DNA- and RNA- Derived Fungal Communities in Subsurface Aquifers Only Partly Overlap but React Similarly to Environmental Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Groundwater Sample Collection
2.3. Physico-Chemical Analysis of Groundwater Samples
2.4. Nucleic Acid Extraction, Amplicon Library Preparation and Pair-End Illumina Sequencing
2.5. Bioinformatics Processing of Sequence Data
2.6. Statistical Processing
2.7. Functional Analysis
3. Results
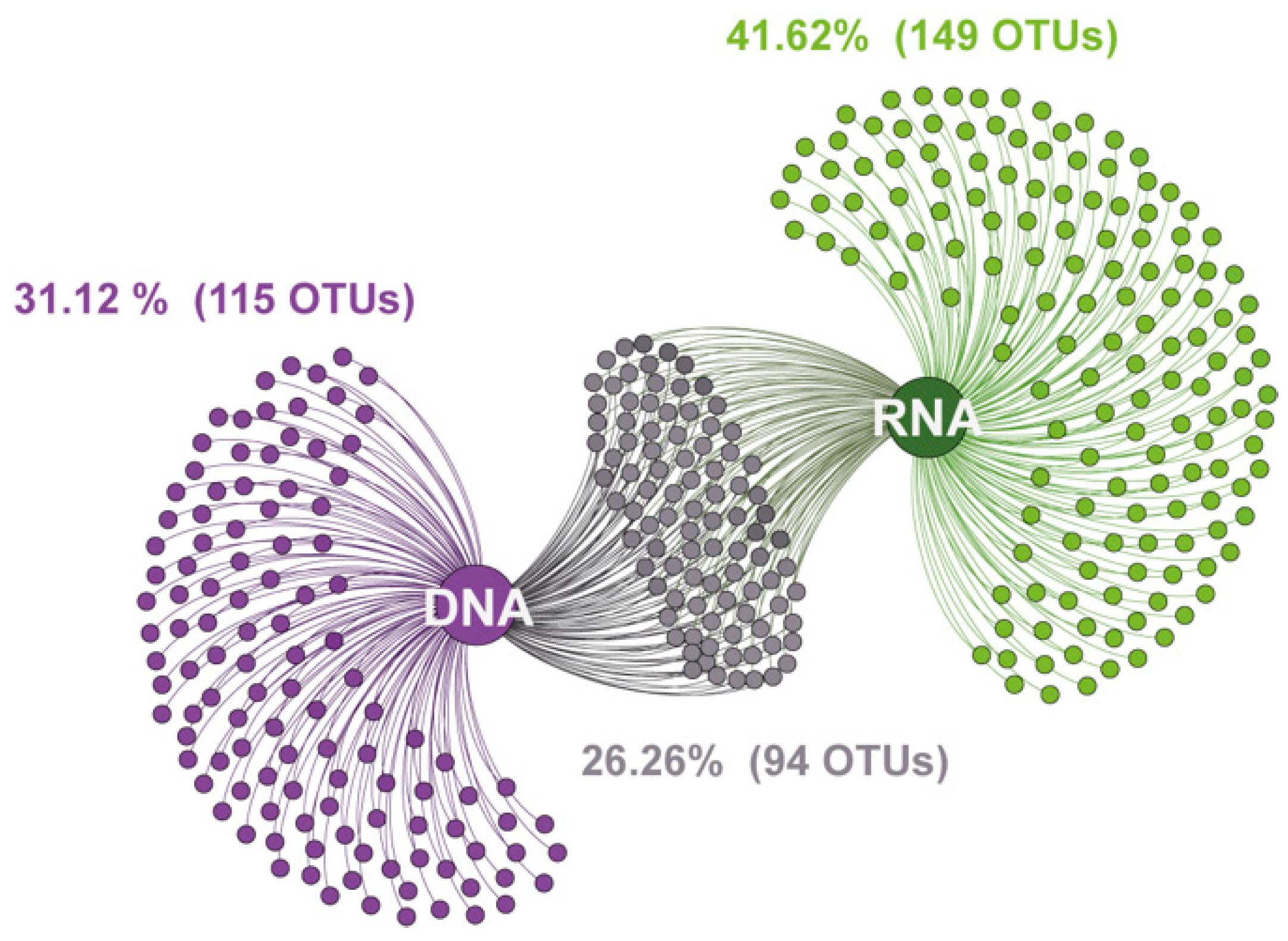
3.1. Illumina Sequencing of DNA- and RNA-Derived Fungal Communities
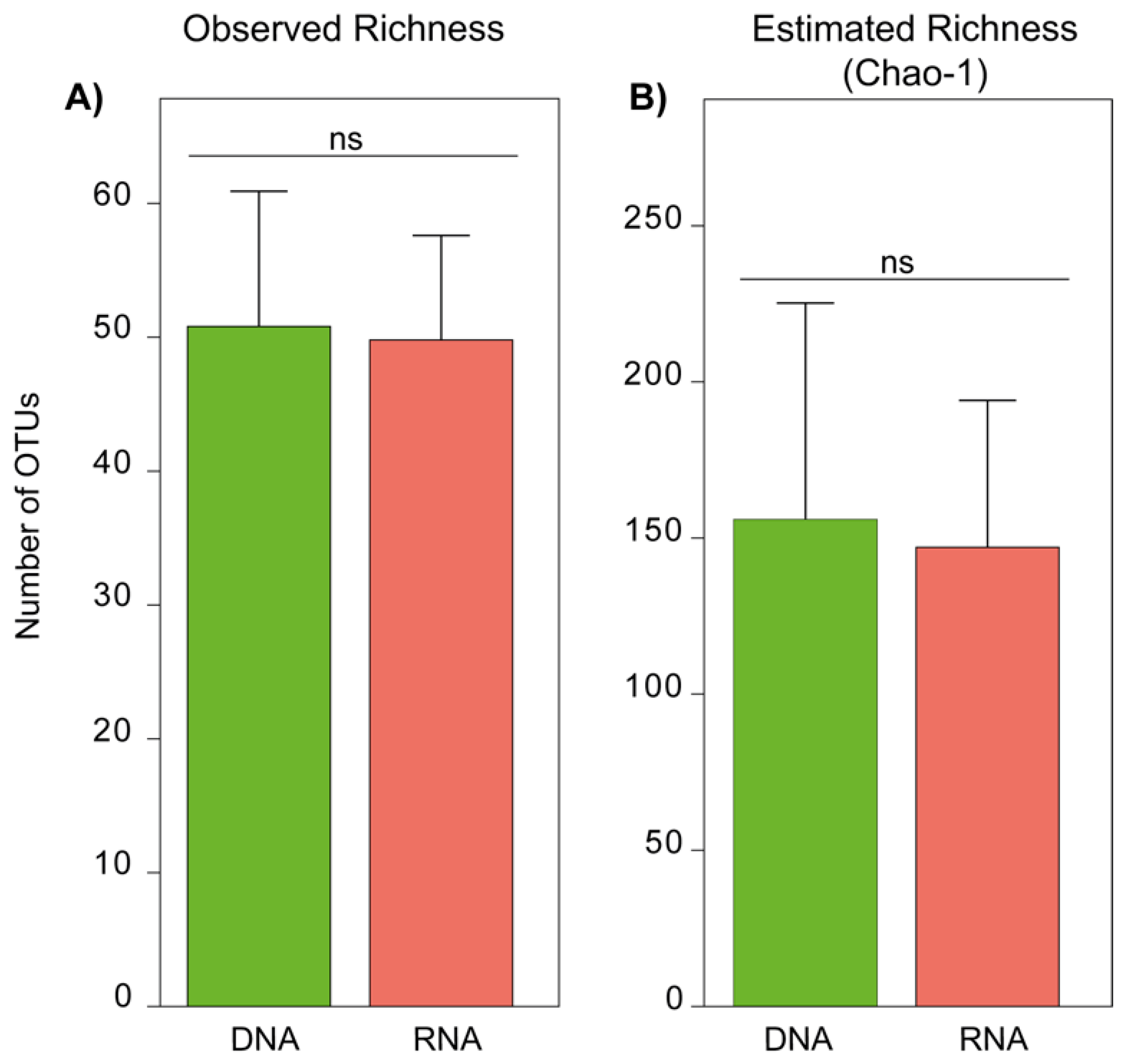
3.2. Fungal Species Richness of DNA- and RNA-Derived Communities
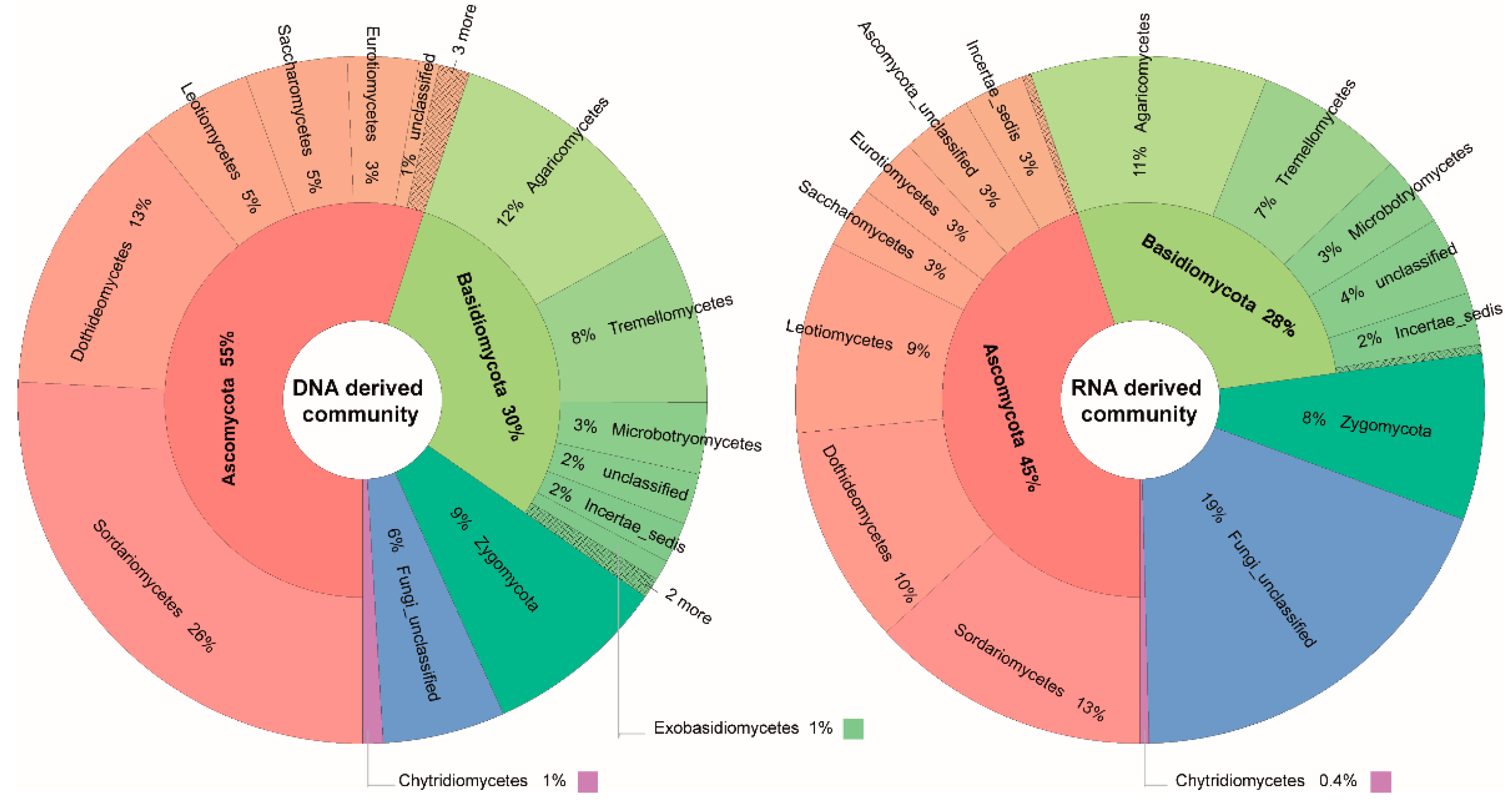
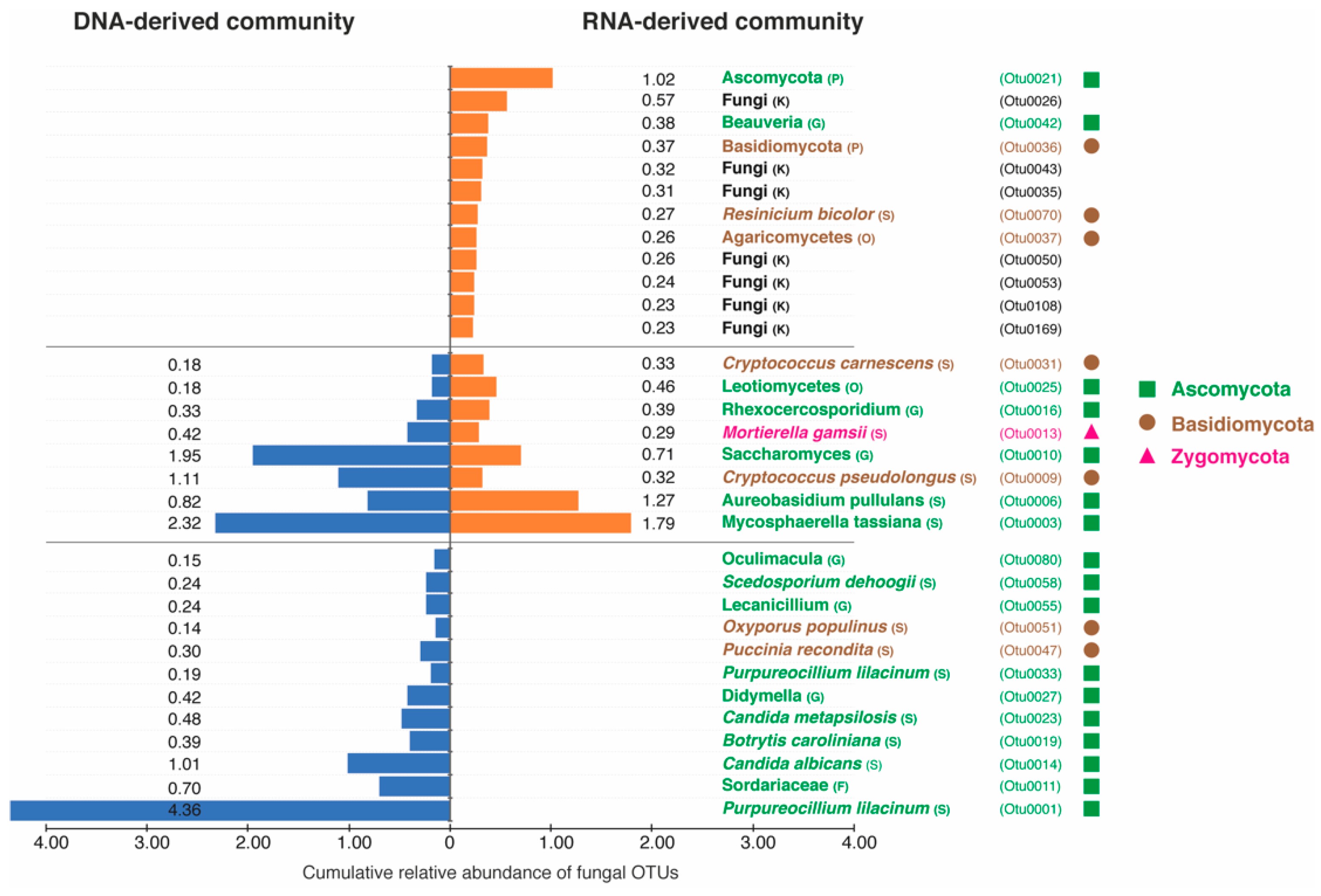
3.3. Taxonomic Differences between DNA- and RNA-Derived Fungal Communities
3.4. DNA- and RNA-Derived Fungal Communities and Significant Environmental Factors
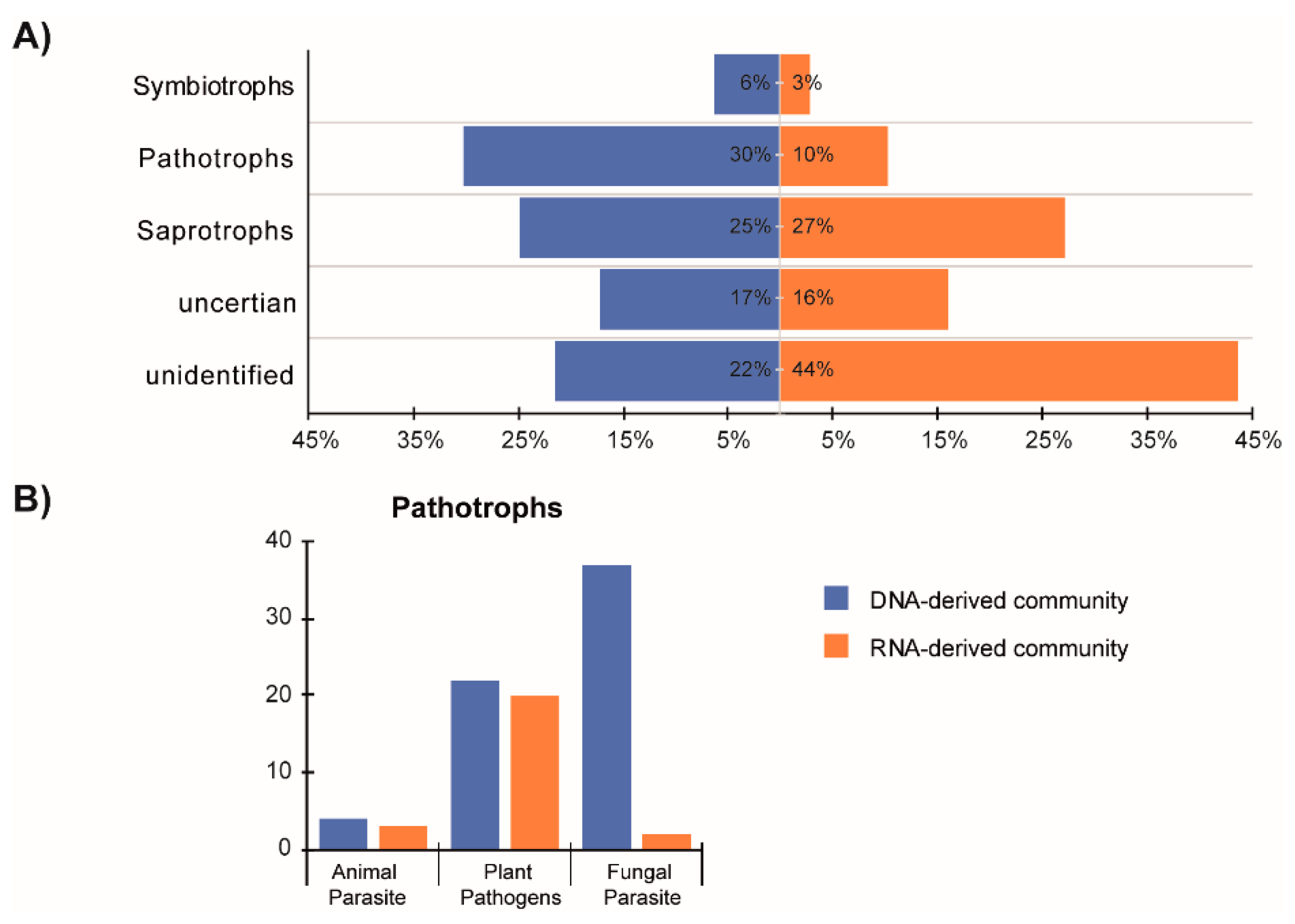
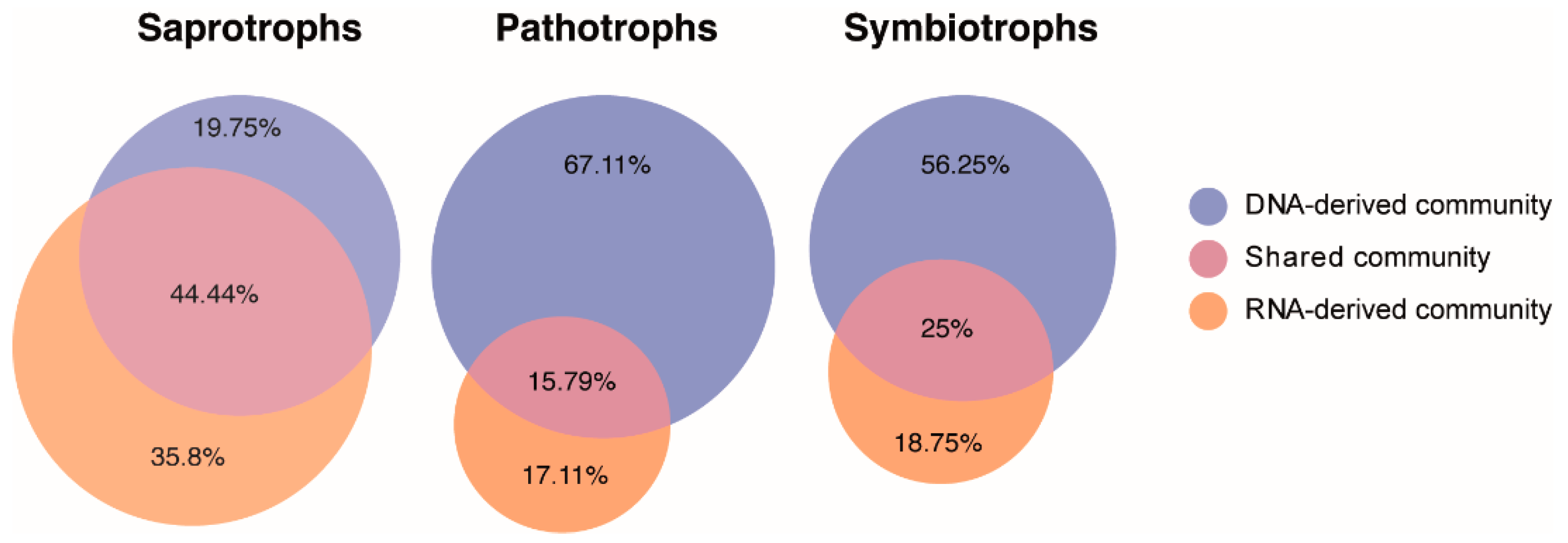
3.5. Distinct Functional Guilds of DNA- and RNA-Derived Fungal Community
4. Discussion
4.1. The Total and Active Fungal Communities Have Similar Structure but Different Community Composition
4.2. Does the DNA-Derived Fungal Community Represent the Total Fungal Community?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hawksworth, D.L.; Lucking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Index Fungorum. 2019. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 27 August 2019).
- Hyde, K.D. Where are the missing fungi? Does Hong Kong have any answers? Mycol. Res. 2001, 105, 1514–1518. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Ford, D.; Williams, P.D. Karst Hydrogeology and Geomorphology; John Wiley & Sons Ltd.: West Suxxex, UK, 2013. [Google Scholar]
- Martin, J.B.; White, W.B. (Eds.) Frontiers of Karst Research, Special Publication Martin; Karst Water Institute: Lessburg, VA, USA, 2008; p. 118. [Google Scholar]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, A.; Purahong, W.; Lehmann, R.; Herrmann, M.; Kusel, K.; Totsche, K.U.; Buscot, F.; Wubet, T. Superimposed pristine limestone aquifers with marked hydrochemical differences exhibit distinct fungal communities. Front. Microbiol. 2016, 7, 666. [Google Scholar] [CrossRef] [PubMed]
- Lategan, M.J.; Torpy, F.R.; Newby, S.; Stephenson, S.; Hose, G.C. Fungal diversity of shallow aquifers in Southeastern Australia. Geomicrobiol. J. 2012, 29, 352–361. [Google Scholar] [CrossRef]
- Panzer, K.; Yilmaz, P.; Weiss, M.; Reich, L.; Richter, M.; Wiese, J.; Schmaljohann, R.; Labes, A.; Imhoff, J.F.; Glockner, F.O.; et al. Identification of habitat-specific biomes of aquatic fungal communities using a comprehensive nearly full-length 18s rrna dataset enriched with contextual data. PLoS ONE 2015, 10, e0134377. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Adams, R.I.; Roman, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrodinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2016, 2, 16242. [Google Scholar] [CrossRef]
- Anderson, I.C.; Parkin, P.I. Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J. Microbiol. Methods 2007, 68, 248–253. [Google Scholar] [CrossRef]
- Baldrian, P.; Kolarik, M.; Stursova, M.; Kopecky, J.; Valaskova, V.; Vetrovsky, T.; Zifcakova, L.; Snajdr, J.; Ridl, J.; Vlcek, C.; et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Cox, F.; Newsham, K.K.; Robinson, C.H. Endemic and cosmopolitan fungal taxa exhibit differential abundances in total and active communities of Antarctic soils. Environ. Microbiol. 2019, 21, 1586–1596. [Google Scholar] [CrossRef] [PubMed]
- Romanowicz, K.J.; Freedman, Z.B.; Upchurch, R.A.; Argiroff, W.A.; Zak, D.R. Active microorganisms in forest soils differ from the total community yet are shaped by the same environmental factors: the influence of pH and soil moisture. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Womack, A.M.; Artaxo, P.E.; Ishida, F.Y.; Mueller, R.C.; Saleska, S.R.; Wiedemann, K.T.; Bohannan, B.J.M.; Green, J.L. Characterization of active and total fungal communities in the atmosphere over the Amazon rainforest. Biogeosci. Discuss. 2015, 12, 7177–7207. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Verbruggen, E.; Hillekens, R.; de Hollander, M.; Roling, W.F.; van der Heijden, M.G.; Kowalchuk, G.A. Tracking fungal community responses to maize plants by DNA- and RNA-based pyrosequencing. PLoS ONE 2013, 8, e69973. [Google Scholar] [CrossRef]
- Wutkowska, M.; Vader, A.; Mundra, S.; Cooper, E.J.; Eidesen, P.B. Dead or alive; or does it really matter? Level of congruency between trophic modes in total and active fungal communities in high arctic soil. Front. Microbiol. 2019, 9, 3243. [Google Scholar] [CrossRef]
- McGee, C.F.; Byrne, H.; Irvine, A.; Wilson, J. Diversity and dynamics of the DNA- and cDNA-derived compost fungal communities throughout the commercial cultivation process for Agaricus bisporus. Mycologia 2017, 109, 475–484. [Google Scholar] [CrossRef]
- Küsel, K.; Totsche, K.U.; Trumbore, S.E.; Lehmann, R.; Steinhäuser, C.; Herrmann, M. How deep can surface signals be traced in the critical zone? Merging biodiversity with biogeochemistry research in a Central German Muschelkalk Landscape. Front. Earth Sci. 2016, 4, 32. [Google Scholar] [CrossRef]
- Kohlhepp, B.; Lehmann, R.; Seeber, P.; Kusel, K.; Trumbore, S.E.; Totsche, K.U. Aquifer configuration and geostructural links control the groundwater quality in thin-bedded carbonate-siliciclastic alternations of the Hainich CZE, central Germany. Hydrol. Earth Syst. Sci. 2017, 21, 6091–6116. [Google Scholar] [CrossRef]
- Nawaz, A.; Purahong, W.; Lehmann, R.; Herrmann, M.; Totsche, K.U.; Kusel, K.; Wubet, T.; Buscot, F. First insights into the living groundwater mycobiome of the terrestrial biogeosphere. Water Res. 2018, 145, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Ihrmark, K.; Bodeker, I.T.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region--evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Mercier, C.; Bonin, A.; Le Bras, Y.; Taberlet, P.; Coissac, E. OBITOOLS: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philosophical Transactions of the Royal Society B-Biological Sciences 2005, 360, 1935–1943. [Google Scholar] [CrossRef]
- Fu, L.M.; Niu, B.F.; Zhu, Z.W.; Wu, S.T.; Li, W.Z. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glockner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sanchez-Garcia, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org (accessed on 11 September 2019).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Hurlbert, S.H. The nonconcept of species diversity: A critique and alternative parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jackson, J.A. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Weblogs and Social Media, Paris, France, 7 July 2019. [Google Scholar]
- Oksanen, J.; Blanchet, F.; Kindt, R.; Legendre, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.3–1. 2015. Available online: https://cran.r-project.org/web/packages/vegan/ (accessed on 11 September 2019).
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.W.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- De Vrieze, J.; Pinto, A.J.; Sloan, W.T.; Ijaz, U.Z. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome 2018, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- Wegner, C.E.; Gaspar, M.; Geesink, P.; Herrmann, M.; Marz, M.; Kusel, K. Biogeochemical regimes in shallow aquifers reflect the metabolic coupling of the elements nitrogen, sulfur, and carbon. Appl. Environ. Microbiol. 2019, 85, e02346-18. [Google Scholar] [CrossRef]
- Li, H.; Su, J.Q.; Yang, X.R.; Zhu, Y.G. Distinct rhizosphere effect on active and total bacterial communities in paddy soils. Sci. Total Environ. 2019, 649, 422–430. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.H.; Dai, M.H.; Jiao, N.Z.; Herndl, G.J. Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol. Ecol. 2014, 23, 2260–2274. [Google Scholar] [CrossRef]
- Franchi, O.; Bovio, P.; Ortega-Martinez, E.; Rosenkranz, F.; Chamy, R. Active and total microbial community dynamics and the role of functional genes bamA and mcrA during anaerobic digestion of phenol and p-cresol. Bioresour. Technol. 2018, 264, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.; Cassio, F.; Pascoal, C. Environmental drivers are more important for structuring fungal decomposer communities than the geographic distance between streams. Limnetica 2017, 36, 491–506. [Google Scholar]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutierrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. mBio 2019, 10, e01189-18. [Google Scholar] [CrossRef]
- Tian, J.Q.; Zhu, D.; Wang, J.Z.; Wu, B.; Hussain, M.; Liu, X.Z. Environmental factors driving fungal distribution in freshwater lake sediments across the Headwater Region of the Yellow River. China Sci. Rep. 2018, 8, 3768. [Google Scholar] [CrossRef] [PubMed]
- Krauss, G.J.; Sole, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Barlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. Fems Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef] [PubMed]
- Tisthammer, K.H.; Cobian, G.M.; Amend, A.S. Global biogeography of marine fungi is shaped by the environment. Fungal Ecol. 2016, 19, 39–46. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.Y.; Gohl, D.M.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the information content of shallow shotgun metagenomics. Msystems 2018, 3, e00069-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Fuhrman, J.A.; Sun, F.; Zhu, S. Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences. Brief. Bioinform 2019. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, I.; Fulci, V.; Stronati, L.; Carissimi, C. Next-generation metagenomics: Methodological challenges and opportunities. OMICS: J. Integr. Biol. 2019, 23, 327–333. [Google Scholar] [CrossRef] [PubMed]
| Factors | DNA-derived Community | RNA-derived Community | ||
|---|---|---|---|---|
| R2 | P | R2 | P | |
| Extraction °T | 0.16 | 0.10 | 0.15 | 0.11 |
| EC25 | 0.14 | 0.14 | 0.15 | 0.11 |
| pH | 0.02 | 0.86 | 0.03 | 0.82 |
| ORP | 0.11 | 0.24 | 0.10 | 0.28 |
| O2 | 0.09 | 0.33 | 0.10 | 0.27 |
| NH4+ | 0.24 | 0.01 * | 0.26 | 0.01 * |
| PO4 | 0.04 | 0.68 | 0.04 | 0.72 |
| TIC | 0.03 | 0.81 | 0.01 | 0.91 |
| NO3− | 0.06 | 0.49 | 0.06 | 0.50 |
| SO42− | 0.10 | 0.27 | 0.11 | 0.22 |
| Cl− | 0.22 | 0.03 * | 0.24 | 0.02 * |
| Ca2+ | 0.18 | 0.05 | 0.20 | 0.04 * |
| Fet | 0.08 | 0.41 | 0.08 | 0.38 |
| K+ | 0.21 | 0.03 * | 0.23 | 0.03 * |
| Mg2+ | 0.19 | 0.06 | 0.21 | 0.03 * |
| Na+ | 0.16 | 0.10 | 0.18 | 0.07 |
| St | 0.10 | 0.28 | 0.10 | 0.23 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, A.; Purahong, W.; Herrmann, M.; Küsel, K.; Buscot, F.; Wubet, T. DNA- and RNA- Derived Fungal Communities in Subsurface Aquifers Only Partly Overlap but React Similarly to Environmental Factors. Microorganisms 2019, 7, 341. https://doi.org/10.3390/microorganisms7090341
Nawaz A, Purahong W, Herrmann M, Küsel K, Buscot F, Wubet T. DNA- and RNA- Derived Fungal Communities in Subsurface Aquifers Only Partly Overlap but React Similarly to Environmental Factors. Microorganisms. 2019; 7(9):341. https://doi.org/10.3390/microorganisms7090341
Chicago/Turabian StyleNawaz, Ali, Witoon Purahong, Martina Herrmann, Kirsten Küsel, François Buscot, and Tesfaye Wubet. 2019. "DNA- and RNA- Derived Fungal Communities in Subsurface Aquifers Only Partly Overlap but React Similarly to Environmental Factors" Microorganisms 7, no. 9: 341. https://doi.org/10.3390/microorganisms7090341
APA StyleNawaz, A., Purahong, W., Herrmann, M., Küsel, K., Buscot, F., & Wubet, T. (2019). DNA- and RNA- Derived Fungal Communities in Subsurface Aquifers Only Partly Overlap but React Similarly to Environmental Factors. Microorganisms, 7(9), 341. https://doi.org/10.3390/microorganisms7090341






