Pepper Bacterial Spot Control by Bacillus velezensis: Bioprocess Solution
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. 16S rDNA Sequencing and Identification of Bacillus IP22
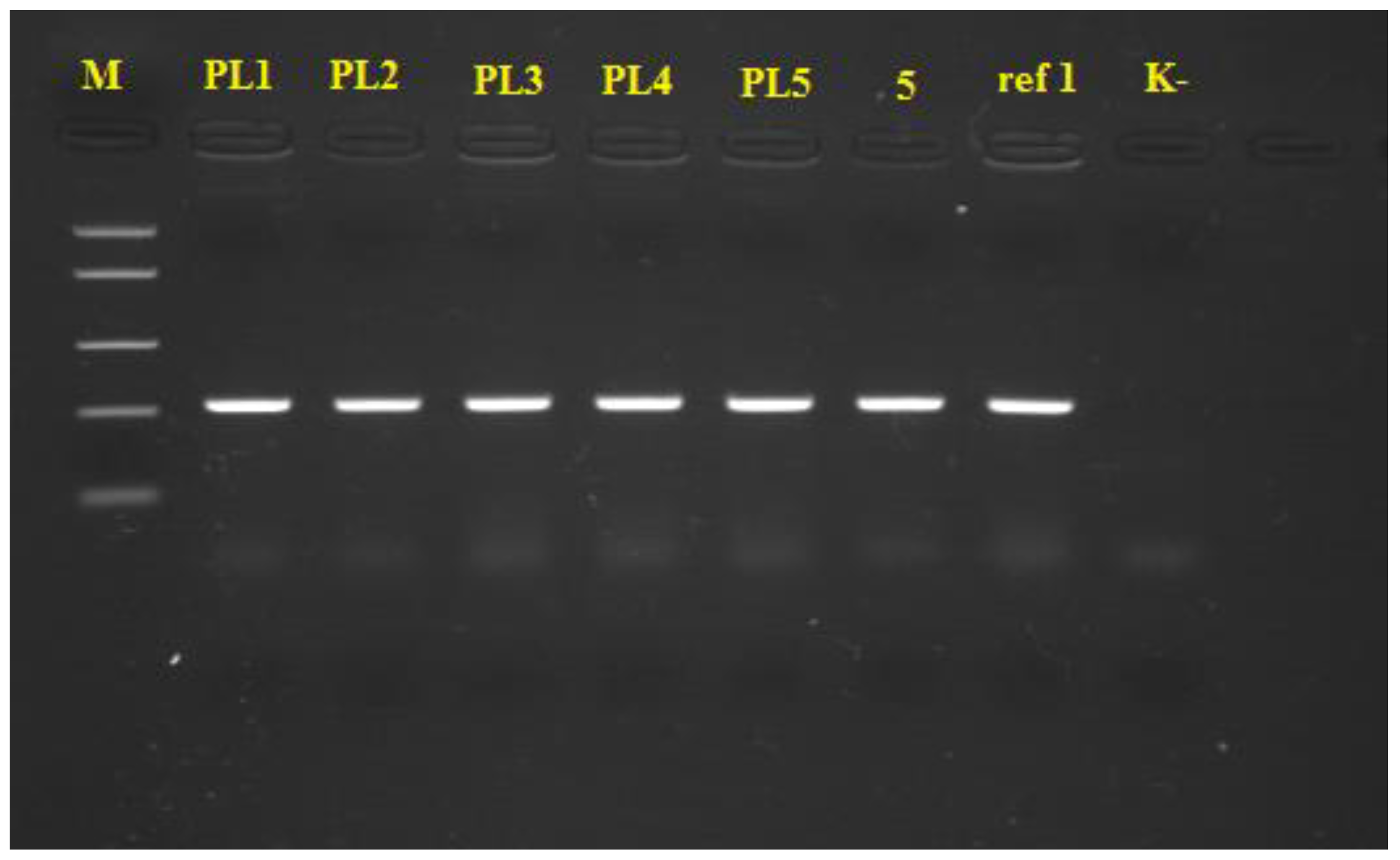
2.3. PCR Identification of Xanthomonas spp.
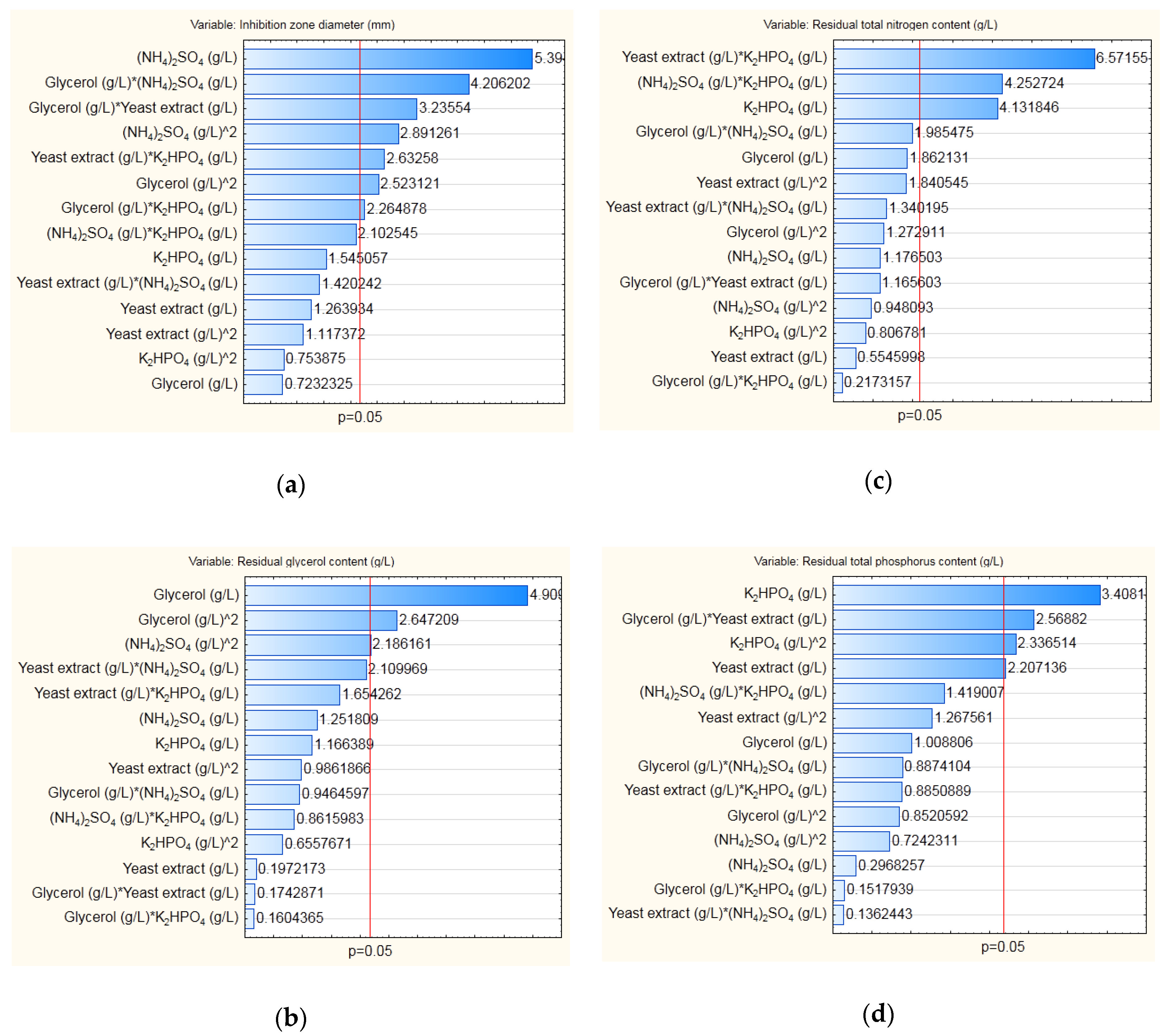
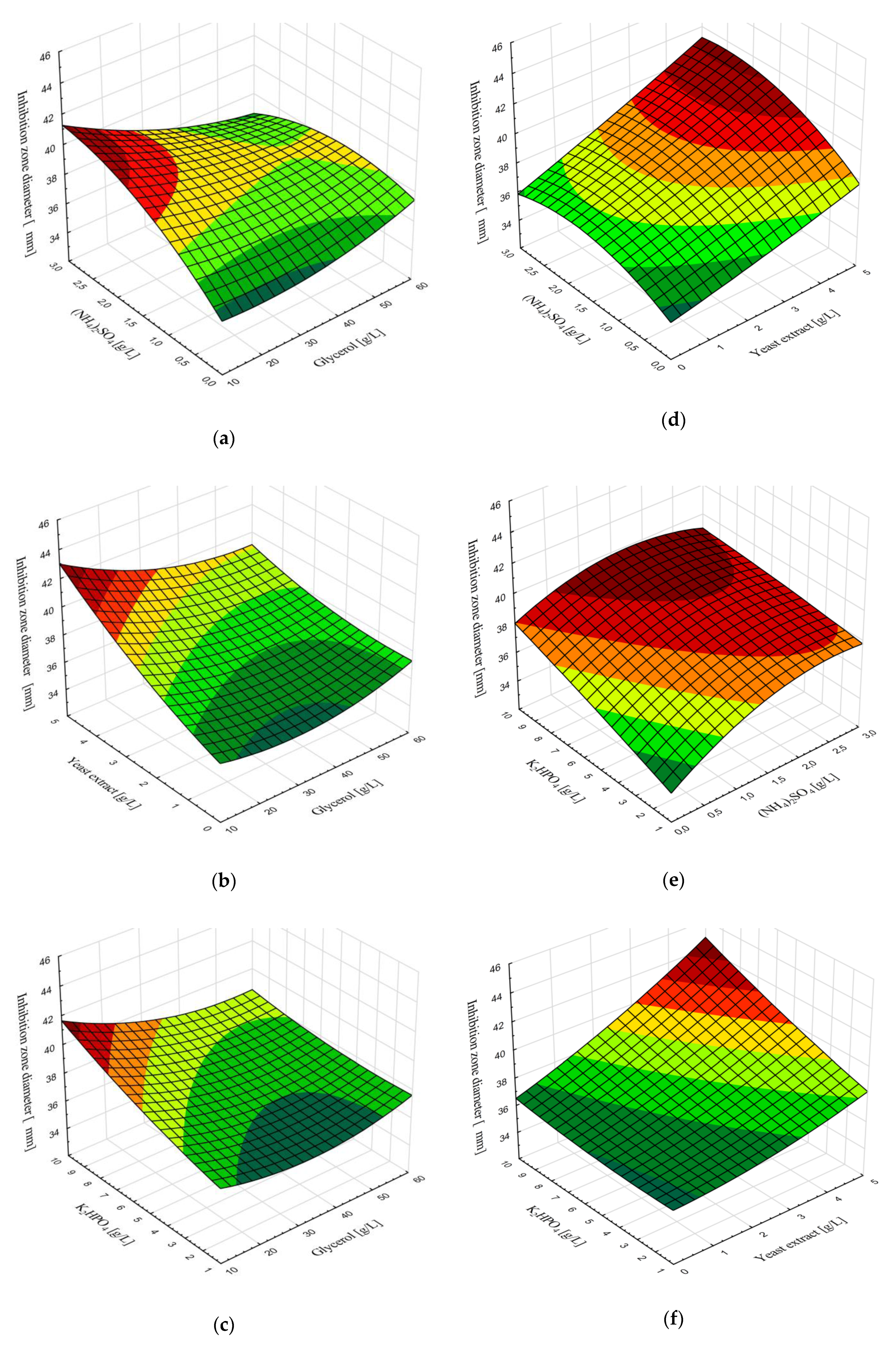
2.4. Modeling and Optimization of Cultivation Medium Composition
2.5. Validation Experiment
2.6. In Planta Experiments
2.7. Analytical Methods
2.7.1. In Vitro Antimicrobial Activity Assaying
2.7.2. Determination of Residual Content of Nutrients
2.7.3. Determination of Bacillus IP22 Biomass Content
2.7.4. HPLC-MS Analysis of the Antimicrobial Lipopeptides Produced by Bacillus IP22
2.8. Statistical Analysis of the Experimental Data
3. Results
3.1. 16S rDNA Sequencing and Identification of Bacillus IP22
3.2. PCR Identification of Xanthomonas spp.
3.3. Modeling and Optimization of Medium Composition for Cultivation of B. velezensis IP22
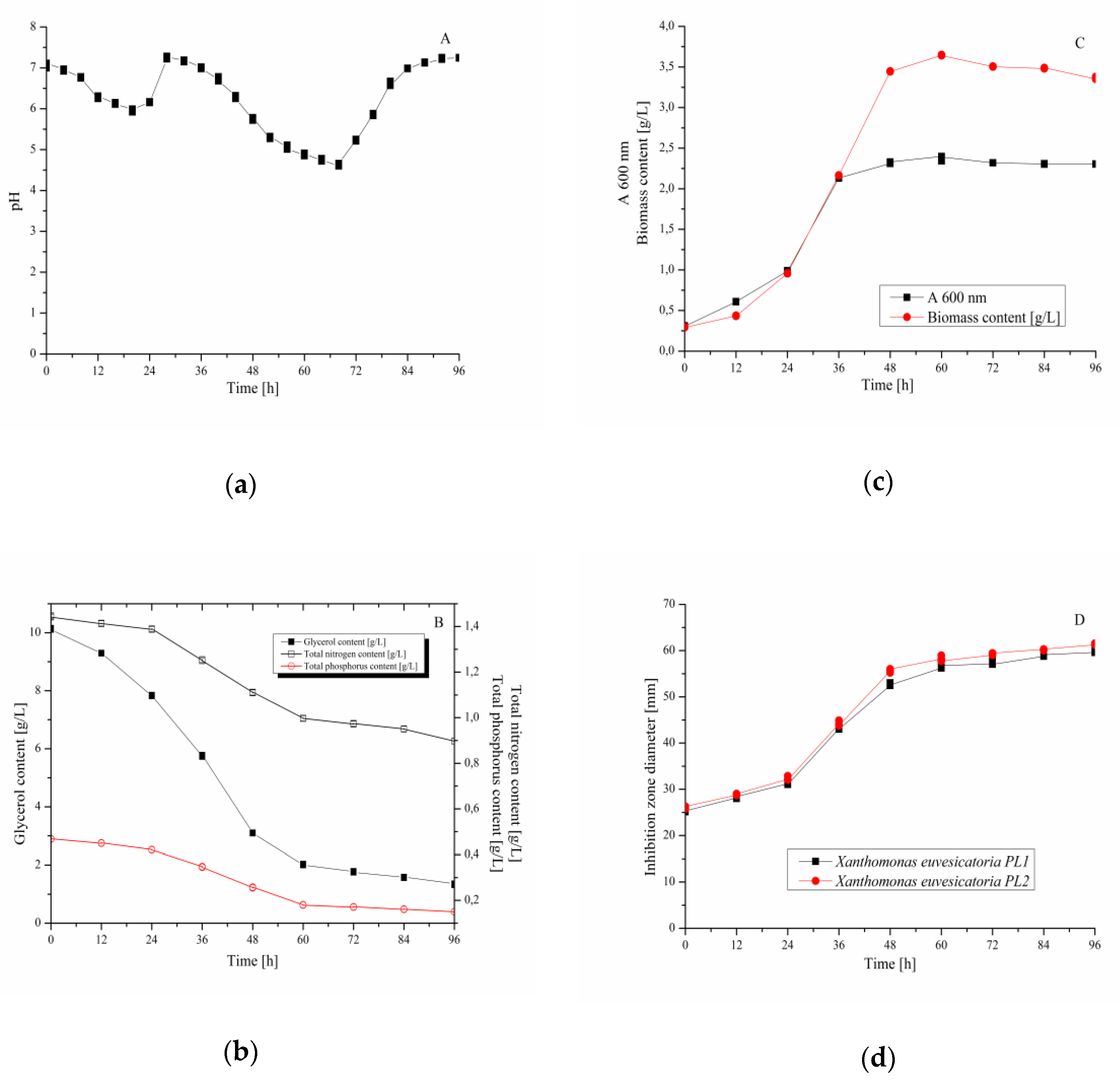
3.4. Validation Experiment–Cultivation of B. velezensis IP22 in a Laboratory-Scale Bioreactor
3.5. HPLC-MS Analysis of Antimicrobial Compounds Produced by B. velezensis IP22
3.6. In Planta Experiments with Pepper Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Potnis, N.; Timilsina, S.; Strayer, A.; Shantharaj, D.; Barak, J.D.; Paret, M.L.; Vallad, G.E.; Jones, J.B. Bacterial spot of tomato and pepper: Diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 2015, 16, 907–920. [Google Scholar] [CrossRef] [PubMed]
- EPPO. PM 7/110 (1) Xanthomonas spp. (Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. Oepp/Eppo Bull. 2013, 43, 7–20. [Google Scholar] [CrossRef]
- Bashan, Y.; Diab, S.; Okon, Y. Survival of Xanthomonas campestris pv. vesicatoria in pepper seeds and roots, in symptomless and dry leaves in non-host plants and in the soil. Plant Soil. 1982, 68, 161–170. [Google Scholar] [CrossRef]
- Diab, S.; Bashan, Y.; Okon, Y. Studies on infection with Xanthomonas campestris pv. vesicatoria, causal agent of bacterial scab of pepper in Israel. Phytoparasitica 1982, 10, 183–191. [Google Scholar] [CrossRef]
- Diab, S.; Bashan, Y.; Okon, Y.; Henis, Y. Effects of relative humidity on bacterial scab caused by Xanthomonas campestris pv. vesicatoria on pepper. Phytopathology 1982, 72, 1257–1260. [Google Scholar] [CrossRef]
- Zhang, Y.; Callaway, E.; Jones, J.B.; Wilson, M. Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur. J. Plant Pathol. 2009, 124, 379–390. [Google Scholar] [CrossRef]
- Hassan, E.O.; Zyton, M.A. Management of bacterial spot of pepper caused by Xanthomonas campestris pv. vesicatoria. Am. J. Biosci. Bioeng. 2017, 5, 41–49. [Google Scholar] [CrossRef]
- Ignjatov, M. Diversity of Xanthomonas spp. Population-Pepper Pathogens in Serbia. PhD Thesis, Faculty of Agriculture, University of Belgrade, Belgrade, Serbia, 2013. [Google Scholar]
- Aysan, Y.; Sahin, F. Occurrence of bacterial spot disease, caused by Xanthomonas axonopodis pv. vesicatoria on pepper in the eastern Mediterranean region of Turkey. Plant Pathol. 2003, 52, 781. [Google Scholar] [CrossRef]
- Balaž, J.; Delibašić, T. Devising methods for isolation of Xanthomonas campestris pv. vesicatoria from pepper seed. Pestic. Phytomed. 2005, 20, 51–60. [Google Scholar]
- Griffin, K.; Gambley, C.; Brown, P.; Li, Y. Copper-tolerance in Pseudomonas syringae pv. tomato and Xanthomonas spp. and the control of diseases associated with these pathogens in tomato and pepper. A systematic literature review. Crop. Prot. 2017, 96, 144–150. [Google Scholar] [CrossRef]
- Vallad, G.E.; Pernezny, K.L.; Balogh, B.; Wen, A.; Figueiredo, J.F.L.; Jones, J.B.; Momol, T.; Muchovej, R.M.; Havranek, N.; Abdallah, N.; et al. Comparison of kasugamycin to traditional bactericides for the management of bacterial spot on tomato. HortScience 2010, 45, 1834–1840. [Google Scholar] [CrossRef]
- Buonaurio, R.; Scarponi, L.; Ferrara, M.; Sidoti, P.; Bertona, A. Induction of systemic acquired resistance in pepper plants by acibenzolar-S-methyl against bacterial spot disease. Eur. J. Plant Pathol. 2002, 108, 41–49. [Google Scholar] [CrossRef]
- Gašić, K.; Kuzmanović, N.; Ivanović, M.; Prokić, A.; Šević, M.; Obradović, A. Complete genome of the Xanthomonas euvesicatoria specific bacteriophage KΦ1, its survival and potential in control of pepper bacterial spot. Front. Microbiol. 2018, 9, 2021. [Google Scholar] [CrossRef] [PubMed]
- Sević, M.; Gašić, K.; Đorđevic, M.; Ignjatov, M.; Mijatović, M.; Zečević, B.; Obradović, A. Efficacy of biocontrol agents and bactericides in control of pepper bacterial spot. Acta Hortic. 2016, 1142, 147–150. [Google Scholar] [CrossRef]
- Gotor-Vila, A.; Usall, J.; Torres, R.; Abadias, M.; Teixidó, N. Formulation of the biocontrol agent Bacillus amyloliquefaciens CPA-8 using different approaches: Liquid, freeze-drying and fluid-bed spray-drying. BioControl 2017, 62, 545–555. [Google Scholar] [CrossRef]
- Zhao, P.; Xue, Y.; Gao, W.; Li, J.; Zu, X.; Fu, D.; Bai, X.; Zuo, Y.; Hu, Z.; Zhang, F. Bacillaceae-derived peptide antibiotics since 2000. Peptides 2018, 101, 10–16. [Google Scholar] [CrossRef]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Antimicrobial activity of a biosurfactant produced by Bacillus licheniformis strain M104 grown on whey. Braz. Arch. Biol. Technol. 2013, 56, 259–268. [Google Scholar] [CrossRef]
- Abiala, M.; Odebode, A.; Hsu, S.; Blackwood, C. Phytobeneficial properties of bacteria isolated from the rhizosphere of maize in southwestern Nigerian soils. Appl. Environ. Microbiol. 2015, 81, 4736–4743. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R. Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant-Microbe Interact. 2018, 5, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, H.; Heng, J.; Wang, D.; Bian, K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 2019, 218, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-H.; Liao, M.-J.; Wang, H.-K.; Zheng, M.-Z.; Xu, J.-J.; Guo, J.-H. Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 2018, 126, 147–157. [Google Scholar] [CrossRef]
- Guo, R.J.; Wang, J.Q.; Li, S.D.; Jing, Y.L.; Gao, Y.H.; Sun, R.L. A Bacillus velezensis Strain and Its Application with Mutifunctions in Suppressing Disease Occurrence, Promoting Plant Growth and Drought Resistance. Patent application number CH20181001.8 2018. [Google Scholar]
- Wang, J.; Guo, R.; Wang, W.; Ma, G.; Li, S. Insight into the surfactin production of Bacillus velezensis B006 through metabolomics analysis. J. Ind. Microbiol. Biotechnol. 2018, 45, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, B.; Liu, H.; Han, J.; Zhang, Y. Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control 2017, 105, 27–39. [Google Scholar] [CrossRef]
- Meng, Q.; Jiang, H.; Hao, J.J. Effects of Bacillus velezensis strain BAC03 in promoting plant growth. Biol. Control 2016, 98, 18–26. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Moretti, C.; Amatulli, M.T.; Buonaurio, R. PCR-based assay for the detection of Xanthomonas euvesicatoria causing pepper and tomato bacterial spot. Appl. Microbiol. 2009, 49, 466–471. [Google Scholar] [CrossRef]
- Nikodinovic, J.; Barrow, K.D.; Chuck, J.A. High yield preparation of genomic DNA from Streptomyces. BioTechniques 2003, 35, 932–934. [Google Scholar] [CrossRef]
- Reysenbach, A.L.; Wickham, G.S.; Pace, N.R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 1994, 60, 2113–2119. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Herlich, K. Official Methods of Analysis of the Association of Official Analytical Chemists, 15th ed.; Association of Official Analytical Chemists: Arlington, VA, USA, 1990. [Google Scholar]
- Gales, M.E.J.; Julian, E.C.; Kroner, R.C. Method for quantitative determination of total phosphorus in water. J. Am. Water Work. Ass. 1966, 58, 1363–1368. [Google Scholar] [CrossRef]
- Smyth, T.J.; Perfumo, A.; McClean, S.; Marchant, R.; Banat, I.M. Isolation and analysis of lipopeptides and high molecular weight biosurfactants. In Handbook of Hydrocarbon and Lipid Microbiology: Biochemical Methods; McGenity, T.J., Timmis, K.N., Nogales, B., Eds.; Springer: Heidelberg, Germany, 2016; pp. 3687–3704. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Pajčin, I.; Rončević, Z.; Dodić, J.; Dodić, S.; Grahovac, M.; Jokić, A.; Grahovac, J. Effect of different inoculum preparation conditions on the biomass growth and antimicrobial activity of Bacillus sp. J. Process. Energy Agric. 2019, 23, 96–100. [Google Scholar] [CrossRef]
- Mejri, S.; Siah, A.; Coutte, F.; Magnin-Robert, M.; Randoux, B.; Tisserant, B.; Krier, F.; Jacques, P.; Reignault, P.; Halama, P. Biocontrol of the wheat pathogen Zymoseptoria tritici using cyclic lipopeptides from Bacillus subtilis. Environ. Sci. Pollut. Res. 2018, 25, 29822–29833. [Google Scholar] [CrossRef]
- Toral, L.; Rodríguez, M.; Béjar, V.; Sampedro, I. Antifungal activity of lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea. Front. Microbiol. 2018, 9, 1315. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Guo, Y.; Sun, W.; Zhang, Q. Co-production of multiple antimicrobial compounds by Bacillus amyloliquefaciens WY047, a strain with broad-spectrum activity. Trans. Tianjin Univ. 2018, 24, 160–171. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 2009, 8, 63. [Google Scholar] [CrossRef]
- De Filippi, S.; Groulx, E.; Megalla, M.; Mohamed, R.; Avis, T.J. Fungal competitors affect production of antimicrobial lipopeptides in Bacillus subtilis strain B9–5. J. Chem. Ecol. 2018, 44, 374–383. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Aremu, O.S.; Babalola, O.O. Selecting lipopeptide-producing, Fusarium-suppressing Bacillus spp.: Metabolomic and genomic probing of Bacillus velezensis NWUMFkBS10.5. Microbiol. Open 2018, 8, e742. [Google Scholar] [CrossRef]
- Luo, C.; Liu, X.; Zhou, H.; Wang, X.; Chen, Z. Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl. Environ. Microbiol. 2015, 81, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Pajčin, I.; Grahovac, J.; Dodić, J.; Vlajkov, V.; Dodić, S.; Jokić, A.; Grahovac, M. Biocontrol of Xanthomonas spp. by Bacillus velezensis cultivated on commercial and raw glycerol. In Proceedings of the 4th International Symposium on Biological Control of Bacterial Plant Diseases (Biocontrol 2019), Viterbo, Italy, 9–11 July 2019. [Google Scholar]
- Sanchez, S.; Demain, A.L. Metabolic regulation of fermentation processes. Enzym. Microb. Technol. 2002, 31, 895–906. [Google Scholar] [CrossRef]
- Rončević, Z.; Pajčin, I.; Cvetković, D.; Dodić, S.; Grahovac, J.; Dodić, J. Optimization of cultivation medium composition for production of bioactive compounds effective against Penicillium sp. Pestic. Phytomed. 2018, 33, 27–37. [Google Scholar] [CrossRef][Green Version]
- Ngatia, L.; Taylor, R. Phosphorus eutrophication and mitigation strategies. In Phosphorus—Recovery and Recycling; Zhang, T., Ed.; InTech Open: London, UK, 2018; pp. 45–61. [Google Scholar] [CrossRef]
- Patel, H.; Tscheka, C.; Edwards, K.; Karlsson, G.; Heerklotz, H. All-or-none membrane permeabilization by fengycin-type lipopeptides from Bacillus subtilis QST713. Biochim. Biophys. Acta 2011, 1808, 2000–2008. [Google Scholar] [CrossRef]
- Medeot, D.B.; Fernandez, M.; Morales, G.M.; Jofré, E. Fengycins from Bacillus amyloliquefaciens MEP218 exhibit antibacterial activity by producing alterations on the cell surface of the pathogens Xanthomonas axonopodis pv. vesicatoria and Pseudomonas aeruginosa PA01. Front. Microbiol. 2020, 10, 3107. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Cheffi, M.; Gharbi, Y.; Medhioub, I.; Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M.A. The endophytic strain Bacillus velezensis OEE1: An efficient biocontrol agent against Verticillium Wilt of Olive and a potential plant growth promoting bacteria. Biol. Control 2019, 142, 104168. [Google Scholar] [CrossRef]
- Dhouib, H.; Zouari, I.; Abdallah, D.B.; Belbahri, D.; Taktak, W.; Triki, M.A.; Tounsi, S. Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol. Control 2019, 139, 104092. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]



| Variant | Xanthomonas Isolate | Treatment | Plant Mark |
|---|---|---|---|
| Uninoculated untreated control–negative control | - | - | 1 |
| Inoculated untreated control–positive control | PL1 | - | 2A |
| Inoculated untreated control–positive control | PL2 | - | 2B |
| Inoculated treated plant | PL1 | Bacillus IP22 cultivation broth | 3A |
| Inoculated treated plant | PL2 | Bacillus IP22 cultivation broth | 3B |
| Response | SS | DF | MS | F | p-Value | R2 (%) |
|---|---|---|---|---|---|---|
| Inhibition zone diameter (mm) | 39076.79 a 7.57 b | 15 a 12 b | 2605.12 a 0.63 b | 4127.72 | <0.01 | 94.94 |
| Residual glycerol content (g/L) | 17270.84 a 218.02 b | 15 a 12 b | 1151.39 a 18.17 b | 63.37 | <0.01 | 94.89 |
| Residual total nitrogen content (g/L) | 27.56 a 0.12 b | 15 a 12 b | 1.84 a 0.01 b | 191.00 | <0.01 | 95.68 |
| Residual total phosphorus content (g/L) | 89.67 a 2.18 b | 15 a 12 b | 5.98 a 0.18 b | 32.91 | <0.01 | 90.39 |
| First Set | Second Set | |||
|---|---|---|---|---|
| Factor | Goal | Optimized Value | Goal | Optimized Value |
| Glycerol content (g/L) | in range | 12.01 | in range | 10.00 |
| Yeast extract content (g/L) | in range | 4.06 | in range | 2.83 |
| (NH4)2SO4 content (g/L) | in range | 2.69 | in range | 3.00 |
| K2HPO4 content (g/L) | in range | 8.11 | in range | 1.07 |
| Response | Goal | Predicted Value | Goal | Predicted Value |
| Inhibition zone diameter (mm) | maximize | 66.75 | Maximize | 60.43 |
| Residual glycerol content (g/L) | in range | 2.05 | Minimize | 1.41 |
| Residual total nitrogen content (g/L) | in range | 1.60 | Minimize | 0.92 |
| Residual total phosphorus content (g/L) | in range | 2.78 | Minimize | 0.13 |
| Desirability | 1.00 | 0.77 | ||
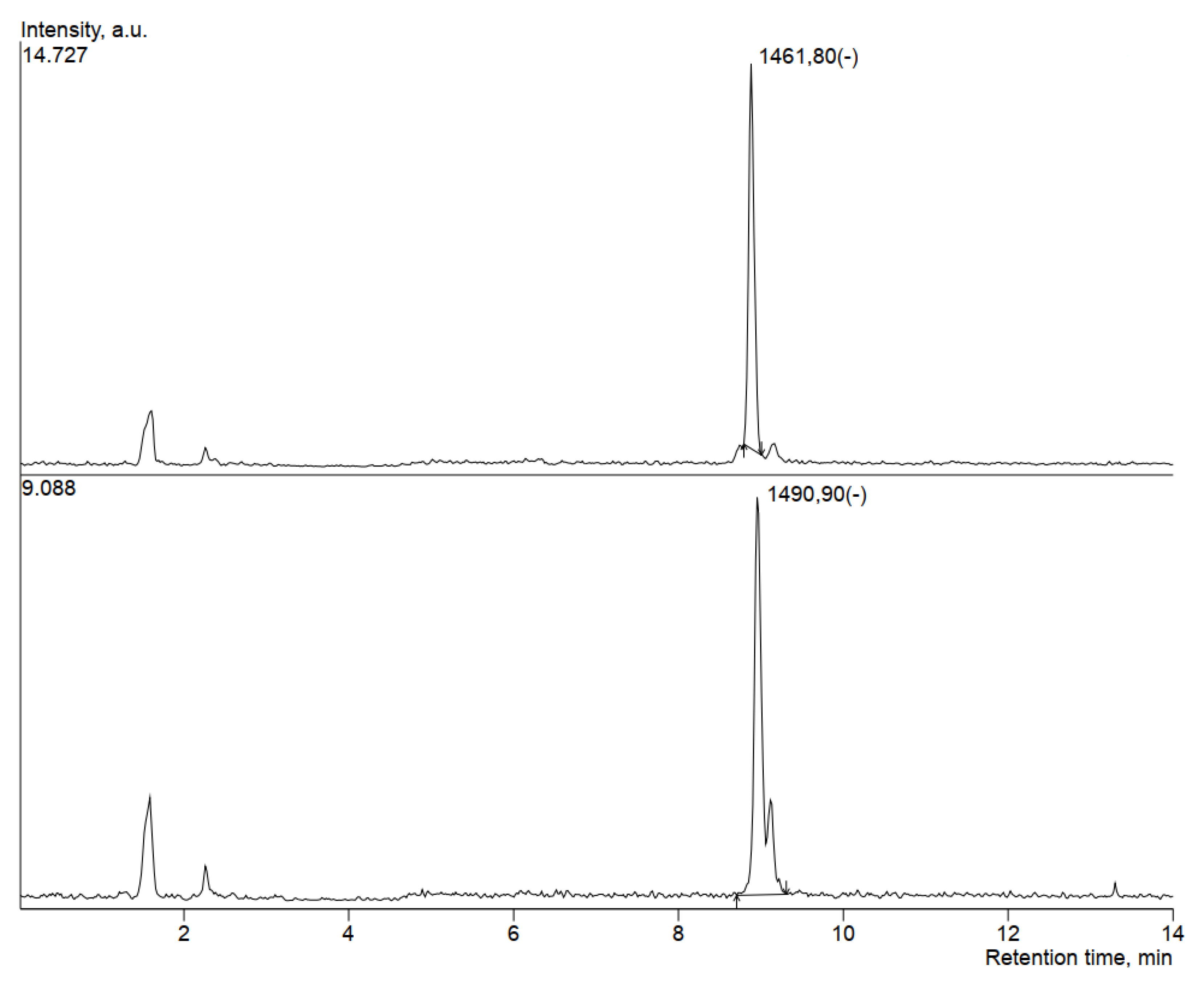
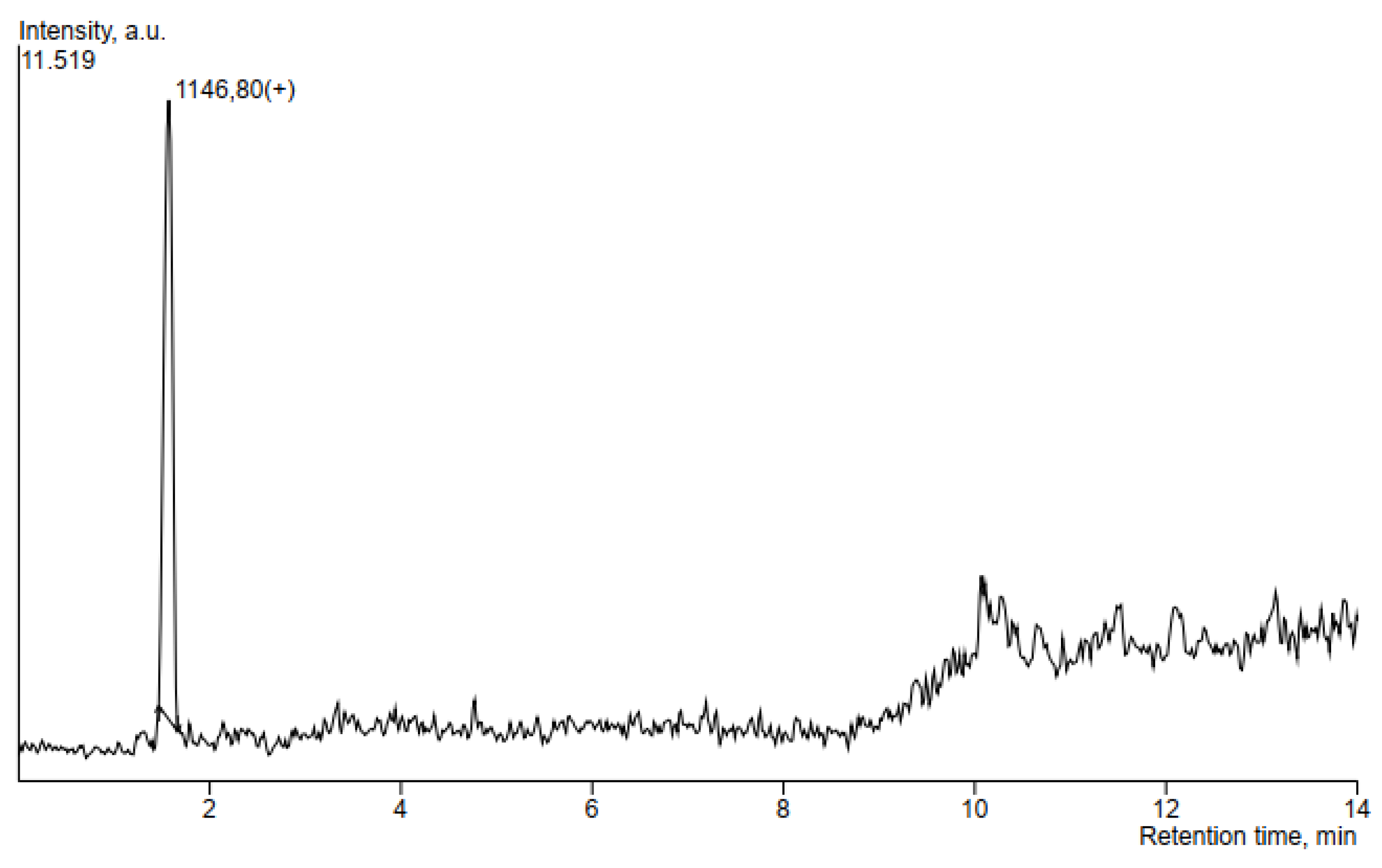
| Putatively Identified Compound | Retention Time (min) | Peak Area | m/z |
|---|---|---|---|
| Fengycin | 8.883 | 62897 | 1461.8 |
| Fengycin | 1.603 | 12026 | 1489.7 |
| Fengycin | 8.96 | 60938 | 1490.9 |
| Locillomycin | 1.574 | 44514 | 1146.8 |
| Plant | Infected Leaves (%) | Leaf Necrosis (%) |
|---|---|---|
| 1 | 0.00 ± 0.00 a | 0.00 ± 0.00 a |
| 2A | 68.33 ± 1.67 b | 31.97 ± 0.14 b |
| 2B | 80.95 ± 2.38 c | 44.43 ± 3.34 c |
| 3A | 14.84 ± 0.55 d | 7.51 ± 1.76 d |
| 3B | 14.84 ± 0.55 d | 5.09 ± 0.09 e |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pajčin, I.; Vlajkov, V.; Frohme, M.; Grebinyk, S.; Grahovac, M.; Mojićević, M.; Grahovac, J. Pepper Bacterial Spot Control by Bacillus velezensis: Bioprocess Solution. Microorganisms 2020, 8, 1463. https://doi.org/10.3390/microorganisms8101463
Pajčin I, Vlajkov V, Frohme M, Grebinyk S, Grahovac M, Mojićević M, Grahovac J. Pepper Bacterial Spot Control by Bacillus velezensis: Bioprocess Solution. Microorganisms. 2020; 8(10):1463. https://doi.org/10.3390/microorganisms8101463
Chicago/Turabian StylePajčin, Ivana, Vanja Vlajkov, Marcus Frohme, Sergii Grebinyk, Mila Grahovac, Marija Mojićević, and Jovana Grahovac. 2020. "Pepper Bacterial Spot Control by Bacillus velezensis: Bioprocess Solution" Microorganisms 8, no. 10: 1463. https://doi.org/10.3390/microorganisms8101463
APA StylePajčin, I., Vlajkov, V., Frohme, M., Grebinyk, S., Grahovac, M., Mojićević, M., & Grahovac, J. (2020). Pepper Bacterial Spot Control by Bacillus velezensis: Bioprocess Solution. Microorganisms, 8(10), 1463. https://doi.org/10.3390/microorganisms8101463








