An M29 Aminopeptidase from Listeria Monocytogenes Contributes to In Vitro Bacterial Growth but not to Intracellular Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Cell Fractionation and Protein Localization Analysis
2.3. Generation of In-Frame Gene Deletion Mutant and Complemented Strain
2.4. Heterologous Overexpression and Protein Purification
2.5. Site-Directed Mutagenesis
2.6. Generation of Polyclonal Antibodies
2.7. Enzymatic Activity Assays
2.8. The Michaelis–Menten Kinetic Parameters
2.9. In Vitro Growth of L. monocytogenes in BHI Broth and Defined Medium
2.10. Intracellular Growth in J774A.1 and RAW264.7 Macrophages
2.11. Adhesion and Invasion in Caco-2 Cells
2.12. Plaque Assay on L929 Fibroblast Cells
2.13. Virulence in Mouse Model
2.14. Statistical Analysis
3. Results
3.1. LmAmpII is an Active M29 Aminopeptidase with a Wide Substrate Specificity
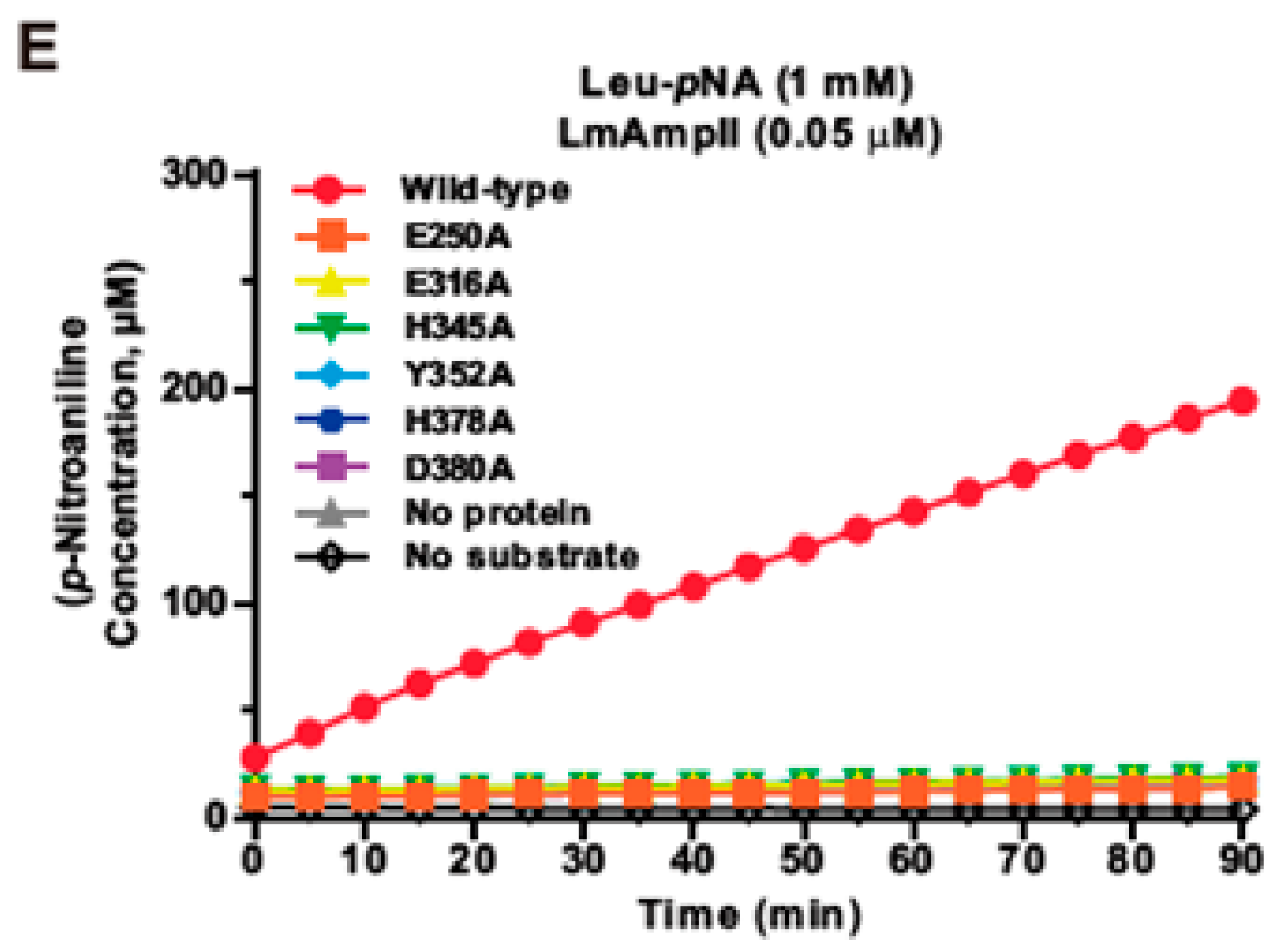
3.2. Site-Directed Mutagenesis Reveals That Key Residues in LmAmpII are Involved in Aminopeptidase Activity
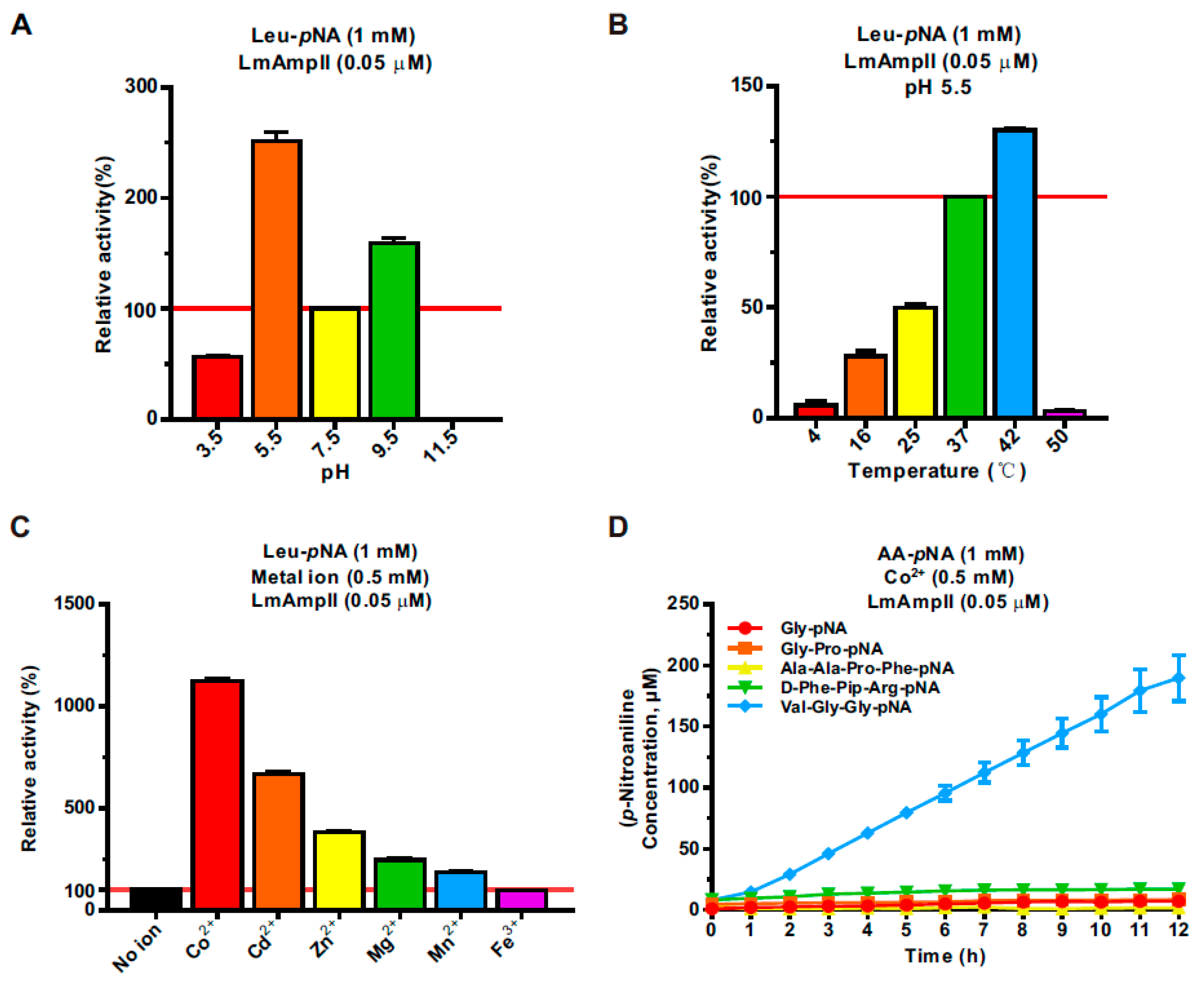
3.3. LmAmpII Is Active in a Wide Range of pHs and Temperatures and can be Strongly Activated by Metal Ions
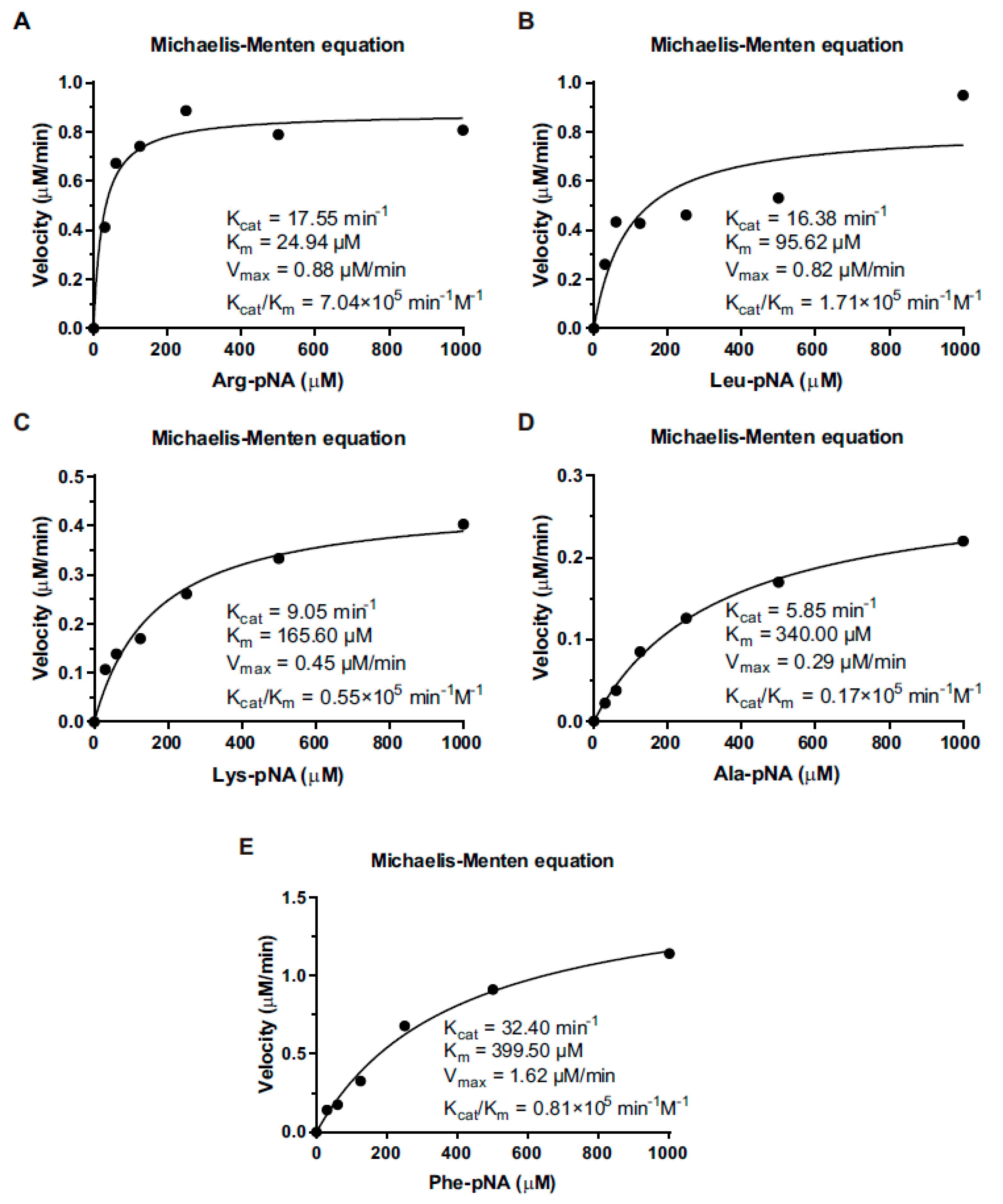
3.4. Kinetic Properties of LmAmpII in the Presence of Various Substrates
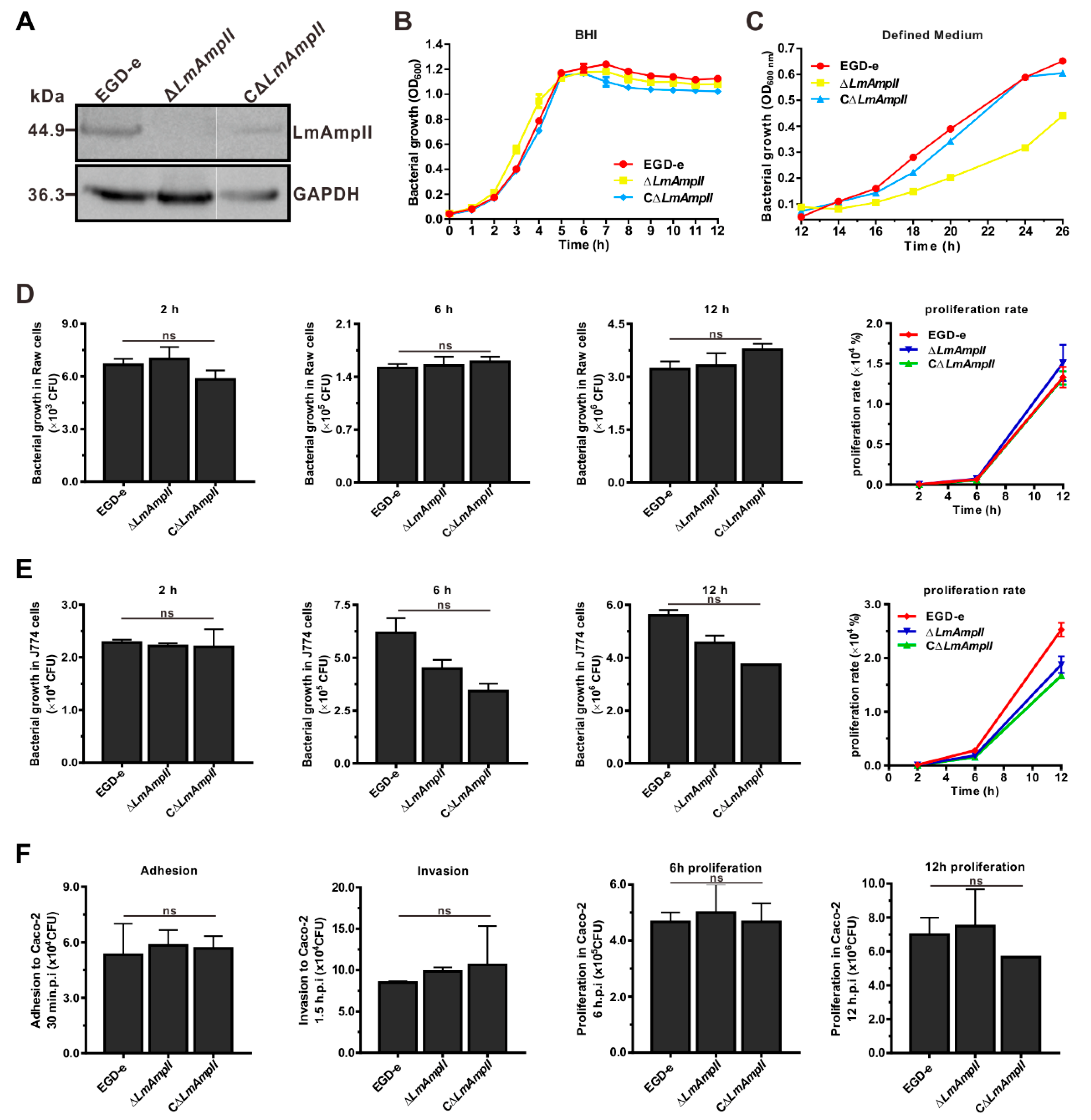
3.5. LmAmpII is Required for In Vitro Growth in Synthetic Media, but not in Rich Media
3.6. LmAmpII Is Not Necessary for Bacterial Infection and Virulence
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AmpII | Aminopeptidase type II |
| Mpl | Metalloprotease |
| AmpS | Aminopeptidase S |
| PepP | X-prolyl aminopeptidase |
| BHI | Brain heart infusion |
| LB | Luria–Bertani broth |
| SP | Secreted proteins |
| TCA | Trichloroacetic acid |
| WCL | Whole cell lysates |
| MP | Membrane proteins |
| WP | Cell wall proteins |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| AAs-pNA | Amino acids-p-nitroaniline |
| DMEM | Dulbecco’s modified eagle medium |
| FBS | Fetal bovine serum |
| AmpT | Aminopeptidase T |
| LAP | Leucine aminopeptidase |
References
- Kljujev, I.; Raicevic, V.; Jovicic-Petrovic, J.; Vujovic, B.; Mirkovic, M.; Rothballer, M. Listeria monocytogenes-Danger for health safety vegetable production. Microb. Pathog. 2018, 120, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Bitar, A.P.; Cao, M.; Marquis, H. The metalloprotease of Listeria monocytogenes is activated by intramolecular autocatalysis. J. Bacteriol. 2008, 190, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.M.; Bitar, A.P.; Slepkov, E.R.; Kota, K.J.; Sondermann, H.; Marquis, H. The metalloprotease of Listeria monocytogenes is regulated by pH. J. Bacteriol. 2011, 193, 5090–5097. [Google Scholar] [CrossRef][Green Version]
- Thomas, S.; Besset, C.; Courtin, P.; Rul, F. The role of aminopeptidase PepS in the growth of Streptococcus thermophilus is not restricted to nitrogen nutrition. J. Appl. Microbiol. 2010, 108, 148–157. [Google Scholar] [CrossRef]
- Sierra, E.M.; Pereira, M.R.; Maester, T.C.; Gomes-Pepe, E.S.; Mendoza, E.R.; Lemos, E.G.M. Halotolerant aminopeptidase M29 from Mesorhizobium SEMIA 3007 with biotechnological potential and its impact on biofilm synthesis. Sci. Rep. 2017, 7, 10684. [Google Scholar] [CrossRef]
- Huang, W.Q.; Zhong, L.F.; Meng, Z.Z.; You, Z.J.; Li, J.Z.; Luo, X.C. The Structure and Enzyme Characteristics of a Recombinant Leucine Aminopeptidase rLap1 from Aspergillus sojae and Its Application in Debittering. Appl. Biochem. Biotechnol. 2015, 177, 190–206. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, X.; Dong, Z.; Shao, C.; Yang, Y.; Fang, W.; Fang, C.; Wang, H.; Yang, M.; Jiang, L.; et al. Aminopeptidase T of M29 Family Acts as A Novel Intracellular Virulence Factor for Listeria monocytogenes Infection. Sci. Rep. 2015, 5, 17370. [Google Scholar] [CrossRef]
- Luckett, J.C.; Darch, O.; Watters, C.; Abuoun, M.; Wright, V.; Paredes-Osses, E.; Ward, J.; Goto, H.; Heeb, S.; Pommier, S.; et al. A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog. 2012, 8, e1002854. [Google Scholar] [CrossRef]
- Nganje, C.N.; Haynes, S.A.; Qabar, C.M.; Lent, R.C.; Bou Ghanem, E.N.; Shainheit, M.G. PepN is a non-essential, cell wall-localized protein that contributes to neutrophil elastase-mediated killing of Streptococcus pneumoniae. PLoS ONE 2019, 14, e0211632. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of peptidases. Biochem. J. 1993, 290, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed]
- Odintsov, S.G.; Sabala, I.; Bourenkov, G.; Rybin, V.; Bochtler, M. Staphylococcus aureus aminopeptidase S is a founding member of a new peptidase clan. J. Biol. Chem. 2005, 280, 27792–27799. [Google Scholar] [CrossRef] [PubMed]
- Myrin, P.A.; Hofsten, B.V. Purification and metal ion activation of an aminopeptidase (aminopeptidase II) from Bacillus stearothermophilus. Biochim. Biophys. Acta 1974, 350, 13–25. [Google Scholar] [CrossRef]
- Motoshima, H.; Azuma, N.; Kaminogawa, S.; Ono, M.; Minagawa, E.; Matsuzawa, H.; Ohta, T.; Yamauchi, K. Molecular cloning and nucleotide sequence of the aminopeptidase T gene of Thermus aquaticus YT-1 and its high-level expression in Escherichia coli. Agric. Biol. Chem. 1990, 54, 2385–2392. [Google Scholar] [CrossRef]
- Ta, H.M.; Bae, S.; Han, S.; Song, J.; Ahn, T.K.; Hohng, S.; Lee, S.; Kim, K.K. Structure-based elucidation of the regulatory mechanism for aminopeptidase activity. Acta Cryst. D Biol. Cryst. 2013, 69, 1738–1747. [Google Scholar] [CrossRef]
- Friedman, S.; Linsky, M.; Lobel, L.; Rabinovich, L.; Sigal, N.; Herskovits, A.A. Metabolic Genetic Screens Reveal Multidimensional Regulation of Virulence Gene Expression in Listeria monocytogenes and an Aminopeptidase That Is Critical for PrfA Protein Activation. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- He, Z.; Wang, H.; Han, X.; Ma, T.; Hang, Y.; Yu, H.; Wei, F.; Sun, J.; Yang, Y.; Cheng, C.; et al. Characterization of a recombinant aminopeptidase Lmo1711 from Listeria monocytogenes. Sheng Wu Gong Cheng Xue Bao 2018, 34, 685–693. [Google Scholar] [CrossRef]
- Camilli, A.; Tilney, L.G.; Portnoy, D.A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993, 8, 143–157. [Google Scholar] [CrossRef]
- Smith, K.; Youngman, P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 1992, 74, 705–711. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, J.; Fang, C.; Xia, Y.; Shan, Y.; Liu, Y.; Wen, G.; Song, H.; Fang, W. Listeria monocytogenes aguA1, but not aguA2, encodes a functional agmatine deiminase: Biochemical characterization of its catalytic properties and roles in acid tolerance. J. Biol. Chem. 2013, 288, 26606–26615. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Yang, Y.; Dong, Z.; Wang, X.; Fang, C.; Yang, M.; Sun, J.; Xiao, L.; Fang, W.; Song, H. Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometry. Front. Microbiol. 2015, 6, 15. [Google Scholar] [CrossRef]
- Park, S.F.; Stewart, G.S. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 1990, 94, 129–132. [Google Scholar] [CrossRef]
- Kothari, H.; Kumar, P.; Singh, N. Prokaryotic expression, purification, and polyclonal antibody production against a novel drug resistance gene of Leishmania donovani clinical isolate. Protein Expr. Purif. 2006, 45, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, S.Y.; Pan, F.Y.; Geng, H.X.; Gong, Y.; Lou, D.; Shu, Y.Q.; Li, C.J. Prokaryotic expression and polyclonal antibody preparation of a novel Rab-like protein mRabL5. Protein Expr. Purif. 2007, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cahan, R.; Axelrad, I.; Safrin, M.; Ohman, D.E.; Kessler, E. A secreted aminopeptidase of Pseudomonas aeruginosa. Identification, primary structure, and relationship to other aminopeptidases. J. Biol. Chem. 2001, 276, 43645–43652. [Google Scholar] [CrossRef]
- Phan-Thanh, L.; Gormon, T. A chemically defined minimal medium for the optimal culture of Listeria. Int. J. Food Microbiol. 1997, 35, 91–95. [Google Scholar] [CrossRef]
- O’Riordan, M.; Moors, M.A.; Portnoy, D.A. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 2003, 302, 462–464. [Google Scholar] [CrossRef]
- Sun, J.; Hang, Y.; Han, Y.; Zhang, X.; Gan, L.; Cai, C.; Chen, Z.; Yang, Y.; Song, Q.; Shao, C.; et al. Deletion of glutaredoxin promotes oxidative tolerance and intracellular infection in Listeria monocytogenes. Virulence 2019, 10, 910–924. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Carroll, R.K.; Veillard, F.; Gagne, D.T.; Lindenmuth, J.M.; Poreba, M.; Drag, M.; Potempa, J.; Shaw, L.N. The Staphylococcus aureus leucine aminopeptidase is localized to the bacterial cytosol and demonstrates a broad substrate range that extends beyond leucine. Biol. Chem. 2013, 394, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Fowler, J.H.; Walling, L.L. Leucine aminopeptidases: Diversity in structure and function. Biol. Chem. 2006, 387, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.K.; Robison, T.M.; Rivera, F.E.; Davenport, J.E.; Jonsson, I.M.; Florczyk, D.; Tarkowski, A.; Potempa, J.; Koziel, J.; Shaw, L.N. Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus. Microbes Infect. 2012, 14, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Odintsov, S.G.; Sabala, I.; Bourenkov, G.; Rybin, V.; Bochtler, M. Substrate access to the active sites in aminopeptidase T, a representative of a new metallopeptidase clan. J. Mol. Biol. 2005, 354, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.Y.; Hwang, G.Y.; Lai, Y.J.; Yang, S.L.; Lin, L.L. Overexpression, purification, and characterization of the recombinant leucine aminopeptidase II of Bacillus stearothermophilus. Curr. Microbiol. 2003, 47, 40–45. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef]
- Zhu, X.; Barman, A.; Ozbil, M.; Zhang, T.; Li, S.; Prabhakar, R. Mechanism of peptide hydrolysis by co-catalytic metal centers containing leucine aminopeptidase enzyme: A DFT approach. J. Biol. Inorg. Chem. 2012, 17, 209–222. [Google Scholar] [CrossRef]
- Mierau, I.; Kunji, E.R.; Leenhouts, K.J.; Hellendoorn, M.A.; Haandrikman, A.J.; Poolman, B.; Konings, W.N.; Venema, G.; Kok, J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J. Bacteriol. 1996, 178, 2794–2803. [Google Scholar] [CrossRef]
- Correa, A.F.; Bastos, I.M.; Neves, D.; Kipnis, A.; Junqueira-Kipnis, A.P.; de Santana, J.M. The Activity of a Hexameric M17 Metallo-Aminopeptidase Is Associated with Survival of Mycobacterium tuberculosis. Front. Microbiol. 2017, 8, 504. [Google Scholar] [CrossRef]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes-from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef] [PubMed]


| Name | Sequence (5’-3’) | Description |
|---|---|---|
| LmAmpII-exp-F | CGGGGTACCATGACAGTATTTAGTGAAAAGTTAGAAAAGTATGC | Recombinant LmAmpII expression |
| LmAmpII-exp-R | CGCGGATCCTTAGAACGCCCAGTCGCCTTTA | |
| LmAmpII-a-front | AGTTTTTCTGAAGACATTTATAATAGAAGGTATCAG | Construction of LmAmpII null mutant |
| LmAmpII-a | CGCGGATCCTAATACACAAGAAATTGCCGACATTTTAG | |
| LmAmpII-b | TTAAAAAAAGGAAATTAATCACTCCAATCTTTTTATTCAATACG | |
| LmAmpII-c | GAGTGATTAATTTCCTTTTTTTAAACTTATGCCTTTTGC | |
| LmAmpII-d | CCCAAGCTTATCATATAGAACACCGAATAAATATGTGTCC | |
| LmAmpII-g | GACGAGCTCCGAAAGCTACGCGAAAACTTTCGTTG | Complementation of the LmAmpII deletion |
| LmAmpII-h | CGCGGATCCTTAGAACGCCCAGTCGCCTTTACG | |
| E250A-fwd | TCCGCACAACAGAAAACTGCTTCTGTTGGCATATTGG | Recombinant LmAmpII E250A expression |
| E250A-rev | CCAATATGCCAACAGAAGCAGTTTTCTGTTGTGCGGA | |
| E316A-fwd | CTGGAACTAGCGCCACTGCACCTAAATAGTGTGAG | Recombinant LmAmpII E316A expression |
| E316A-rev | CTCACACTATTTAGGTGCAGTGGCGCTAGTTCCAG | |
| H345A-fwd | CATACGCACTACCAATTGCTAAGGCGTTAGAAGCATTTTCGTCAAATA | Recombinant LmAmpII H345A expression |
| H345A-rev | TATTTGACGAAAATGCTTCTAACGCCTTAGCAATTGGTAGTGCGTATG | |
| Y352A-fwd | GCCACCTTTAACATTAAATGCAGCCGCACTACCAATTGCTAAGTGG | Recombinant LmAmpII Y352A expression |
| Y352A-rev | CCACTTAGCAATTGGTAGTGCGGCTGCATTTAATGTTAAAGGTGGC | |
| H378A-fwd | AATCATGAAATCAACGGCTGTTAAACTATTGTTCACACCAGCTGCTTCT | Recombinant LmAmpII H378A expression |
| H378A-rev | AGAAGCAGCTGGTGTGAACAATAGTTTAACAGCCGTTGATTTCATGATT | |
| D380A-fwd | TTTCAGAAGAACCAATCATGAAAGCAACGTGTGTTAAACTATTGTTC | Recombinant LmAmpII D380A expression |
| D380A-rev | GAACAATAGTTTAACACACGTTGCTTTCATGATTGGTTCTTCTGAAA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Guan, C.; Hang, Y.; Liu, F.; Sun, J.; Yu, H.; Gan, L.; Zeng, H.; Zhu, Y.; Chen, Z.; et al. An M29 Aminopeptidase from Listeria Monocytogenes Contributes to In Vitro Bacterial Growth but not to Intracellular Infection. Microorganisms 2020, 8, 110. https://doi.org/10.3390/microorganisms8010110
Zhang X, Guan C, Hang Y, Liu F, Sun J, Yu H, Gan L, Zeng H, Zhu Y, Chen Z, et al. An M29 Aminopeptidase from Listeria Monocytogenes Contributes to In Vitro Bacterial Growth but not to Intracellular Infection. Microorganisms. 2020; 8(1):110. https://doi.org/10.3390/microorganisms8010110
Chicago/Turabian StyleZhang, Xian, Chiyu Guan, Yi Hang, Fengdan Liu, Jing Sun, Huifei Yu, Li Gan, Huan Zeng, Yiran Zhu, Zhongwei Chen, and et al. 2020. "An M29 Aminopeptidase from Listeria Monocytogenes Contributes to In Vitro Bacterial Growth but not to Intracellular Infection" Microorganisms 8, no. 1: 110. https://doi.org/10.3390/microorganisms8010110
APA StyleZhang, X., Guan, C., Hang, Y., Liu, F., Sun, J., Yu, H., Gan, L., Zeng, H., Zhu, Y., Chen, Z., Song, H., & Cheng, C. (2020). An M29 Aminopeptidase from Listeria Monocytogenes Contributes to In Vitro Bacterial Growth but not to Intracellular Infection. Microorganisms, 8(1), 110. https://doi.org/10.3390/microorganisms8010110





