Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Article
2.2. In Vitro Dissolution Analyses
2.3. Animals and Test Article Administration
2.4. Amoxicillin Serum Measurement
2.5. Fecal DNA Extraction, Whole Genome Shotgun Sequencing and Metagenomic Analyses
2.6. Data Availability
2.7. Ethics Approval
3. Results
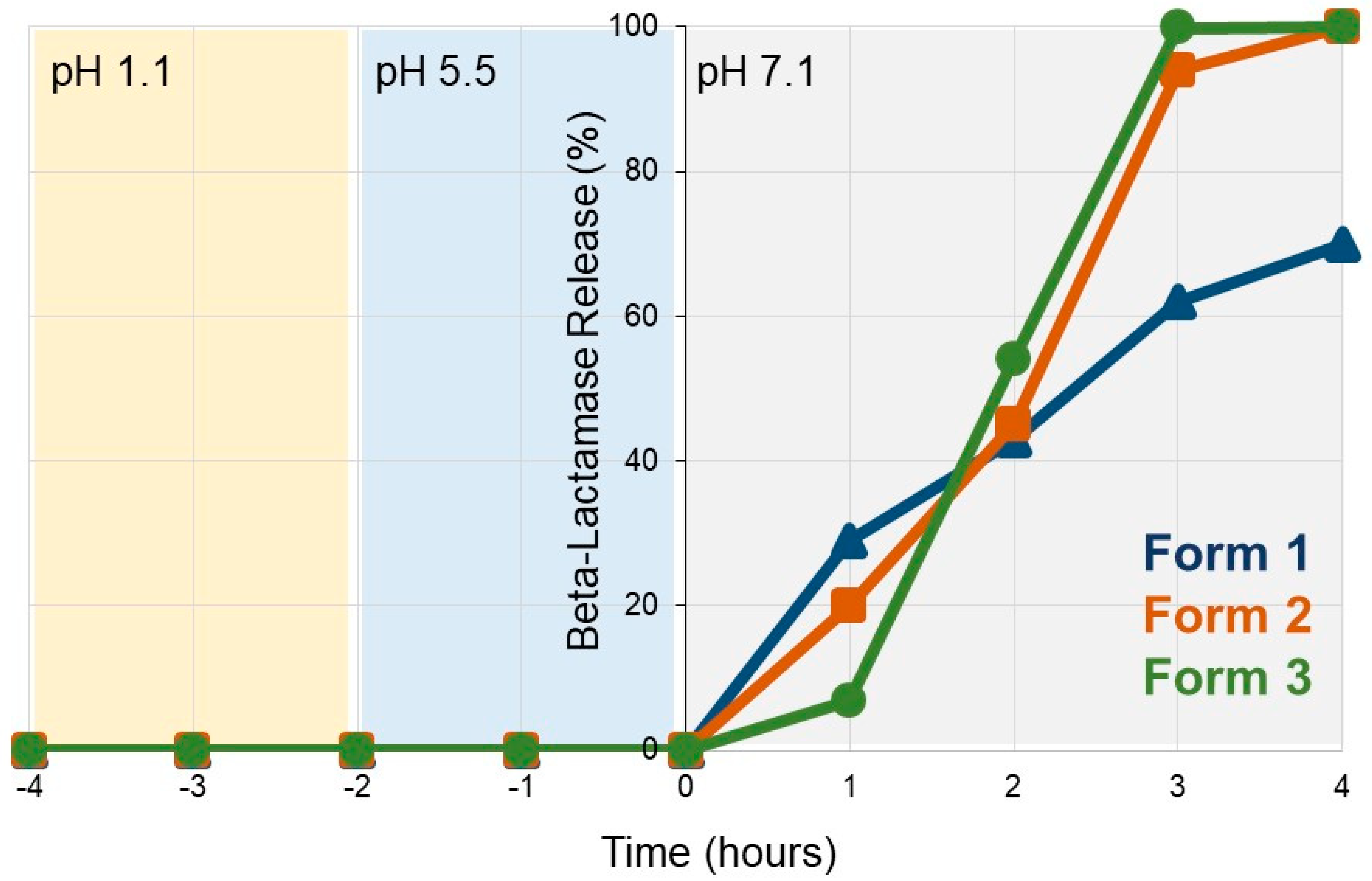
3.1. SYN-007 Exhibits pH-Dependent Dissolution In Vitro
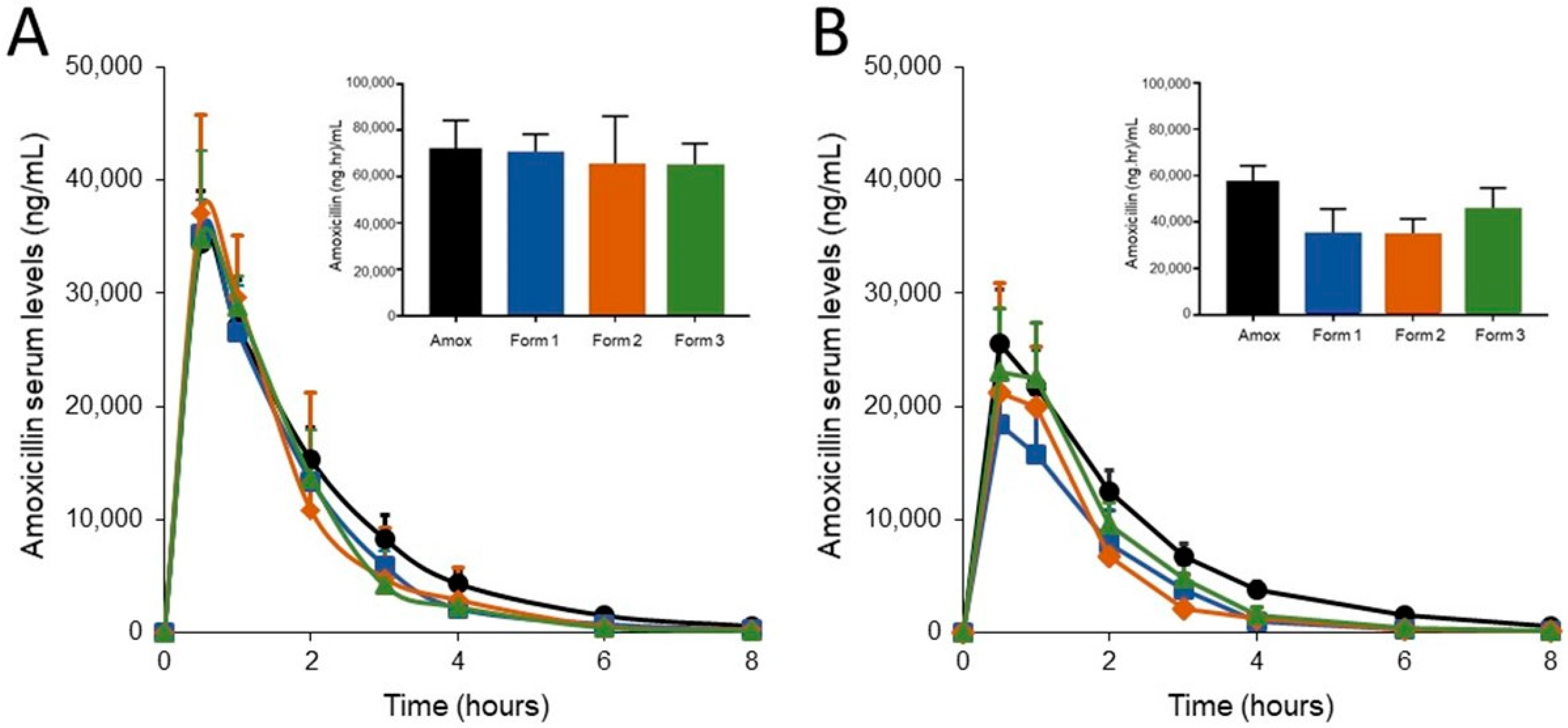
3.2. SYN-007 Formulation 3 Did Not Significantly Affect Oral Amoxicillin Systemic Absorption
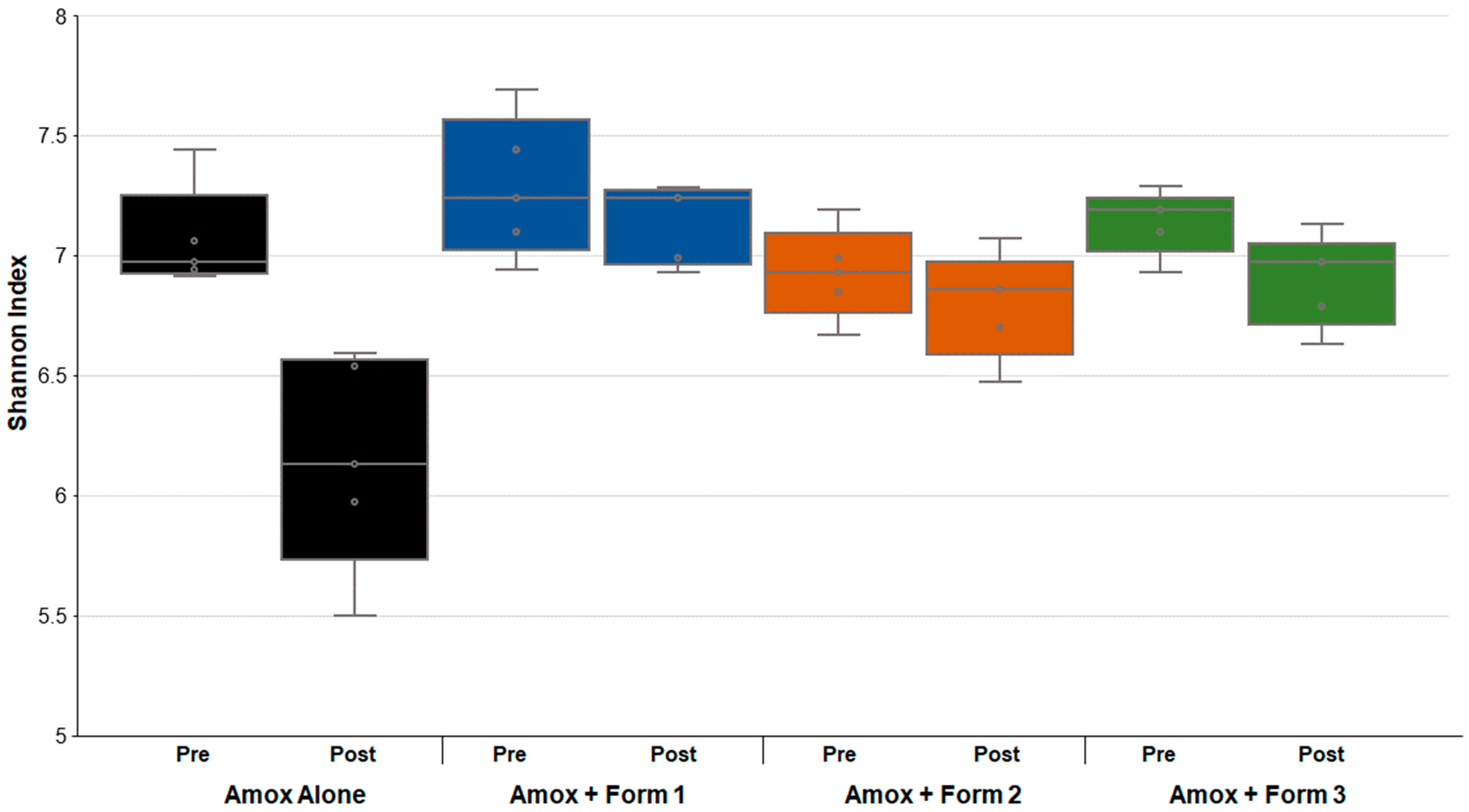
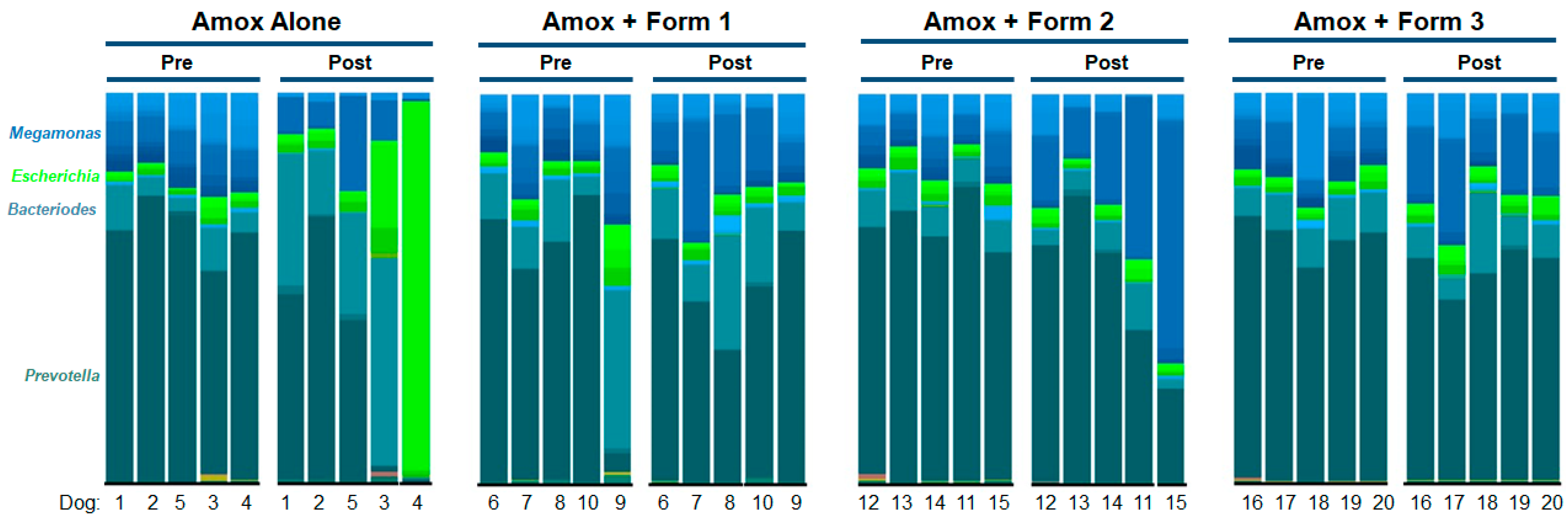
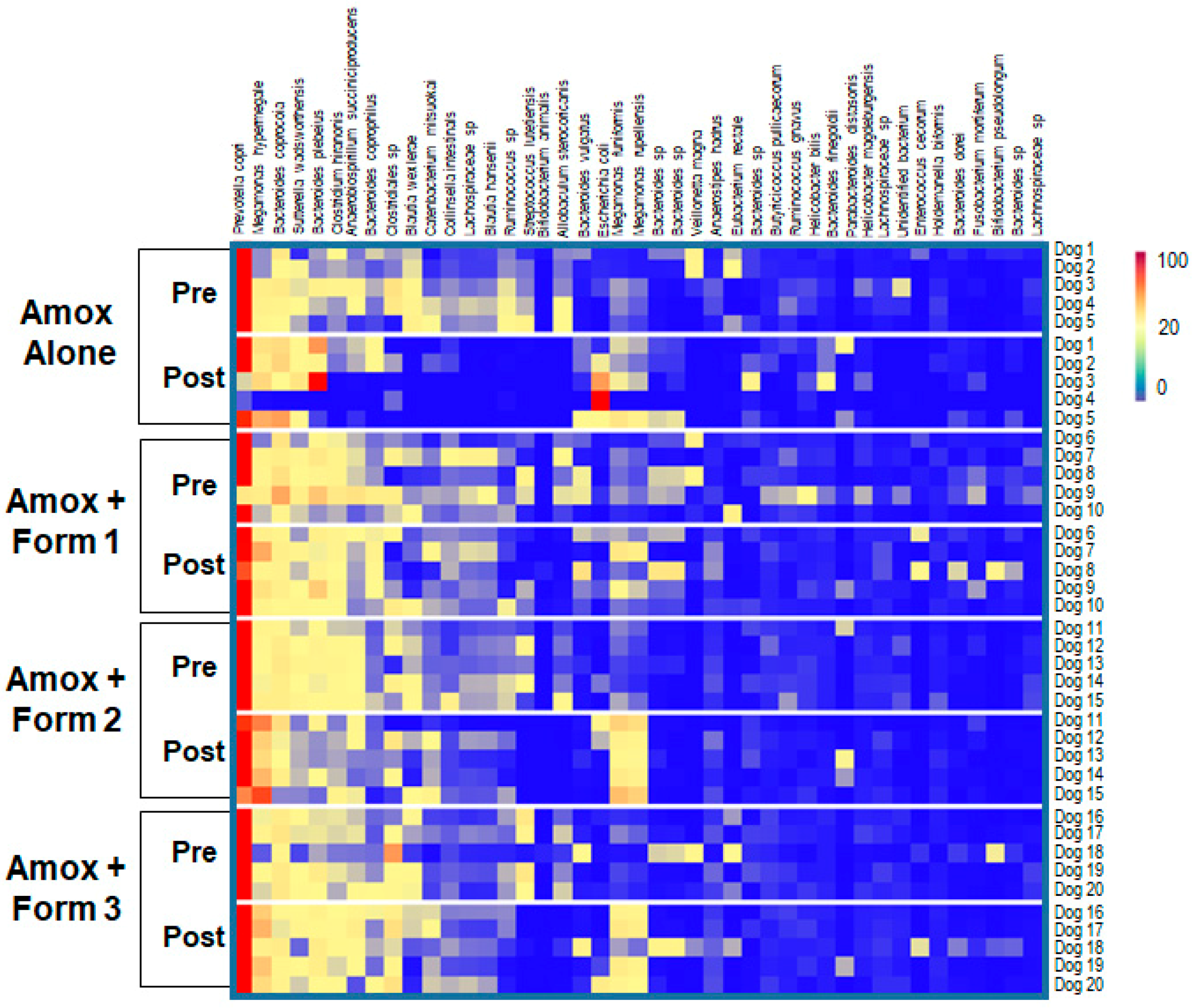
3.3. SYN-007 Mitigated Oral Amoxicillin-Mediated Microbiome Damage
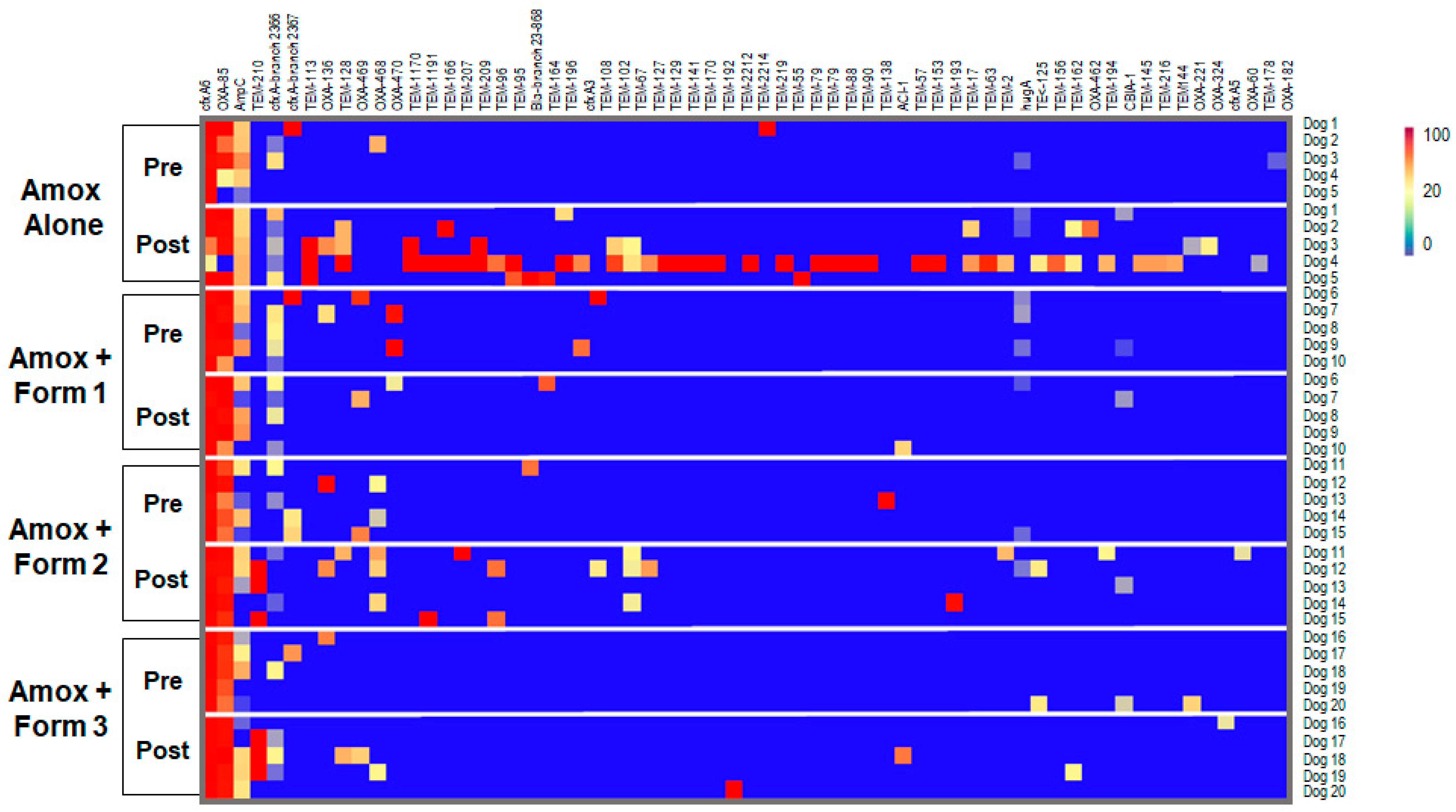
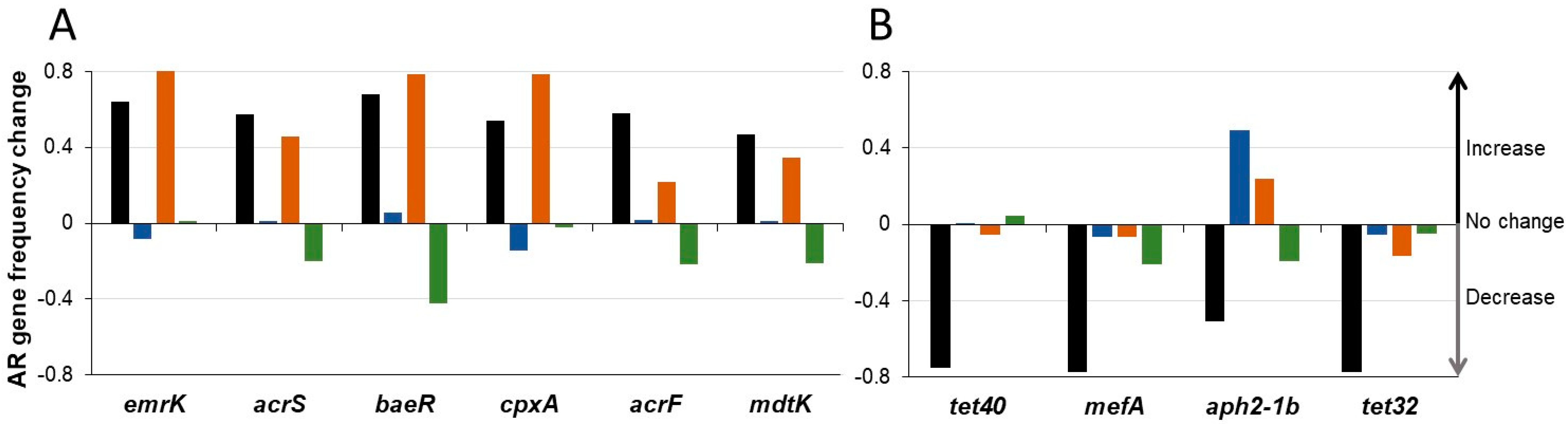
3.4. SYN-007 Reduced Antibiotic Resistance Gene Propagation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crandon, J.L.; Nicolau, D.P. Pharmacodynamic approaches to optimizing beta-lactam therapy. Crit. Care Clin. 2011, 27, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.; Zahar, J.R.; Lesprit, P.; Ruppe, E.; Leone, M.; Chastre, J.; Lucet, J.C.; Paugam-Burtz, C.; Brun-Buisson, C.; Timsit, J.F.; et al. Elaboration of a consensual definition of de-escalation allowing a ranking of beta-lactams. Clin. Microbiol. Infect. 2015, 21, 649.e1–649.e10. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Vardakas, K.Z.; Trigkidis, K.K.; Boukouvala, E.; Falagas, M.E. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2016, 48, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Watson, T.; Hickok, J.; Fraker, S.; Korwek, K.; Poland, R.E.; Septimus, E. Evaluating the Risk Factors for Hospital-Onset Clostridium difficile Infections in a Large Healthcare System. Clin. Infect. Dis. 2018, 66, 1957–1959. [Google Scholar] [CrossRef]
- Connelly, S.; Bristol, J.A.; Hubert, S.; Subramanian, P.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. SYN-004 (ribaxamase), an oral beta-lactamase, mitigates antibiotic-mediated dysbiosis in a porcine gut microbiome model. J. Appl Microbiol. 2017, 123, 66–79. [Google Scholar] [CrossRef]
- Kokai-Kun, J.; Robets, T.; Coughlin, O.; Le, C.; Whalen, H.; Stevenson, R.; Wacher, V.J.; Sliman, J. Use of ribaxamase (SYN-004), a beta-lactamase, to prevent Clostridium difficile infection in beta-lactam-treated patients: a double-blind, phase 2b, randomised placebo-controlled trial. Lancet Infect. Dis. 2019, 19, 487–496. [Google Scholar] [CrossRef]
- Kokai-Kun, J.F.; Roberts, T.; Coughlin, O.; Sicard, E.; Rufiange, M.; Fedorak, R.; Carter, C.; Adams, M.H.; Longstreth, J.; Whalen, H.; Sliman, J. The Oral beta-Lactamase SYN-004 (Ribaxamase) Degrades Ceftriaxone Excreted into the Intestine in Phase 2a Clinical Studies. Antimicrob Agents Chemother. 2017, 61, e02197-16. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.gov. A study of SYN-004 for the prevention of C. diff in patients with a LRTI. Available online: https://clinicaltrials.gov/ct2/show/NCT02563106 (accessed on 1 May 2019).
- Kaleko, M.; Bristol, J.A.; Hubert, S.; Parsley, T.; Widmer, G.; Tzipori, S.; Subramanian, P.; Hasan, N.; Koski, P.; Kokai-Kun, J.; et al. Development of SYN-004, an oral beta-lactamase treatment to protect the gut microbiome from antibiotic-mediated damage and prevent Clostridium difficile infection. Anaerobe 2016, 41, 58–67. [Google Scholar] [CrossRef]
- Bristol, A.; Hubert, S.; Hofmann, F.; Baer, H. Formulation development of SYN-004 (ribaxamase) oral solid dosage form, a beta-lactamase to prevent intravenous antibiotic-associated dysbiosis of the colon. Int J. Pharm. 2017, 534, 25–34. [Google Scholar] [CrossRef]
- Prevention CfDCa: Outpatient antibiotic prescriptions—United States, 2015. 2015.
- Barr, W.H.; Zola, E.M.; Candler, E.L.; Hwang, S.M.; Tendolkar, A.V.; Shamburek, R.; Parker, B.; Hilty, M.D. Differential absorption of amoxicillin from the human small and large intestine. Clin. Pharm. 1994, 56, 279–285. [Google Scholar] [CrossRef]
- Philip, A.K.; Philip, B. Colon targeted drug delivery systems: a review on primary and novel approaches. Oman. Med. J. 2010, 25, 79–87. [Google Scholar] [CrossRef]
- Yoshida, T.; Lai, T.C.; Kwon, G.S.; Sako, K. pH- and ion-sensitive polymers for drug delivery. Expert. Opin. Drug Deliv. 2013, 10, 1497–1513. [Google Scholar] [CrossRef] [PubMed]
- Hubert, S.; Chadwick, A.; Wacher, V.; Coughlin, O.; Kokai-Kun, J.; Bristol, A. Development of a Modified-Release Formulation of Lovastatin Targeted to Intestinal Methanogens Implicated in Irritable Bowel Syndrome With Constipation. J. Pharm. Sci. 2018, 107, 662–671. [Google Scholar] [CrossRef]
- Nugent, S.G.; Kumar, D.; Rampton, D.S.; Evans, D.F. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 2001, 48, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Bae, J.; Oshi, M.A.; Kim, M.S.; Moon, H.R.; Lee, B.L.; Im, E.; Jung, Y.; Yoo, J.W. Colon-targeted delivery of cyclosporine A using dual-functional Eudragit® FS30D/PLGA nanoparticles ameliorates murine experimental colitis. Int J. Nanomed. 2018, 13, 1225–1240. [Google Scholar] [CrossRef]
- Evonik Industries AG, Darmstadt, Germany. Technical Information: EUDRAGIT L 30 D-55 and EUDRAGIT L 100–55, Summary of Safety Data. vol. TOX.LD/E. 2012, pp. 1–15. Available online: https://www.stobec.com/DATA/PRODUIT/1598~v~data_8595.pdf (accessed on 1 May 2019).
- Arndt, M.; Chokshi, H.; Tang, K.; Parrott, N.J.; Reppas, C.; Dressman, J.B. Dissolution media simulating the proximal canine gastrointestinal tract in the fasted state. Eur. J. Pharm. Biopharm. 2013, 84, 633–641. [Google Scholar] [CrossRef]
- Coelho, L.P.; Kultima, J.R.; Costea, P.I.; Fournier, C.; Pan, Y.; Czarnecki-Maulden, G.; Hayward, M.R.; Forslund, S.K.; Schmidt, T.S.B.; Descombes, P.; et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome 2018, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.A.; Young, B.A.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS ONE 2014, 9, e97699. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Kozlova, E.V.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA 2016, 113, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Hourigan, S.K.; Subramanian, P.; Hasan, N.A.; Ta, A.; Klein, E.; Chettout, N.; Huddleston, K.; Deopujari, V.; Levy, S.; Baveja, R.; et al. Comparison of Infant Gut and Skin Microbiota, Resistome and Virulome Between Neonatal Intensive Care Unit (NICU) Environments. Front. Microbiol. 2018, 9, 1361. [Google Scholar] [CrossRef]
- Roy, M.A.; Arnaud, J.M.; Jasmin, P.M.; Hamner, S.; Hasan, N.A.; Colwell, R.R.; Ford, T.E. A Metagenomic Approach to Evaluating Surface Water Quality in Haiti. Int. J. Environ. Res. Public Health 2018, 15, 2211. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell. Syst. Tech. J. 1948, 27, 44. [Google Scholar] [CrossRef]
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010, 11, 367. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Bourgeois, S.; Laham, A.; Besnard, M.; Andremont, A.; Fattal, E. In vitro and in vivo evaluation of pectin beads for the colon delivery of beta-lactamases. J. Drug Target. 2005, 13, 277–284. [Google Scholar] [CrossRef]
- Bourgeois, S.; Tsapis, N.; Honnas, H.; Andremont, A.; Shakweh, M.; Besnard, M.; Fattal, E. Colonic delivery of beta-lactamases does not affect amoxicillin pharmacokinetics in rats. J. Pharm. Sci. 2008, 97, 1853–1863. [Google Scholar] [CrossRef]
- Martinez, M.N.; Papich, M.G. Factors influencing the gastric residence of dosage forms in dogs. J. Pharm. Sci. 2009, 98, 844–860. [Google Scholar] [CrossRef]
- Park, H.M.; Chernish, S.M.; Rosenek, B.D.; Brunelle, R.L.; Hargrove, B.; Wellman, H.N. Gastric emptying of enteric-coated tablets. Dig. Dis. Sci. 1984, 29, 207–212. [Google Scholar] [CrossRef]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef]
- Gronvold, A.M.; L’Abee-Lund, T.M.; Sorum, H.; Skancke, E.; Yannarell, A.C.; Mackie, R.I. Changes in fecal microbiota of healthy dogs administered amoxicillin. FEMS Microbiol. Ecol. 2010, 71, 313–326. [Google Scholar] [CrossRef]
- Beloshapka, A.N.; Dowd, S.E.; Suchodolski, J.S.; Steiner, J.M.; Duclos, L.; Swanson, K.S. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol. Ecol. 2013, 84, 532–541. [Google Scholar] [CrossRef]
- Li, M.; van Esch, B.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharm. 2018, 831, 52–59. [Google Scholar] [CrossRef]
- Schmidt, V.M.; Pinchbeck, G.; McIntyre, K.M.; Nuttall, T.; McEwan, N.; Dawson, S.; Williams, N.J. Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J. Antimicrob Chemother. 2018, 73, 3305–3316. [Google Scholar] [CrossRef]
- Connelly, S.; Subramanian, P.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Distinct consequences of amoxicillin and ertapenem exposure in the porcine gut microbiome. Anaerobe 2018. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef]

| Formulation | Enzyme Pellets | Capsule | Pellets per Capsule | Capsules per dose | Delivery Package | Schematic |
|---|---|---|---|---|---|---|
| 1 | FS30D | FS30D-coated Size 9h | 12 | 5 | Uncoated Size 0 |  |
| 2 | FS30D | Uncoated Size 5 | 64 | 1 | Uncoated Size 5 |  |
| 3 | L30-D55 | FS30D-coated Size 9h | 8 | 8 | Uncoated Size 0 |  |
| Group | Bacterial Species (Mean ± SD) | Significance | |
|---|---|---|---|
| Pretreatment | Post-Treatment | p value | |
| Amox Alone | 307 ± 42 | 202 ± 3 | 0.006 |
| Amox + Form 1 | 337 ± 61 | 303 ± 21 | > 0.999 |
| Amox + Form 2 | 274 ± 26 | 258 ± 29 | > 0.999 |
| Amox + Form 3 | 310 ± 27 | 259 ± 30 | 0.170 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Connelly, S.; Fanelli, B.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage. Microorganisms 2019, 7, 150. https://doi.org/10.3390/microorganisms7050150
Connelly S, Fanelli B, Hasan NA, Colwell RR, Kaleko M. Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage. Microorganisms. 2019; 7(5):150. https://doi.org/10.3390/microorganisms7050150
Chicago/Turabian StyleConnelly, Sheila, Brian Fanelli, Nur A. Hasan, Rita R. Colwell, and Michael Kaleko. 2019. "Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage" Microorganisms 7, no. 5: 150. https://doi.org/10.3390/microorganisms7050150
APA StyleConnelly, S., Fanelli, B., Hasan, N. A., Colwell, R. R., & Kaleko, M. (2019). Oral Beta-Lactamase Protects the Canine Gut Microbiome from Oral Amoxicillin-Mediated Damage. Microorganisms, 7(5), 150. https://doi.org/10.3390/microorganisms7050150





