Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Locations and Sample Collection
2.2. Bacterial Identification and Enumeration
2.3. Antimicrobial Susceptibility Testing
2.4. Antimicrobial Consumption
2.5. Statistical Analyses
2.6. Contribution of Hospital Wastewater to AMRB in Influent
2.7. Antimicrobial Residues and Chemical Concentrations
3. Results and Discussion
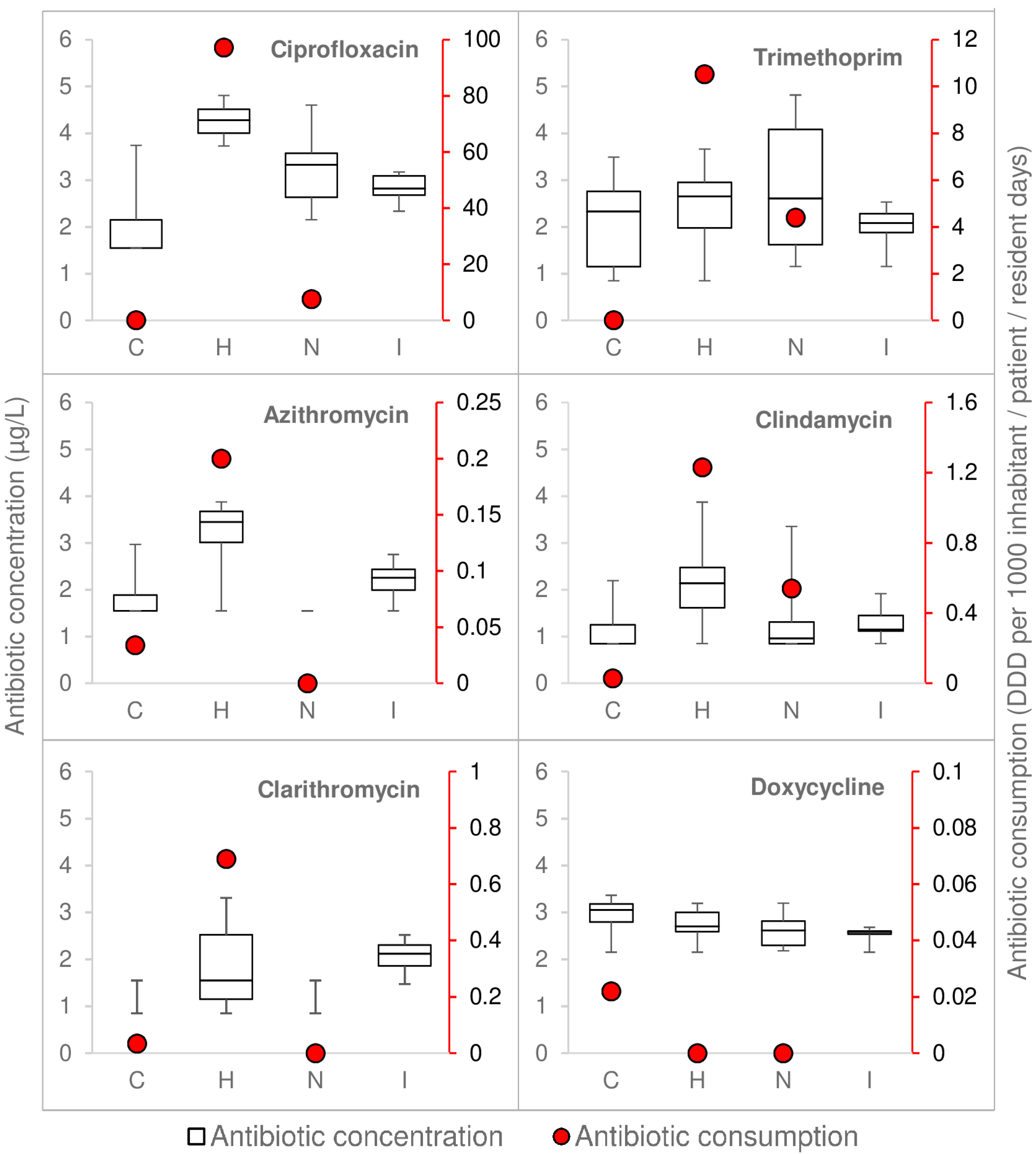
3.1. Antibiotic Consumption and Measured Concentrations Were Highest in Hospital
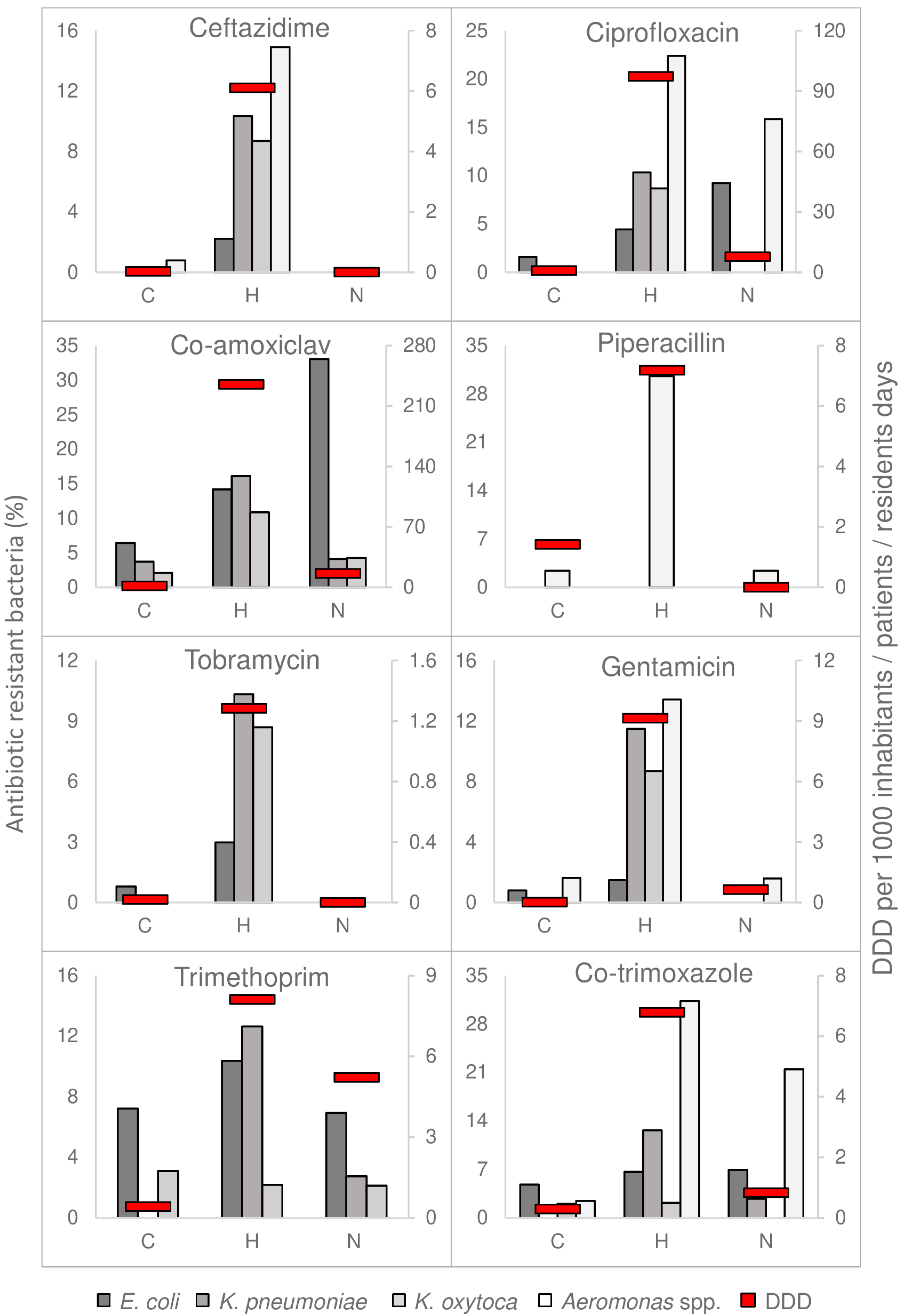
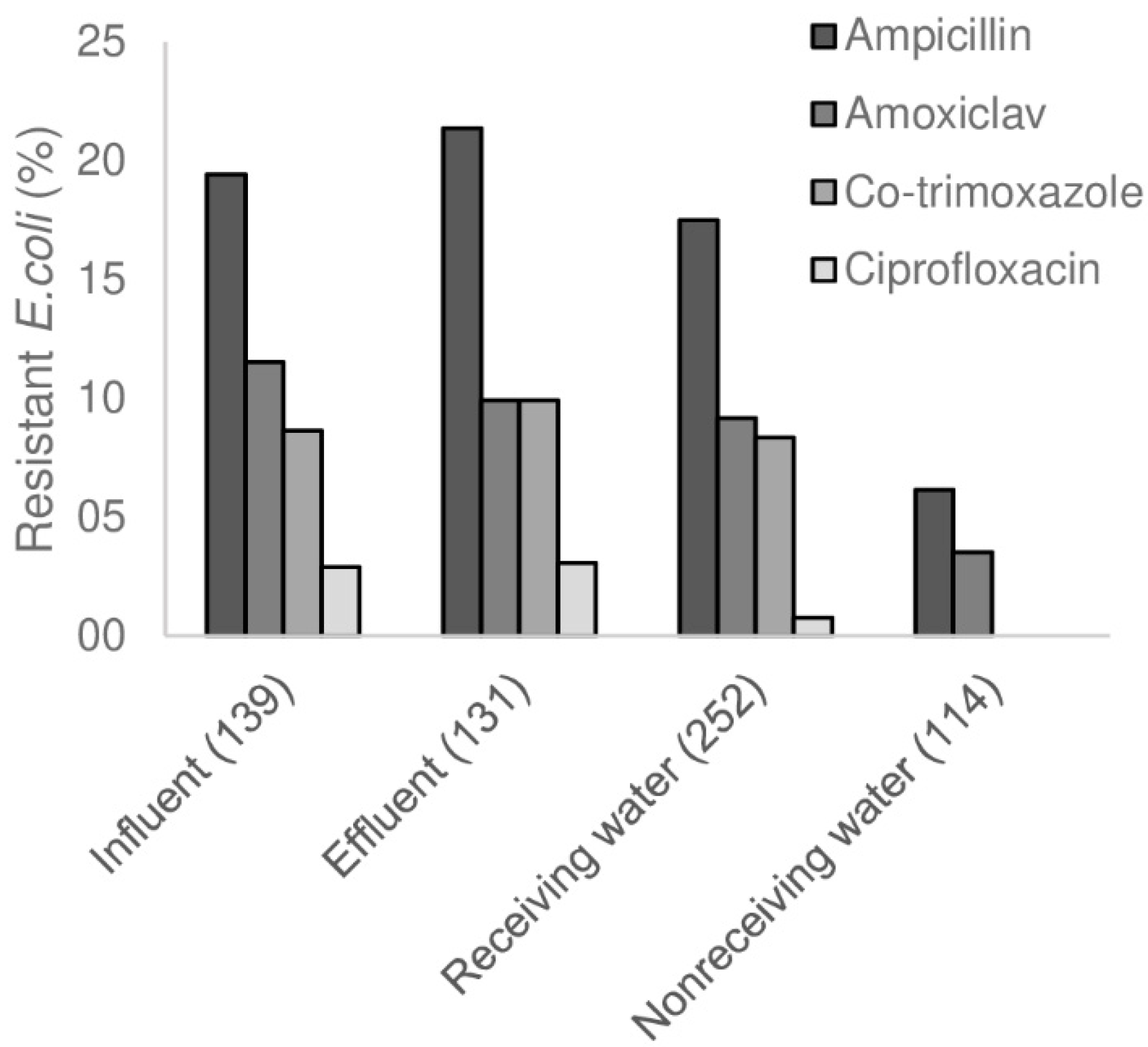
3.2. Concentrations of Resistant Bacteria Mirrored Antimicrobial Consumption
3.3. ESBL in Wastewater
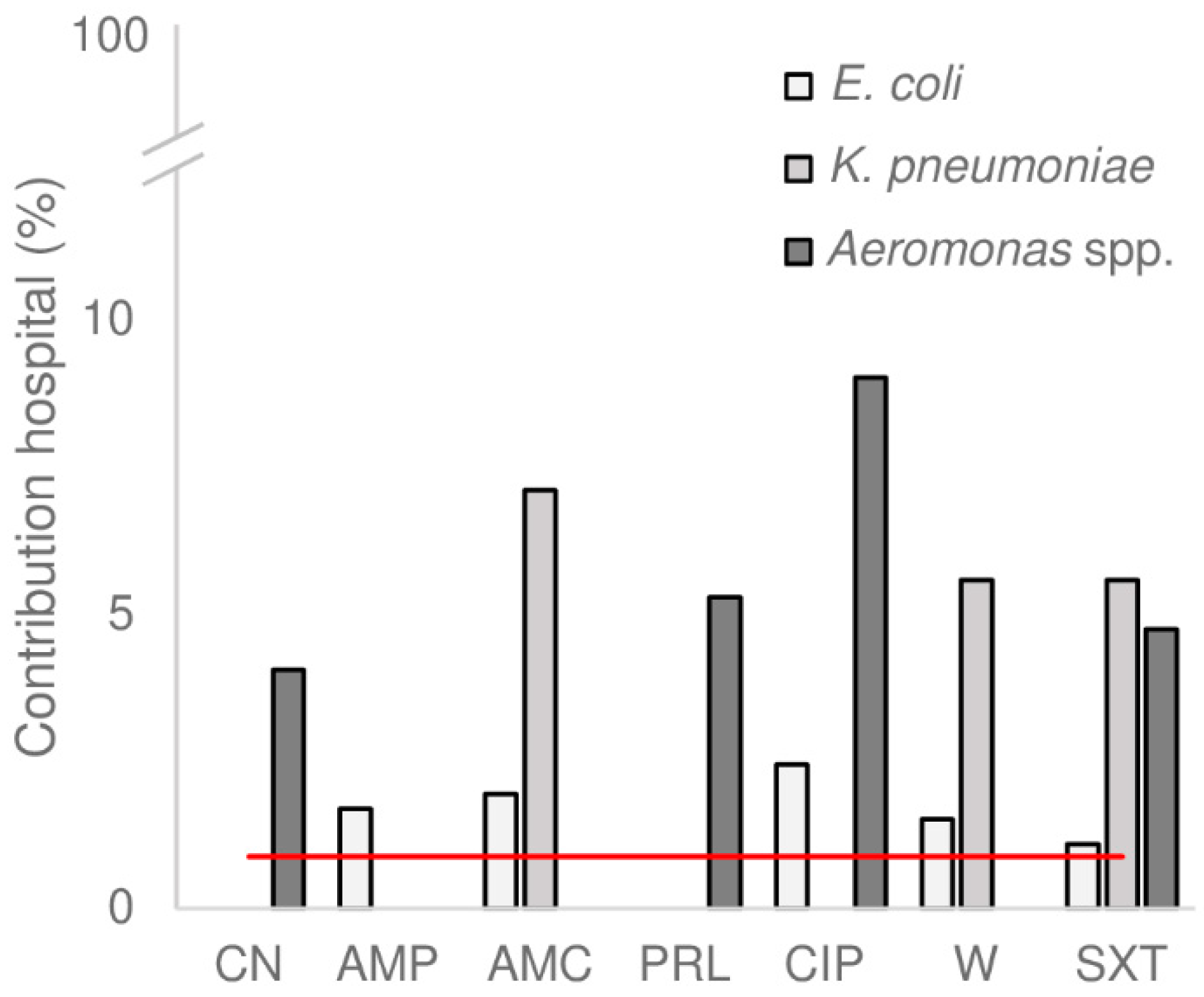
3.4. Hospital Wastewater as Source for Antimicrobial Resistance in the WWTP
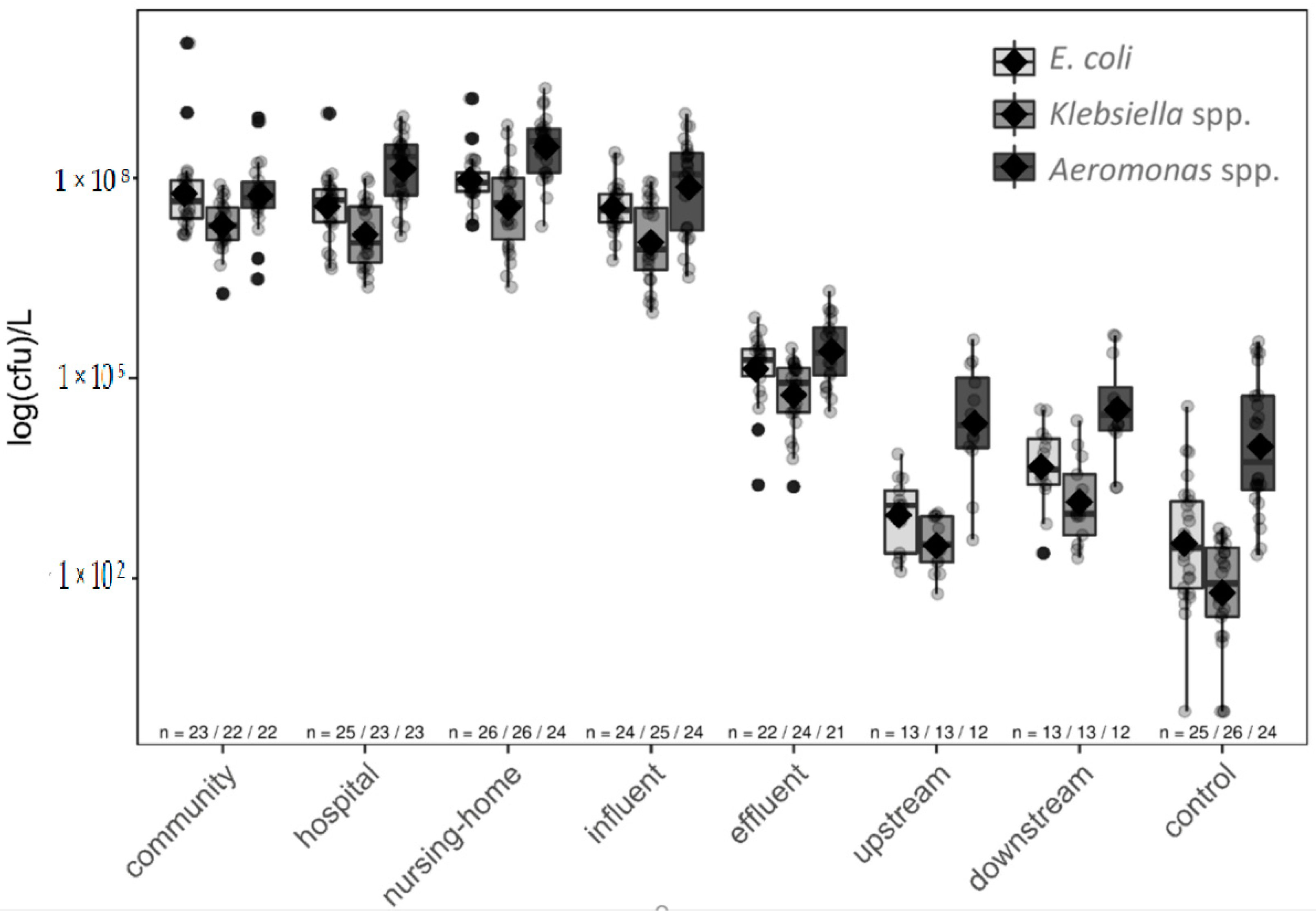
3.5. All Studied Species Survived WWTP Treatment and Impacted the Receiving River
3.6. Changes in AMR in WWTP, and Influence of WWTP on Surface Water
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Antimicrobial Resistance Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Shallcross, L.; Beckely, N.; Rait, G.; Hayward, A.; Petersen, I. Antibiotic prescribing frequency amongst patients in primary care: A cohort study using electronic health records. J. Am. Chem. 2017, 72, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Summary of the Latest Data on Antibiotic Consumption in the European Union; European Centre for Disease Prevention and Control: Solna Municipality, Sweden, 2017.
- Buelow, E.; Bayjanov, J.R.; Majoor, E.; Willems, R.J.; Bonten, M.J.; Schmitt, H.; van Schaik, W. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.K.; Colque, P.; Byfors, S.; Giske, C.G.; Mollby, R.; Kuhn, I. Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: Does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents 2015, 45, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef]
- Tello, A.; Austin, B.; Telfer, T.C. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 2012, 120, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Ferreira da Silva, M.; Vaz-Moreira, I.; Gonzalez-Pajuelo, M.; Nunes, O.C.; Manaia, C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007, 60, 166–176. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef]
- Amador, P.P.; Fernandes, R.M.; Prudencio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of class A and class C beta-lactamases. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2015, 50, 26–39. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sanchez-Melsio, A.; Borrego, C.M.; Barcelo, D.; Balcazar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Flach, C.F.; Genheden, M.; Fick, J.; Joakim Larsson, D.G. A Comprehensive Screening of Escherichia coli Isolates from Scandinavia’s Largest Sewage Treatment Plant Indicates No Selection for Antibiotic Resistance. Environ. Sci. Technol. 2018, 52, 11419–11428. [Google Scholar] [CrossRef] [PubMed]
- Reece, J.B.; Urry, L.A.; Cain, M.L.; Wasserman, S.A.; Minorsky, P.V.; Jackson, R.B.; Campbell, N.A. Campbell Biology; Pearson: London, UK, 2014. [Google Scholar]
- Odonkor, S.T.; Ampofo, J.K. Escherichia coli as an indicator of bacteriological quality of water: An overview. Microb. Res. 2013, 4, e2. [Google Scholar] [CrossRef]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Burgmann, H.; Sorum, H.; Norstrom, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Piotrowska, M.; Popowska, M. The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 2014, 64, 921–934. [Google Scholar] [CrossRef]
- Veeningen, R.; van der Meer, F. Geneesmiddelen in het oppervlaktewater: Standaarden bij de monitoring en prioritering van ‘hotspots’: [thema] nieuwe verontreinigingen. Bodem: Kwartaalblad Voor Informatie-Uitwisseling en Discussie over Bodembescherming en Bodemsanering 2015, 25, 31–34. [Google Scholar]
- ISO. Water Quality—General Guidance on the Enumeration of Micro-Organisms by Culture (ISO 8199:2005); ISO: Geneva, Switzerland, 2007. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters; The European Committee on Antimicrobial Susceptibility Testing, 2016. Version 6.0. Available online: http://www.eucast.org (accessed on 14 December 2016).
- Lamy, B.; Laurent, F.; Kodjo, A.; Roger, F.; Jumas-Bilak, E.; Marchandin, H. Which antibiotics and breakpoints should be used for Aeromonas susceptibility testing? Considerations from a comparison of agar dilution and disk diffusion methods using Enterobacteriaceae breakpoints. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2369–2377. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- WHO. Collaborating Centre for Drug Statistics Methodology; ATC Index with DDDs 2018; WHO Collaborating Centre: Oslo, Norway, 2018. [Google Scholar]
- Antonius-Zorggroep. Maatschappelijk Jaarverslag 2017; Antonius-Zorggroep: Sneek, The Netherlands, 2018. [Google Scholar]
- Liem, T.B.; Filius, F.M.; van der Linden, P.D.; Janknegt, R.; Natsch, S.; Vulto, A.G. Changes in antibiotic use in Dutch hospitals over a six-year period: 1997 to 2002. Neth. J. Med. 2005, 63, 354–360. [Google Scholar] [PubMed]
- SWAB (Dutch Working Party on Antibiotic Policy). Nethmap 2018—Consumption on Antimicrobial Agents and Antimicrobial Resistance among Medically Important Bacteria in The Netherlands; RIVM: Bilthoven, The Netherlands, 2018. [Google Scholar]
- Van Krimpen, P.C.; Van Bennekom, W.P.; Bult, A. Penicillins and cephalosporins. Physicochemical properties and analysis in pharmaceutical and biological matrices. Pharm. Weekbl. Sci. 1987, 9, 1–23. [Google Scholar] [PubMed]
- Gorrie, C.L.; Mirceta, M.; Wick, R.R.; Edwards, D.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; Pilcher, D.V.; et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 2017, 65, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Conlan, S.; Kong, H.H.; Segre, J.A. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS ONE 2012, 7, e47075. [Google Scholar] [CrossRef]
- Gray, J. Aeromonas in the gut. J. Infect. 1987, 15, 197–200. [Google Scholar]
- Chen, P.L.; Tsai, P.J.; Chen, C.S.; Lu, Y.C.; Chen, H.M.; Lee, N.Y.; Lee, C.C.; Li, C.W.; Li, M.C.; Wu, C.J.; et al. Aeromonas stool isolates from individuals with or without diarrhea in southern Taiwan: Predominance of Aeromonas veronii. J. Microbiol. Immunol. Infect. 2015, 48, 618–624. [Google Scholar] [CrossRef]
- Talan, D.A.; Takhar, S.S.; Krishnadasan, A.; Abrahamian, F.M.; Mower, W.R.; Moran, G.J.; Emergency ID Net Study Group. Fluoroquinolone-Resistant and Extended-Spectrum beta-Lactamase-Producing Escherichia coli Infections in Patients with Pyelonephritis, United States. Emerg. Infect. Dis. 2016, 22. [Google Scholar] [CrossRef]
- Doi, Y.; Park, Y.S.; Rivera, J.I.; Adams-Haduch, J.M.; Hingwe, A.; Sordillo, E.M.; Lewis, J.S., 2nd; Howard, W.J.; Johnson, L.E.; Polsky, B.; et al. Community-associated extended-spectrum beta-lactamase-producing Escherichia coli infection in the United States. Clin. Infect. Dis. 2013, 56, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Rooney, P.J.; O’Leary, M.C.; Loughrey, A.C.; McCalmont, M.; Smyth, B.; Donaghy, P.; Badri, M.; Woodford, N.; Karisik, E.; Livermore, D.M. Nursing homes as a reservoir of extended-spectrum beta-lactamase (ESBL)-producing ciprofloxacin-resistant Escherichia coli. J. Antimicrob. Chemother. 2009, 64, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Arvand, M.; Ruscher, C.; Bettge-Weller, G.; Goltz, M.; Pfeifer, Y. Prevalence and risk factors for colonization by Clostridium difficile and extended-spectrum beta-lactamase-producing Enterobacteriaceae in rehabilitation clinics in Germany. J. Hosp. Infect. 2018, 98, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; Lynch, G.; Italiaander, R.; Hamidjaja, R.A.; Schets, F.M.; de Roda Husman, A.M. Multidrug-Resistant and Extended Spectrum Beta-Lactamase-Producing Escherichia coli in Dutch Surface Water and Wastewater. PLoS ONE 2015, 10, e0127752. [Google Scholar] [CrossRef] [PubMed]
- Frigon, D.; Biswal, B.K.; Mazza, A.; Masson, L.; Gehr, R. Biological and physicochemical wastewater treatment processes reduce the prevalence of virulent Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef]
- Piotrowska, M.; Przygodzinska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of beta-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Health Aging 2018, 4, 267–285. [Google Scholar] [CrossRef]
- Percival, S.L.; Yates, M.V.; Williams, D.W.; Chalmers, R.; Gray, N.F. Microbiology of Waterborne Diseases: Microbiological Aspects and Risks; Elsevier: New York, NY, USA, 2014. [Google Scholar]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Larsson, D.G. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2017, 136, 25–39. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verburg, I.; García-Cobos, S.; Hernández Leal, L.; Waar, K.; Friedrich, A.W.; Schmitt, H. Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway. Microorganisms 2019, 7, 312. https://doi.org/10.3390/microorganisms7090312
Verburg I, García-Cobos S, Hernández Leal L, Waar K, Friedrich AW, Schmitt H. Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway. Microorganisms. 2019; 7(9):312. https://doi.org/10.3390/microorganisms7090312
Chicago/Turabian StyleVerburg, Ilse, Silvia García-Cobos, Lucia Hernández Leal, Karola Waar, Alex W. Friedrich, and Heike Schmitt. 2019. "Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway" Microorganisms 7, no. 9: 312. https://doi.org/10.3390/microorganisms7090312
APA StyleVerburg, I., García-Cobos, S., Hernández Leal, L., Waar, K., Friedrich, A. W., & Schmitt, H. (2019). Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway. Microorganisms, 7(9), 312. https://doi.org/10.3390/microorganisms7090312




