth Tubercle Bacilli: Neglected Opportunistic Tropical Pathogens. Front. Public Health 2015, 3, 283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- WHO. Global Tuberculosis Report, Geneva. World Health Organization 2019. Available online: https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1 (accessed on 19 July 2020).
- Brites, D.; Gagneux, S. The Nature and Evolution of Genomic Diversity in the Mycobacterium tuberculosis Complex. Adv. Exp. Med. Biol. 2017, 1019, 1–26. [Google Scholar] [PubMed]
- Copin, R.; Wang, X.; Louie, E.; Escuyer, V.; Coscolla, M.; Gagneux, S.; Palmer, G.H.; Ernst, J.D. Within Host Evolution Selects for a Dominant Genotype of Mycobacterium tuberculosis while T Cells Increase Pathogen Genetic Diversity. PLoS Pathog. 2016, 12, e1006111. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.D.; Chiu, C.; Churchyard, G.J.; Esmail, H.; Lewinsohn, D.M.; Gandhi, N.R.; Fennelly, K.P. Tuberculosis Infectiousness and Host Susceptibility. J. Infect. Dis. 2017, 216, S636–S643. [Google Scholar] [CrossRef]
- Bustamante, J.; Aksu, G.; Vogt, G.; de Beaucoudrey, L.; Genel, F.; Chapgier, A.; Orchidée, F.S.; Jacqueline Feinberg, J.; Emile, J.F.; Kutukculer, N.; et al. BCG-osis and tuberculosis in a child with chronic granulomatous disease. J. Allergy Clin. Immunol. 2007, 120, 32–38. [Google Scholar] [CrossRef]
- Abel, L.; El-Baghdadi, J.; Bousfiha, A.A.; Casanova, J.-L.; Schurr, E. Human genetics of tuberculosis: A long and winding road. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 369, 20130428. [Google Scholar] [CrossRef]
- Casanova, J.-L.; Abel, L. Genetic Dissection of Immunity to Mycobacteria: The Human Model. Annu. Rev. Immunol. 2002, 20, 581–620. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X.; Han, Y.; Wang, C.; Zhou, Y.; Zheng, F. Certain Polymorphisms in SP110 Gene Confer Susceptibility to Tuberculosis: A Comprehensive Review and Updated Meta-Analysis. Yonsei Med. J. 2017, 58, 165–173. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Maningi, N.E.; Fourie, P.B. Mycobacterium tuberculosis, antimicrobials, immunity, and lung–gut microbiota crosstalk: Current updates and emerging advances. Ann. N. Y. Acad. Sci. 2020, 1–27. [Google Scholar] [CrossRef]
- Winglee, K.; Eloe-Fadrosh, E.; Gupta, S.; Guo, H.; Fraser, C.; Bishai, W. Aerosol Mycobacterium tuberculosis Infection Causes Rapid Loss of Diversity in Gut Microbiota. PLoS ONE 2014, 9, e97048. [Google Scholar] [CrossRef]
- Luo, M.; Liu, Y.; Wu, P.; Luo, D.-X.; Sun, Q.; Zheng, H.; Hu, R.; Pandol, S.J.; Li, Q.-F.; Han, Y.-P.; et al. Alternation of Gut Microbiota in Patients with Pulmonary Tuberculosis. Front. Physiol. 2017, 8, 822. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Feng, Y.; Wu, J.; Liu, F.; Zhang, Z.; Hao, Y.; Liang, S.; Li, B.; Li, J.; Lv, N.; et al. The Gut Microbiome Signatures Discriminate Healthy From Pulmonary Tuberculosis Patients. Front. Cell. Infect. Microbiol. 2019, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Asmar, S.; Chatellier, S.; Mirande, C.; van Belkum, A.; Canard, I.; Raoult, D.; Drancourt, M. A Novel Solid Medium for Culturing Mycobacterium tuberculosis Isolates from Clinical Specimens. J. Clin. Microbiol. 2015, 53, 2566–2569. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lagier, J.-C.; Hugon, P.; Khelaifia, S.; Fournier, P.-E.; Scola, B.L.; Raoult, D. The Rebirth of Culture in Microbiology through the Example of Culturomics To Study Human Gut Microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raout, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, I.S.; Seck, E.S.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fellag, M.; Loukil, A.; Saad, J.; Lepidi, H.; Bouzid, F.; Brégeon, F.; Drancourt, M. Translocation of Mycobacterium tuberculosis after experimental ingestion. PLoS ONE 2019, 14, e0227005. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Million, M.; Togo, A.H.; Khelaifia, S.; Raoult, D. Culturomics provides critical prokaryote strains for anti-Listeria and anti-cancer probiotics. Int. J. Antimicrob. Agents 2019, 54, 407–409. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Fellag, M.; Saad, J.; Armstrong, N.; Chabrière, E.; Eldin, C.; Lagier, J.-C.; Drancourt, M. Routine Culture-Resistant Mycobacterium tuberculosis Rescue and Shell-Vial Assay, France. Emerging Infect. Dis. 2019, 25, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Lapierre, S.G.; Fellag, M.; Magan, C.; Drancourt, M. Mycobacterium malmoense pulmonary infection in France: A case report. BMC Res. Notes 2017, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Asmar, S.; Drancourt, M. Chlorhexidine decontamination of sputum for culturing Mycobacterium tuberculosis. BMC Microbiol. 2015, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Gouba, N.; Saad, J.; Drancourt, M.; Fellag, M. Genome Sequence of Enorma sp. Strain Marseille-P9525T, a Member of a Human Gut Microbiome. Microbiol. Resour. Announc. 2019, 8, e00785-19. [Google Scholar] [CrossRef]
- Gouba, N.; Hassani, Y.; Saad, J.; Drancourt, M.; Mbogning Fonkou, M.D.; Fellag, M. Enorma burkinafasonensis sp. nov., a new bacterium isolated from a human gut microbiota. New Microbes New Infect. 2020, 36, 100687. [Google Scholar]
- Gouba, N.; Yimagou, E.K.; Hassani, Y.; Drancourt, M.; Fellag, M.; Mbogning Fonkou, M.D. Enterococcus burkinafasonensis sp. nov. isolated from human gut microbiota. New Microbes New Infect. 2020, 36, 100702. [Google Scholar] [CrossRef]
- Drancourt, M. Culturing Stools to Detect Mycobacterium tuberculosis. J. Clin. Microbiol. 2018, 56, e02033-17. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, Q.; Liu, B.; Dong, J.; Sun, L.; Zhu, Y.; Su, H.; Liu, B.; Yang, J.; Yang, F.; et al. Gut microbiota associated with pulmonary tuberculosis and dysbiosis caused by anti-tuberculosis drugs. J. Infect. 2019, 78, 317–322. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Y.; Liao, Q.; Wang, Z.; Wan, C. Characterization of gut microbiota in children with pulmonary tuberculosis. BMC Pediatr. 2019, 19, 445. [Google Scholar] [CrossRef]
- Namasivayam, S.; Kauffman, K.D.; McCulloch, J.A.; Yuan, W.; Thovarai, V.; Mittereder, L.R.; Trinchieri, G.; Barber, D.L.; Sheret, A. Correlation between Disease Severity and the Intestinal Microbiome in Mycobacterium tuberculosis-Infected Rhesus Macaques. mBio 2019, 10, 3. [Google Scholar] [CrossRef]
- Behr, M.A.; Waters, W.R. Is tuberculosis a lymphatic disease with a pulmonary portal? Lancet Infect. Dis. 2014, 14, 250–255. [Google Scholar] [CrossRef]
- Bravo, M.; Combes, T.; Martinez, F.O.; Cerrato, R.; Rey, J.; Garcia-Jimenez, W.; Fernandez-Llario, P.; Risco, D.; Gutierrez-Merinet, J. Lactobacilli Isolated From Wild Boar (Sus scrofa) Antagonize Mycobacterium bovis Bacille Calmette-Guerin (BCG) in a Species-Dependent Manner. Front. Microbiol. 2019, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Sivaraj, A.; Sundar, R.; Manikkam, R.; Parthasarathy, K.; Rani, U.; Kumar, V. Potential applications of lactic acid bacteria and bacteriocins in anti-mycobacterial therapy. Asian Pac. J. Trop. Med. 2018, 11, 453. [Google Scholar] [CrossRef]
- Sosunov, V.; Mischenko, V.; Eruslanov, B.; Svetoch, E.; Shakina, Y.; Stern, N.; Majorov, K.; Sorokoumova, G.; Selishcheva, A.; Apt, A. Antimycobacterial activity of bacteriocins and their complexes with liposomes. J. Antimicrob. Chemother. 2007, 59, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; Stefani, S.; de Niederhausern, S.; Bondi, M.; Sabia, C.; Messi, P. Characterization of Anti-Listeria monocytogenes Properties of two Bacteriocin-Producing Enterococcus mundtii Isolated from Fresh Fish and Seafood. Curr. Microbiol. 2019, 76, 1010–1019. [Google Scholar] [CrossRef]
- Fox, G.J.; Orlova, M.; Schurr, E. Tuberculosis in Newborns: The Lessons of the “Lübeck Disaster” (1929–1933). PLoS Pathog. 2016, 12, e1005271. [Google Scholar] [CrossRef]
- Bouzid, F.; Brégeon, F.; Lepidi, H.; Donoghue, H.D.; Minnikin, D.E.; Drancourt, M. Ready Experimental Translocation of Mycobacterium canettii Yields Pulmonary Tuberculosis. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Zitvogel, L.; Kroemer, G. Immunostimulatory gut bacteria. Science 2019, 366, 1077–1078. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Naidoo, C.C.; Nyawo, G.R.; Wu, B.G.; Walzl, G.; Warren, R.M.; Segal, L.N.; Theron, G. The microbiome and tuberculosis: State of the art, potential applications, and defining the clinical research agenda. Lancet Respir. Med. 2019, 7, 892–906. [Google Scholar] [CrossRef]
- Lachmandas, E.; van den Heuvel, C.N.A.M.; Damen, M.S.M.A.; Cleophas, M.C.P.; Netea, M.G.; van Crevel, R. Diabetes Mellitus and Increased Tuberculosis Susceptibility: The Role of Short-Chain Fatty Acids. J. Diabetes Res. 2016, 2016, e6014631. [Google Scholar] [CrossRef] [PubMed]
- Chingwaru, W.; Mpuchane, S.F.; Gashe, B.A. Enterococcus faecalis and Enterococcus faecium Isolates from Milk, Beef, and Chicken and Their Antibiotic Resistance. J. Food Prot. 2003, 66, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.A.; Pot, B.; Van Damme, L.; Kersters, K.; Haesebrouck, F. Identification of Enterococcus species isolated from foods of animal origin. Int. J. Food Microbiol. 1995, 26, 187–197. [Google Scholar] [CrossRef]

| Patients | Sex | Age | Weight | Size | BMI | Smear Result | Sputum Culture | GeneXpert |
|---|---|---|---|---|---|---|---|---|
| CR017 | M | 45 | 90 | 1.81 | 27 | negative | negative | negative |
| CR018 | M | 65 | 56 | 1.68 | 19 | positive+++ | positive | positive |
| CR019 | F | 28 | 50 | 1.60 | 19.53 | positive+++ | positive | positive |
| CR020 | F | 22 | 73 | 1.66 | 26.54 | negative | negative | negative |
| MTBC Strain | DPBS | Mixture | E. mundtii | p-Value * | E. casseliflavus | p-Value * |
|---|---|---|---|---|---|---|
| M. tuberculosis L 2 | 0 | 8.4 ± 1.2 | 10.83 ± 1.77 | 0.015 | 9.1 ± 1.88 | 0.023 |
| M. tuberculosis L1 | 0 | - | 10.66 ± 3.40 | 0.032 | 9.83 ± 1.75 | 0.0104 |
| M. tuberculosis L 3 | 0 | - | 8.16 ± 1.04 | 0.005 | 6.33 ± 1.52 | 0.01884 |
| M. tuberculosis L 4 | 0 | - | 9.66 ± 3.05 | 0.031 | 8.83 ± 3 | 0.000516 |
| M. africanum | 0 | - | 9.66 ± 1.52 | 0.008 | 8.33 ± 1.52 | 0.011015 |
| M. canettii | 0 | - | 29.33 ± 1.15 | 0.0005 | 27.33 ± 2.08 | 0.00192 |
| M. bovis | 0 | - | 11.33 ± 2.08 | 0.011 | 10.66 ± 2.51 | 0.0180 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fellag, M.; Gouba, N.; Bedotto, M.; Sakana, M.; Zingué, D.; Tarnagda, Z.; Million, M.; Drancourt, M. Culturomics Discloses Anti-Tubercular Enterococci Exclusive of Pulmonary Tuberculosis: A Preliminary Report. Microorganisms 2020, 8, 1544. https://doi.org/10.3390/microorganisms8101544
Fellag M, Gouba N, Bedotto M, Sakana M, Zingué D, Tarnagda Z, Million M, Drancourt M. Culturomics Discloses Anti-Tubercular Enterococci Exclusive of Pulmonary Tuberculosis: A Preliminary Report. Microorganisms. 2020; 8(10):1544. https://doi.org/10.3390/microorganisms8101544
Chicago/Turabian StyleFellag, Mustapha, Nina Gouba, Marielle Bedotto, Moussa Sakana, Dezemon Zingué, Zékiba Tarnagda, Matthieu Million, and Michel Drancourt. 2020. "Culturomics Discloses Anti-Tubercular Enterococci Exclusive of Pulmonary Tuberculosis: A Preliminary Report" Microorganisms 8, no. 10: 1544. https://doi.org/10.3390/microorganisms8101544
APA StyleFellag, M., Gouba, N., Bedotto, M., Sakana, M., Zingué, D., Tarnagda, Z., Million, M., & Drancourt, M. (2020). Culturomics Discloses Anti-Tubercular Enterococci Exclusive of Pulmonary Tuberculosis: A Preliminary Report. Microorganisms, 8(10), 1544. https://doi.org/10.3390/microorganisms8101544





