Two Homologues of the Global Regulator Csr/Rsm Redundantly Control Phaseolotoxin Biosynthesis and Virulence in the Plant Pathogen Pseudomonas amygdali pv. phaseolicola 1448A
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Molecular Procedures
2.3. Bioinformatics Tools
2.4. Mutagenesis and Cloning of rsm Genes
2.5. Pathogenicity Assays and Autoagglutination
2.6. Assays of Motility and Biosynthesis of Phaseolotoxin
2.7. Carbon Source Utilization
3. Results and Discussion
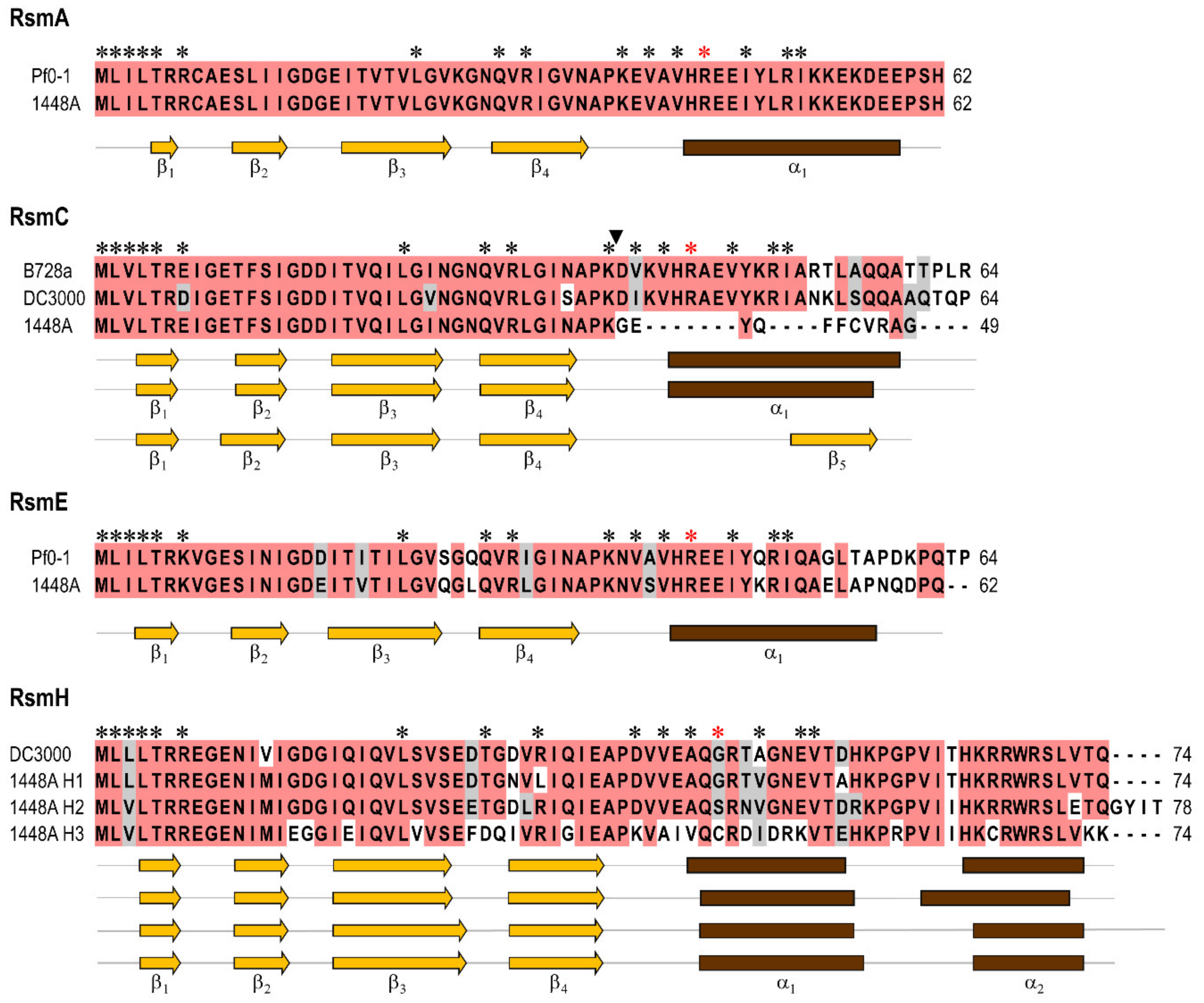
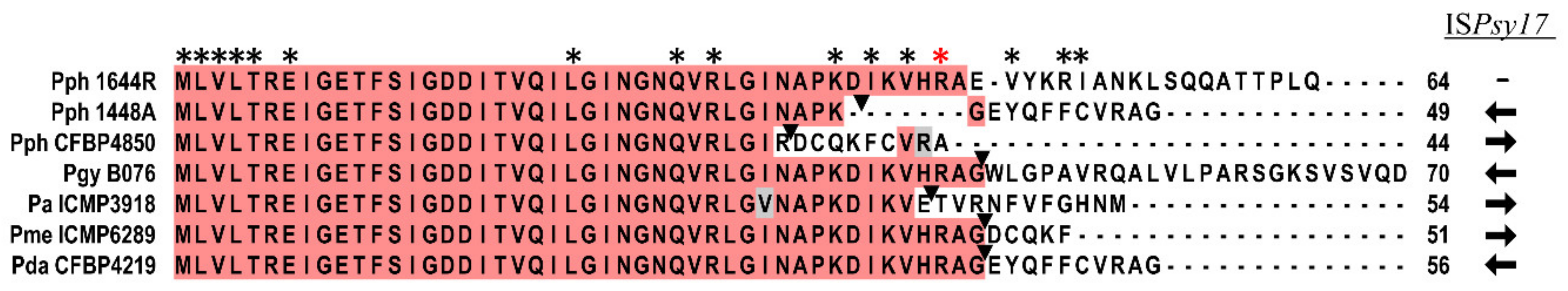
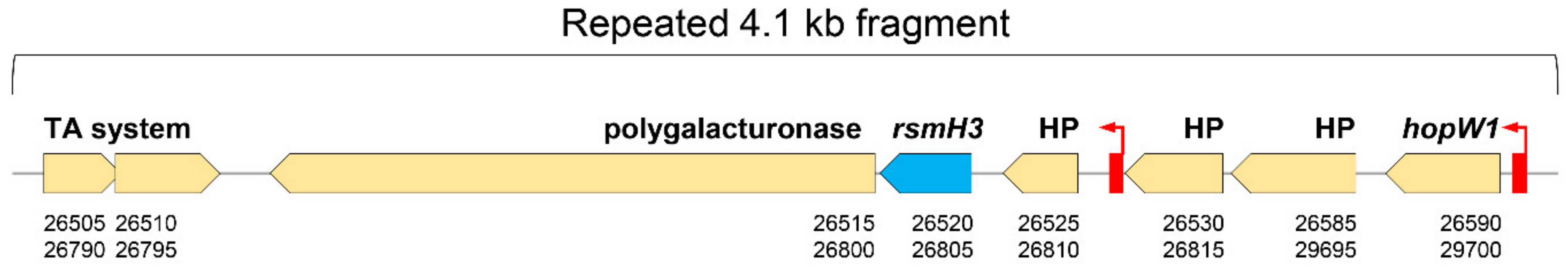
3.1. Pseudomonas amygdali pv. phaseolicola Contains Seven rsm Homologues
3.2. Construction of Single and Multiple rsm Mutants
3.3. Expression Levels of rsm Genes
3.4. Genes rsmA and rsmE Redundantly Control Virulence in Bean and the Expression of the T3SS
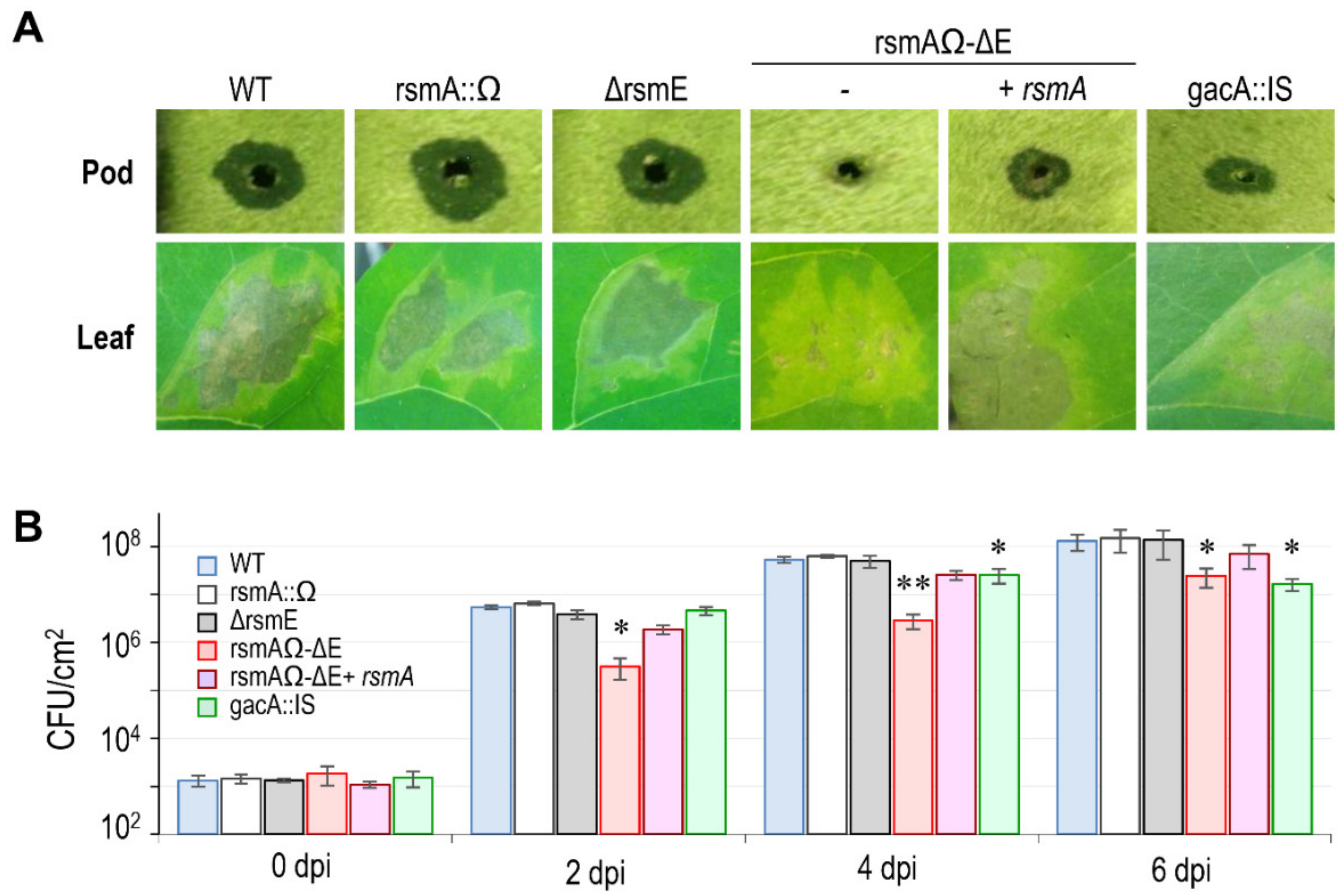
3.4.1. Virulence in bean
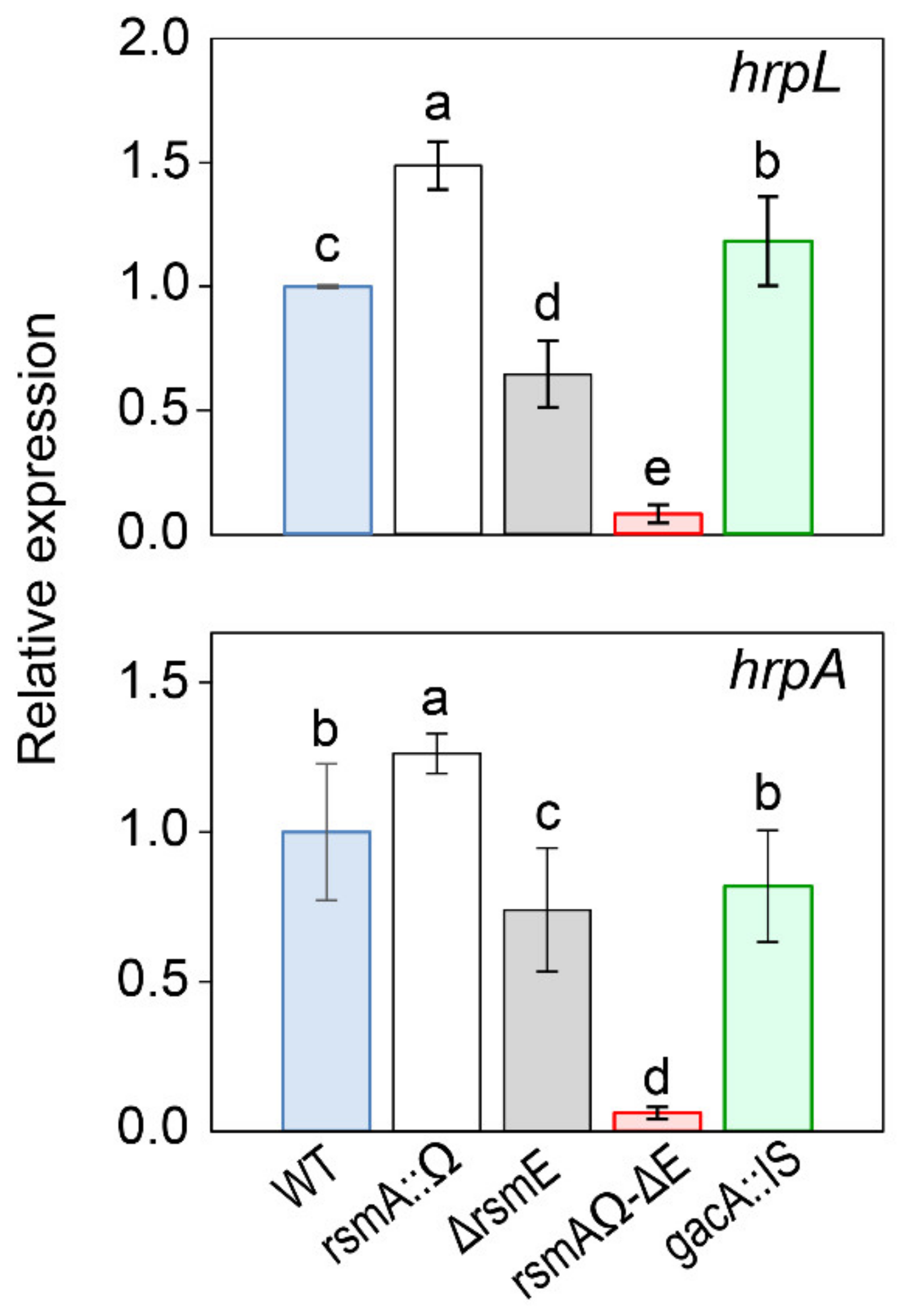
3.4.2. Expression of the T3SS.
3.5. Genes rsmA and rsmE also Redundantly Control Motility and Carbon Source Utilization
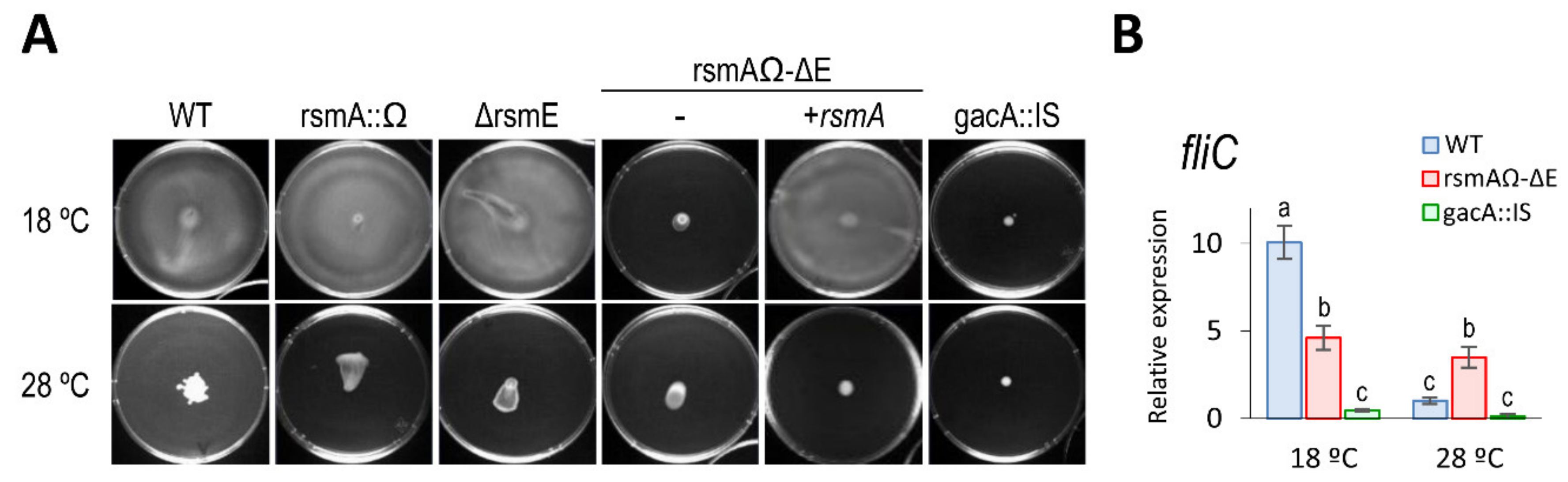
3.5.1. Motility
3.5.2. Carbon Source Utilization
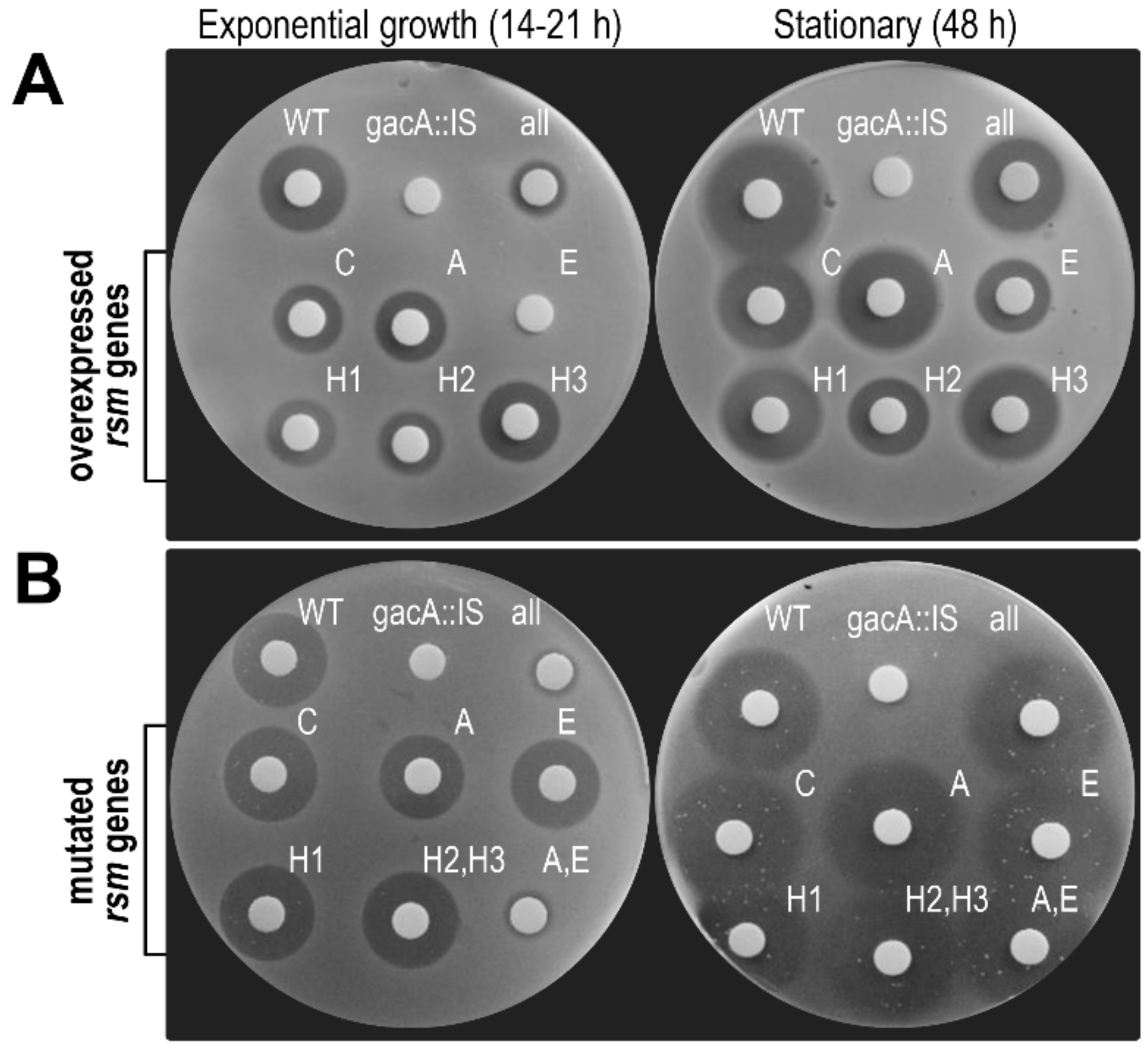
3.6. RsmA and RsmE Redundantly Contribute to the Activation of Phaseolotoxin Biosynthesis at 18 °C
3.6.1. Toxin Biosynthesis by Strain 1448A Depends on GacA and GacS
3.6.2. Overexpression of rsmE Suppresses Phaseolotoxin Synthesis only in Exponentially Growing Cultures
3.6.3. Double rsmA–rsmE Mutants are Unable to Synthesize Phaseolotoxin only in Exponentially Growing Cultures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Romeo, T.; Babitzke, P. Global regulation by CsrA and its RNA antagonists. In Regulating with RNA in Bacteria and Archaea; Storz, G., Papenfort, K., Eds.; ASM Press: Washington, DC, USA, 2019; pp. 341–354. [Google Scholar] [CrossRef]
- Sobrero, P.M.; Valverde, C. Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front. Mol. Biosci. 2020, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant.-Microbe Interact. 2001, 14, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.S.; Morozov, I.; Suzuki, K.; Romeo, T.; Babitzke, P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 2002, 44, 1599–1610. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Potts, A.H.; Babitzke, P.; Ahmer, B.M.M.; Romeo, T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 2015, 79, 193–224. [Google Scholar] [CrossRef] [PubMed]
- Marden, J.N.; Diaz, M.R.; Walton, W.G.; Gode, C.J.; Betts, L.; Urbanowski, M.L.; Redinbo, M.R.; Yahr, T.L.; Wolfgang, M.C. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2013, 110, 15055–15060. [Google Scholar] [CrossRef] [PubMed]
- Huertas-Rosales, Ó.; Ramos-González, M.I.; Espinosa-Urgel, M. Self-regulation and interplay of Rsm family proteins modulate the lifestyle of Pseudomonas putida. Appl. Environ. Microbiol. 2016, 82, 5673–5686. [Google Scholar] [CrossRef]
- Abbott, Z.D.; Flynn, K.J.; Byrne, B.G.; Mukherjee, S.; Kearns, D.B.; Swanson, M.S. csrT represents a new class of csrA-like regulatory genes associated with integrative conjugative elements of Legionella pneumophila. J. Bacteriol. 2016, 198, 553–564. [Google Scholar] [CrossRef]
- Ferreiro, M.D.; Nogales, J.; Farias, G.A.; Olmedilla, A.; Sanjuan, J.; Gallegos, M.T. Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol. Plant.-Microbe Interact. 2018, 31, 525–536. [Google Scholar] [CrossRef]
- Ge, Y.; Lee, J.H.; Liu, J.; Yang, H.-w.; Tian, Y.; Hu, B.; Zhao, Y. Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant. Pathol. 2019, 20, 1217–1236. [Google Scholar] [CrossRef]
- Lapouge, K.; Schubert, M.; Allain, F.H.; Haas, D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef]
- Duss, O.; Michel, E.; Yulikov, M.; Schubert, M.; Jeschke, G.; Allain, F.H.T. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 2014, 509, 588–592. [Google Scholar] [CrossRef]
- Liu, M.Y.; Gui, G.J.; Wei, B.D.; Preston, J.F.; Oakford, L.; Yuksel, U.; Giedroc, D.P.; Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997, 272, 17502–17510. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; Haas, D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 2011, 91, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Janssen, K.H.; Diaz, M.R.; Gode, C.J.; Wolfgang, M.C.; Yahr, T.L. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol. 2018, 200, e00277-00218. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Busquets, A.; Mulet, M.; García-Valdés, E.; Lalucat, J. Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analysis. Front. Microbiol. 2017, 8, 2422. [Google Scholar] [CrossRef] [PubMed]
- Arnold, D.L.; Lovell, H.C.; Jackson, R.W.; Mansfield, J.W. Pseudomonas syringae pv. phaseolicola: From ‘has bean’ to supermodel. Mol. Plant. Pathol. 2011, 12, 617–627. [Google Scholar] [CrossRef]
- Noble, T.J.; Young, A.J.; Douglas, C.A.; Williams, B.; Mundree, S. Diagnosis and management of halo blight in Australian mungbeans: A review. Crop. Pasture Sci. 2019, 70, 195–203. [Google Scholar] [CrossRef]
- Bachmann, A.S.; Matile, P.; Slusarenko, A.J. Inhibition of ornithine decarboxylase activity by phaseolotoxin: Implications for symptom production in halo blight of French bean. Physiol. Mol. Plant. Pathol. 1998, 53, 287–299. [Google Scholar] [CrossRef]
- Arrebola, E.; Cazorla, F.M.; Perez-Garcia, A.; de Vicente, A. Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins 2011, 3, 1089–1110. [Google Scholar] [CrossRef]
- Aguilera, S.; López-López, K.; Nieto, Y.; Garcidueñas-Piña, R.; Hernández-Guzmán, G.; Hernández-Flores, J.L.; Murillo, J.; Alvarez-Morales, A. Functional characterization of the gene cluster from Pseudomonas syringae pv. phaseolicola NPS3121 involved in synthesis of phaseolotoxin. J. Bacteriol. 2007, 189, 2834–2843. [Google Scholar] [CrossRef]
- Arvizu-Gómez, J.L.; Hernández-Morales, A.; Pacheco Aguilar, J.R.; Álvarez-Morales, A. Transcriptional profile of P. syringae pv. phaseolicola NPS3121 at low temperature: Physiology of phytopathogenic bacteria. BMC Microbiol. 2013, 13, 81. [Google Scholar] [CrossRef]
- Rich, J.J.; Hirano, S.S.; Willis, D.K. Pathovar-specific requirement for the Pseudomonas syringae lemA gene in disease lesion formation. Appl. Environ. Microbiol. 1992, 58, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.J.; Kinscherf, T.G.; Kitten, T.; Wilis, D.K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 1994, 176, 7468–7475. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Cui, Y.; Yang, H.; Collmer, A.; Alfano, J.R.; Chatterjee, A.K. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant.-Microbe Interact. 2003, 16, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Lee, D.G.; Lee, J.S.; Oh, J.-I.; Baik, H.S. GacA directly regulates expression of several virulence genes in Pseudomonas syringae pv. tabaci 11528. Biochem. Biophys. Res. Commun. 2012, 417, 665–672. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, Y.; Wu, Z.; Tan, G. Characterization of response regulator GacA involved in phaseolotoxin production, hypersensitive response and cellular processes in Pseudomonas syringae pv. actinidiae A18. Physiol. Mol. Plant. Pathol. 2018, 103, 137–142. [Google Scholar] [CrossRef]
- Ortiz-Martín, I.; Thwaites, R.; Macho, A.P.; Mansfield, J.W.; Beuzón, C.R. Positive regulation of the Hrp Type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant.-Microbe Interact. 2010, 23, 665–681. [Google Scholar] [CrossRef]
- Rowley, K.B.; Xu, R.; Patil, S.S. Molecular analysis of thermoregulation of phaseolotoxin-resistant ornithine carbamoyltransferase (argK) from Pseudomonas syringae pv. phaseolicola. Mol. Plant.-Microbe Interact. 2000, 13, 1071–1080. [Google Scholar] [CrossRef]
- Valverde, C.; Heeb, S.; Keel, C.; Haas, D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 2003, 50, 1361–1379. [Google Scholar] [CrossRef]
- De la Torre-Zavala, S.; Aguilera, S.; Ibarra-Laclette, E.; Hernández-Flores, J.L.; Hernández-Morales, A.; Murillo, J.; Alvarez-Morales, A. Gene expression of Pht cluster genes and a putative non-ribosomal peptide synthetase required for phaseolotoxin production is regulated by GacS/GacA in Pseudomonas syringae pv. phaseolicola. Res. Microbiol. 2011, 162, 488–498. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- King, E.O.; Ward, N.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.S.; Quigley, M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981, 114, 193–197. [Google Scholar] [CrossRef]
- Choi, K.H.; Kumar, A.; Schweizer, H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Meth. 2006, 64, 391–397. [Google Scholar] [CrossRef]
- Joardar, V.; Lindeberg, M.; Jackson, R.W.; Selengut, J.; Dodson, R.; Brinkac, L.M.; Daugherty, S.C.; DeBoy, R.; Durkin, A.S.; Giglio, M.G.; et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 2005, 187, 6488–6498. [Google Scholar] [CrossRef]
- Matas, I.M.; Castañeda-Ojeda, M.P.; Aragón, I.M.; Antúnez-Lamas, M.; Murillo, J.; Rodríguez-Palenzuela, P.; López-Solanilla, E.; Ramos, C. Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant.-Microbe Interact. 2014, 27, 424–436. [Google Scholar] [CrossRef]
- Bardaji, L.; Añorga, M.; Ruiz-Masó, J.A.; del Solar, G.; Murillo, J. Plasmid replicons from Pseudomonas are natural chimeras of functional, exchangeable modules. Front. Microbiol. 2017, 8, 190. [Google Scholar] [CrossRef]
- Huynh, T.V.; Dahlbeck, D.; Staskawicz, B.J. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science 1989, 245, 1374–1377. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/.
- Meyer, J.M.; Abdallah, M.A. The fluorescent pigment of Pseudomonas fluorescens: Biosynthesis, purification and physicochemical properties. Microbiology 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Falgueras, J.; Lara, A.J.; Fernandez-Pozo, N.; Canton, F.R.; Perez-Trabado, G.; Gonzalo Claros, M. SeqTrim: A high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics 2010, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Giddens, S.R.; Jackson, R.W.; Moon, C.D.; Jacobs, M.A.; Zhang, X.-X.; Gehrig, S.M.; Rainey, P.B. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc. Natl. Acad. Sci. USA 2007, 104, 18247–18252. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Prentki, P.; Krisch, H.M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 1984, 29, 303–313. [Google Scholar] [CrossRef]
- Newman, J.R.; Fuqua, C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 1999, 227, 197–203. [Google Scholar] [CrossRef]
- Harper, S.; Zewdie, N.; Brown, I.R.; Mansfield, J.W. Histological, physiological and genetical studies of the responses of leaves and pods of Phaseolus vulgaris to three races of Pseudomonas syringae pv. phaseolicola and to Pseudomonas syringae pv. coronafaciens. Physiol. Mol. Plant. Pathol. 1987, 31, 153–172. [Google Scholar] [CrossRef]
- Yessad, S.; Manceau, C.; Luisetti, J. A detached leaf assay to evaluate virulence and pathogenicity of strains of Pseudomonas syringae pv. syringae on pear. Plant. Dis. 1992, 76, 370–373. [Google Scholar] [CrossRef]
- Bertoni, G.; Mills, D. A simple method to monitor growth of baterial populations in leaf tissue. Phytopathology 1987, 77, 832–835. [Google Scholar] [CrossRef]
- Taira, S.; Tuimala, J.; Roine, E.; Nurmiaho-Lassila, E.L.; Savilahti, H.; Romantschuk, M. Mutational analysis of the Pseudomonas syringae pv. tomato hrpA gene encoding Hrp pilus subunit. Mol. Microbiol. 1999, 34, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Staskawicz, B.; Panopoulos, N.J. A rapid and sensitive microbiological assay for phaseolotoxin. Phytopathology 1979, 69, 663–666. [Google Scholar] [CrossRef]
- Hernández-Guzmán, G.; Alvarez-Morales, A. Isolation and characterization of the gene coding for the amidinotransferase involved in the biosynthesis of phaseolotoxin in Pseudomonas syringae pv. phaseolicola. Mol. Plant.-Microbe Interact. 2001, 14, 545–554. [Google Scholar] [CrossRef]
- Rico, A.; López, R.; Asensio, C.; Aizpún, M.; Asensio-S.-Manzanera, C.; Murillo, J. Nontoxigenic strains of P. syringae pv. phaseolicola are a main cause of halo blight of beans in Spain and escape current detection methods. Phytopathology 2003, 93, 1553–1559. [Google Scholar] [CrossRef]
- Carrión, V.J.; Arrebola, E.; Cazorla, F.M.; Murillo, J.; de Vicente, A. The mbo operon is specific and essential for biosynthesis of mangotoxin in Pseudomonas syringae. PLoS ONE 2012, 7, e36709. [Google Scholar] [CrossRef] [PubMed]
- Lelliott, R.A.; Stead, D.E. Methods for the Diagnosis of Bacterial Diseases of Plants; Blackwell Scientific Publications: Oxford, UK, 1987; Volume 2. [Google Scholar]
- Palmer, D.A.; Bender, C.L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl. Environ. Microbiol. 1993, 59, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Kuehne, S.A.; Bycroft, M.; Crivii, S.; Allen, M.D.; Haas, D.; Cámara, M.; Williams, P. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 2006, 355, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Neubacher, S.; Grossmann, T.N. Protein–RNA interactions: Structural characteristics and hotspot amino acids. RNA 2018, 24, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Lapouge, K.; Duss, O.; Oberstrass, F.C.; Jelesarov, I.; Haas, D.; Allain, F.H.T. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 2007, 14, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Bardaji, L.; Añorga, M.; Jackson, R.W.; Martínez-Bilbao, A.; Yanguas, N.; Murillo, J. Miniature transposable sequences are frequently mobilized in the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola. PLoS ONE 2011, 6, e25773. [Google Scholar] [CrossRef] [PubMed]
- Zumaquero, A.; Macho, A.P.; Rufian, J.S.; Beuzon, C.R. Analysis of the role of the Type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448A in interaction with the plant. J. Bacteriol. 2010, 192, 4474–4488. [Google Scholar] [CrossRef] [PubMed]
- Vencato, M.; Tian, F.; Alfano, J.R.; Buell, C.R.; Cartinhour, S.; DeClerck, G.A.; Guttman, D.S.; Stavrinides, J.; Joardar, V.; Lindeberg, M.; et al. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant.-Microbe Interact. 2006, 19, 1193–1206. [Google Scholar] [CrossRef]
- Mucyn, T.S.; Yourstone, S.; Lind, A.L.; Biswas, S.; Nishimura, M.T.; Baltrus, D.A.; Cumbie, J.S.; Chang, J.H.; Jones, C.D.; Dangl, J.L.; et al. Variable suites of non-effector genes are co-regulated in the Type III Secretion virulence regulon across the Pseudomonas syringae phylogeny. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef][Green Version]
- Hockett, K.L.; Burch, A.Y.; Lindow, S.E. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS ONE 2013, 8, e59850. [Google Scholar] [CrossRef]
- O’Malley, M.R.; Weisberg, A.J.; Chang, J.H.; Anderson, J.C. Re-evaluation of a Tn5::gacA mutant of Pseudomonas syringae pv. tomato DC3000 uncovers roles for uvrC and anmK in promoting virulence. PLoS ONE 2019, 14, e0223637. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, Y.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Ichinose, Y. Quorum-dependent expression of rsmX and rsmY, small non-coding RNAs, in Pseudomonas syringae. Microbiol. Res. 2019, 223-225, 72–78. [Google Scholar] [CrossRef]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009, 73, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Humair, B.; Wackwitz, B.; Haas, D. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 2010, 76, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lund, S.P.; Greenwald, J.W.; Records, A.H.; Scott, R.A.; Nettleton, D.; Lindow, S.E.; Gross, D.C.; Beattie, G.A. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 2014, 5, e01683-01614. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Romero, M.; Karna, S.L.R.; Chen, T.; Heeb, S.; Leung, K.P. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol. 2016, 16, 155. [Google Scholar] [CrossRef]
- Xie, Y.P.; Shao, X.L.; Deng, X. Regulation of type III secretion system in Pseudomonas syringae. Environ. Microbiol. 2019, 21, 4465–4477. [Google Scholar] [CrossRef]
- Oh, H.-S.; Kvitko, B.H.; Morello, J.E.; Collmer, A. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 2007, 189, 8277–8289. [Google Scholar] [CrossRef]
- Kong, H.S.; Roberts, D.P.; Patterson, C.D.; Kuehne, S.A.; Heeb, S.; Lakshman, D.K.; Lydon, J. Effect of overexpressing rsmA from Pseudomonas aeruginosa on virulence of select phytotoxin-producing strains of P. syringae. Phytopathology 2012, 102, 575–587. [Google Scholar] [CrossRef]
- O’Malley, M.R.; Chien, C.-F.; Peck, S.C.; Lin, N.-C.; Anderson, J.C. A revised model for the role of GacS/GacA in regulating type III secretion by Pseudomonas syringae pv. tomato DC3000. Mol. Plant. Pathol. 2020, 21, 139–144. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.M.; Neale, H.C.; Geilfus, C.-M.; Jackson, R.W.; Arnold, D.L.; Preston, G.M. Early changes in apoplast composition associated with defence and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae pv. phaseolicola. Plant. Cell Environ. 2016, 39, 2172–2184. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Willis, D.K. Involvement of the lemA gene in production of syringomycin and protease by Pseudomonas syringae pv. syringae. Mol. Plant.-Microbe Interact. 1993, 6, 368–375. [Google Scholar] [CrossRef]
- Barta, T.M.; Kinscherf, T.G.; Willis, D.K. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J. Bacteriol. 1992, 174, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Williams, F.; Hindle, Z.; Heurlier, K.; Holden, M.T.G.; Cámara, M.; Haas, D.; Williams, P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6676–6683. [Google Scholar] [CrossRef]

| Strains | Main featuresa | Reference or source |
|---|---|---|
| 1448A | Wild type (WT) strain, isolated from Phaseolus in Ethiopia, 1985 | [37] |
| ΔpA | UPN1162; 1448A cured of plasmid p1448A-A; ∆rsmH2 ∆rsmH3-1 ∆rsmH3-2 | D. Ramírez-Zapata, unpublished |
| ∆rsmE | UPN1168; 1448A ∆rsmE | This work |
| ΔrsmC | UPN1187; 1448A ∆rsmC | This work |
| rsmA::Ω | UPN1225; 1448A rsmA::Ω | This work |
| rsmAΩ-∆E | UPN1227; derives from UPN1168 rsmA::Ω ∆rsmE | This work |
| Mut-7-rsm | UPN1229; derives from UPN1185 ∆rsmC rsmA::Ω ∆rsmE rsmH1-fsX ∆rsmH2 ∆rsmH3-1 ∆rsmH3-2 | This work |
| gacA::IS | UPN1230; 1448A gacA::IS-Ω-Km/hah | This work |
| gacS::IS | UPN1362; 1448A gacS::IS-Ω-Km/hah | This work |
| FPKMa | Fold change (log2)b | |||||||
|---|---|---|---|---|---|---|---|---|
| WT | gacA::IS | 18 °C/28 °C | gacA::IS/WT | |||||
| Gene | 18 °C | 28 °C | 18 °C | 28 °C | WT | gacA::IS | 18 °C | 28 °C |
| gacS | 81.3 | 71.5 | 44.1 | 40.9 | 0.2 | 0.1 | −0.9 | −0.8 |
| gacA | 347.3 | 354.7 | 200.3 | 144.9 | 0.0 | 0.5 | 0.8 | −1.3 |
| uvrC | 115.3 | 140.1 | 3.4 | 4.5 | −0.3 | −0.4 | −5.1 | −5.0 |
| rsmA | 932.9 | 565.1 | 1235.0 | 844.0 | 0.7 | 0.6 | 0.4 | 0.6 |
| rsmC | 53.8 | 75.1 | 67.1 | 43.9 | −0.5 | −0.6 | −0.3 | 0.2 |
| rsmE | 614.6 | 241.7 | 179.2 | 204.9 | 1.4 | −0.2 | −1.8 | 0.2 |
| rsmH1 | 179.8 | 228.2 | 168.2 | 280.5 | −0.3 | −0.7 | −0.1 | 0.3 |
| rsmH2 | 88.1 | 193.2 | 93.6 | 155.0 | −1.1 | −0.7 | 0.1 | −0.3 |
| rsmH3-1 | 96.2 | 169.5 | 86.2 | 419.2 | −0.8 | −2.3 | −0.2 | 1.3 |
| rsmH3-2 | 99.6 | 171.4 | 86.5 | 412.0 | −0.8 | −2.3 | −0.2 | 1.3 |
| sRNAs | ||||||||
| rsmX1 | 61440.0 | 181313.0 | 980.2 | 2283.9 | −1.6 | −1.2 | −6.0 | −6.3 |
| rsmX2 | 85990.4 | 129285.0 | 765.7 | 1228.2 | −0.6 | −0.7 | −6.8 | −6.7 |
| rsmX3 | 54830.1 | 51529.8 | 116.8 | 400.4 | 0.1 | −1.8 | −8.9 | −7.0 |
| rsmX4 | 338.7 | 737.1 | 71.4 | 100.1 | −1.1 | −0.5 | −2.3 | −2.9 |
| rsmX5 | 682.1 | 2853.5 | 32.6 | 26.0 | −2.1 | 0.3 | −4.4 | −6.8 |
| rsmY | 15372.1 | 11471.4 | 330.3 | 966.0 | 0.4 | −1.6 | −5.5 | −3.6 |
| rsmZ | 791.7 | 2504.5 | 357.5 | 2712.2 | −1.7 | −2.9 | −1.2 | 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Zapata, D.; Ramos, C.; Aguilera, S.; Bardaji, L.; Martínez-Gil, M.; Murillo, J. Two Homologues of the Global Regulator Csr/Rsm Redundantly Control Phaseolotoxin Biosynthesis and Virulence in the Plant Pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms 2020, 8, 1536. https://doi.org/10.3390/microorganisms8101536
Ramírez-Zapata D, Ramos C, Aguilera S, Bardaji L, Martínez-Gil M, Murillo J. Two Homologues of the Global Regulator Csr/Rsm Redundantly Control Phaseolotoxin Biosynthesis and Virulence in the Plant Pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms. 2020; 8(10):1536. https://doi.org/10.3390/microorganisms8101536
Chicago/Turabian StyleRamírez-Zapata, Diana, Cayo Ramos, Selene Aguilera, Leire Bardaji, Marta Martínez-Gil, and Jesús Murillo. 2020. "Two Homologues of the Global Regulator Csr/Rsm Redundantly Control Phaseolotoxin Biosynthesis and Virulence in the Plant Pathogen Pseudomonas amygdali pv. phaseolicola 1448A" Microorganisms 8, no. 10: 1536. https://doi.org/10.3390/microorganisms8101536
APA StyleRamírez-Zapata, D., Ramos, C., Aguilera, S., Bardaji, L., Martínez-Gil, M., & Murillo, J. (2020). Two Homologues of the Global Regulator Csr/Rsm Redundantly Control Phaseolotoxin Biosynthesis and Virulence in the Plant Pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms, 8(10), 1536. https://doi.org/10.3390/microorganisms8101536







