MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Host Cells
2.3. MycoMarT7 Transposon Library Screening for Nitric Oxide Susceptibility
2.4. Sequencing of Mmt7 Gene Knockout Clones
2.5. Complementation of MAV_4644 Gene Knockout Clone
2.6. Uptake and Survival Assays
2.7. In Silico Analysis
2.8. Expression of MAV_4644, MAV_4643, and MAV_4642 Recombinant Proteins and Immunoprecipitation Assays
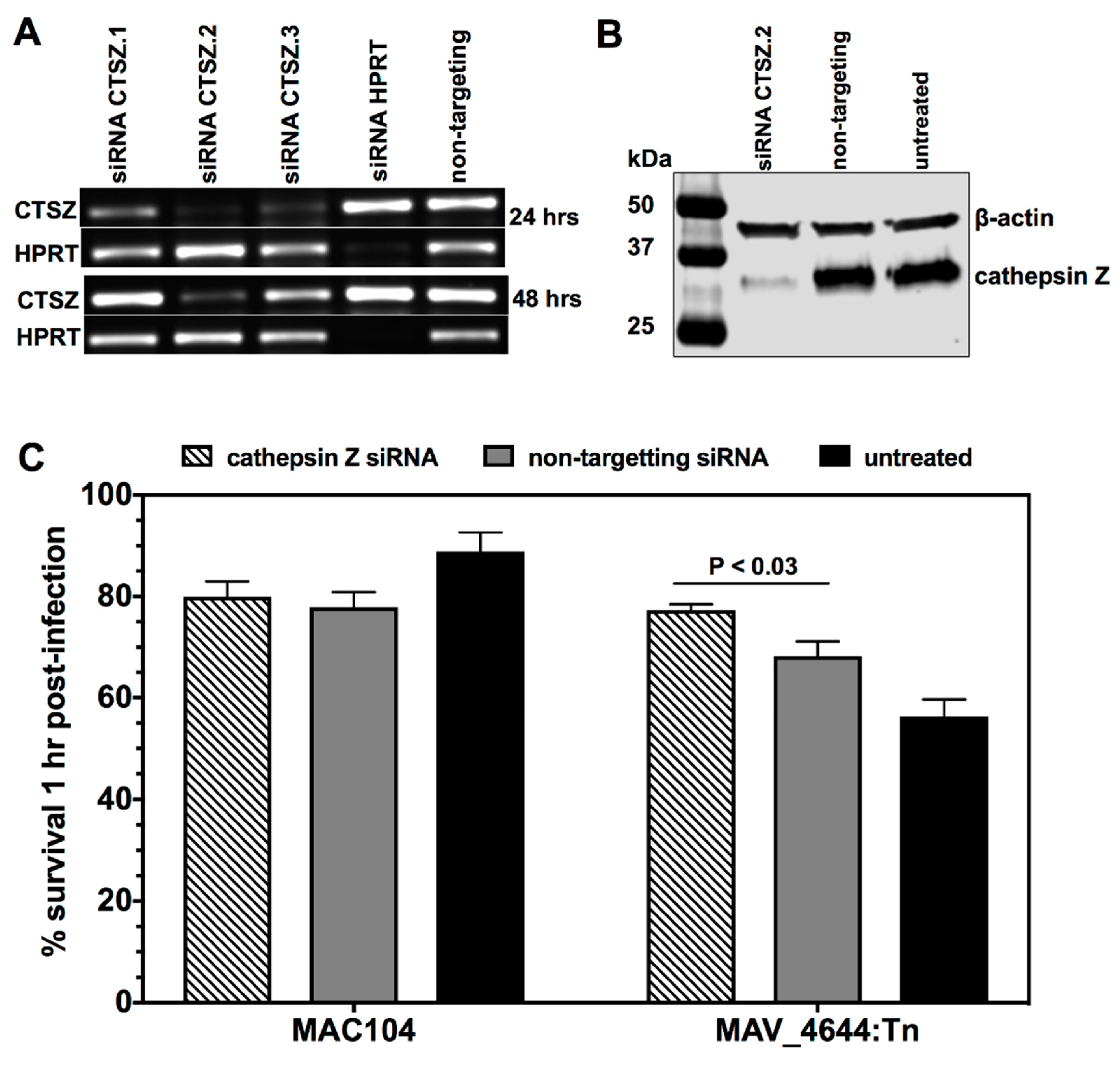
2.9. The siRNA Knock-Down of Cathepsin Z
2.10. Cathepsin Z Activity against MAC104 and MAV_4644:Tn
2.11. Statistical Analysis
3. Results
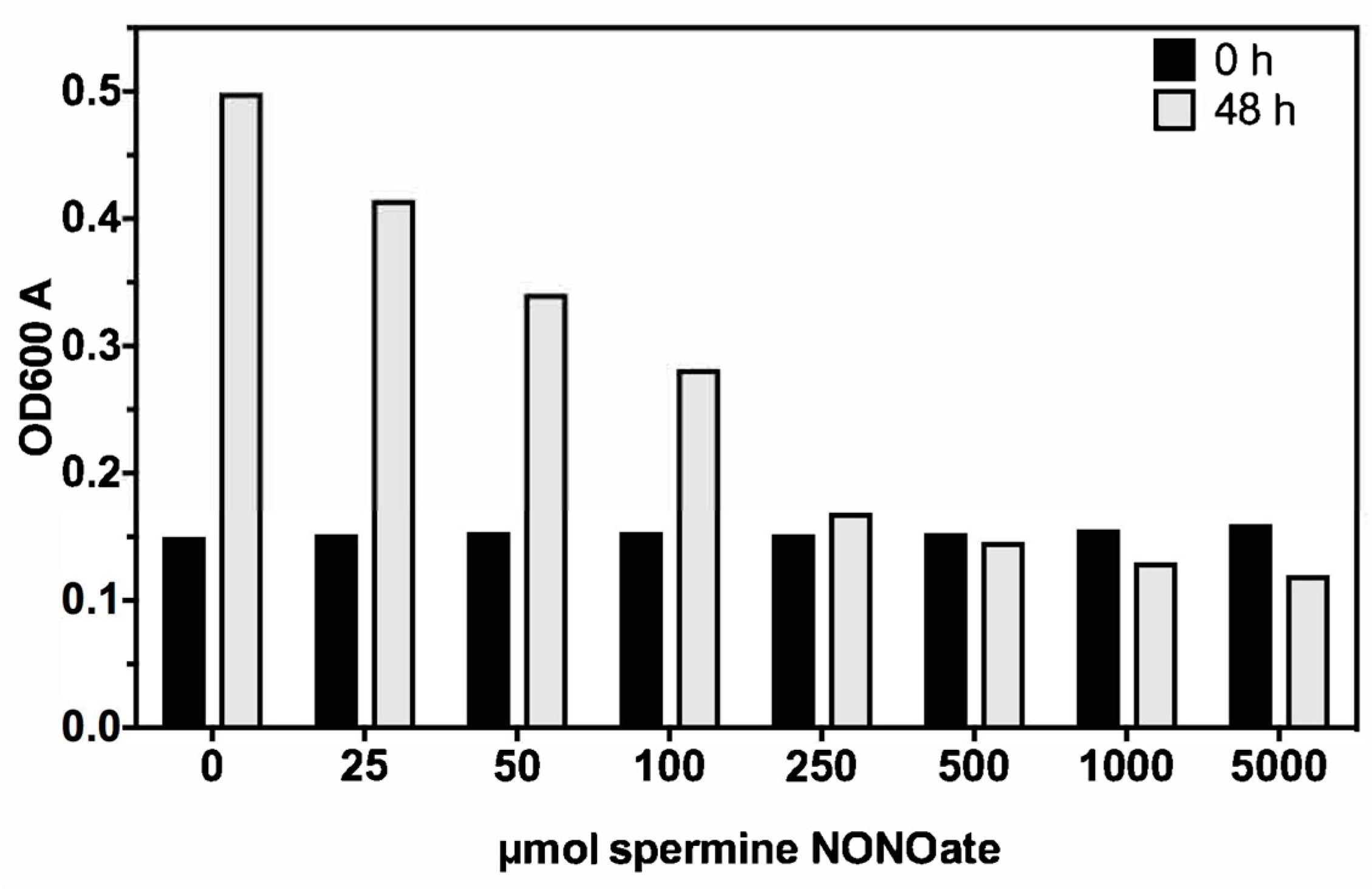
3.1. Nitric Oxide Susceptibility of MAH
3.2. Isolation of MAH Mmt7 Transposon Mutants Susceptible to Low Bacteriostatic Concentration of Nitric Oxide
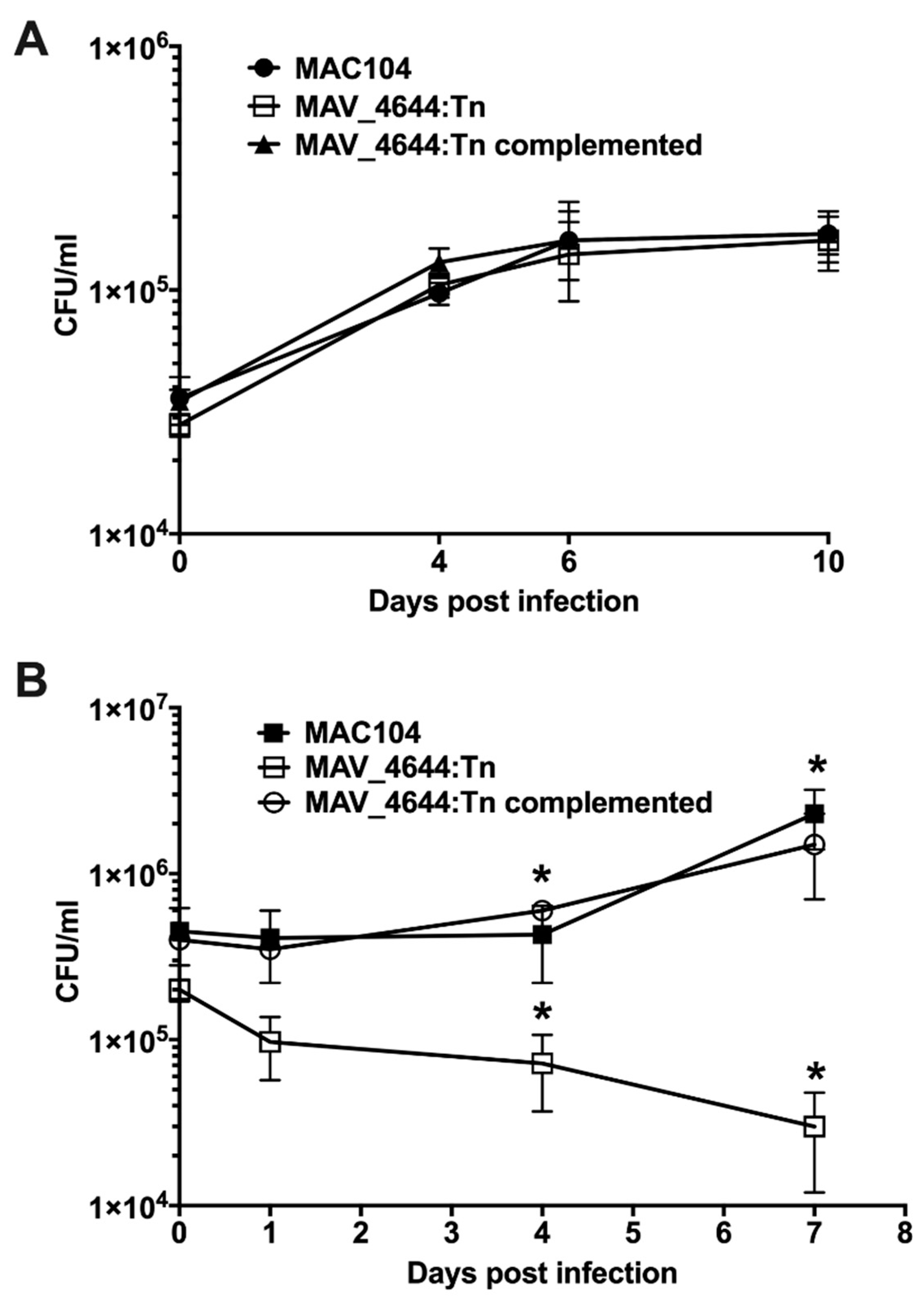
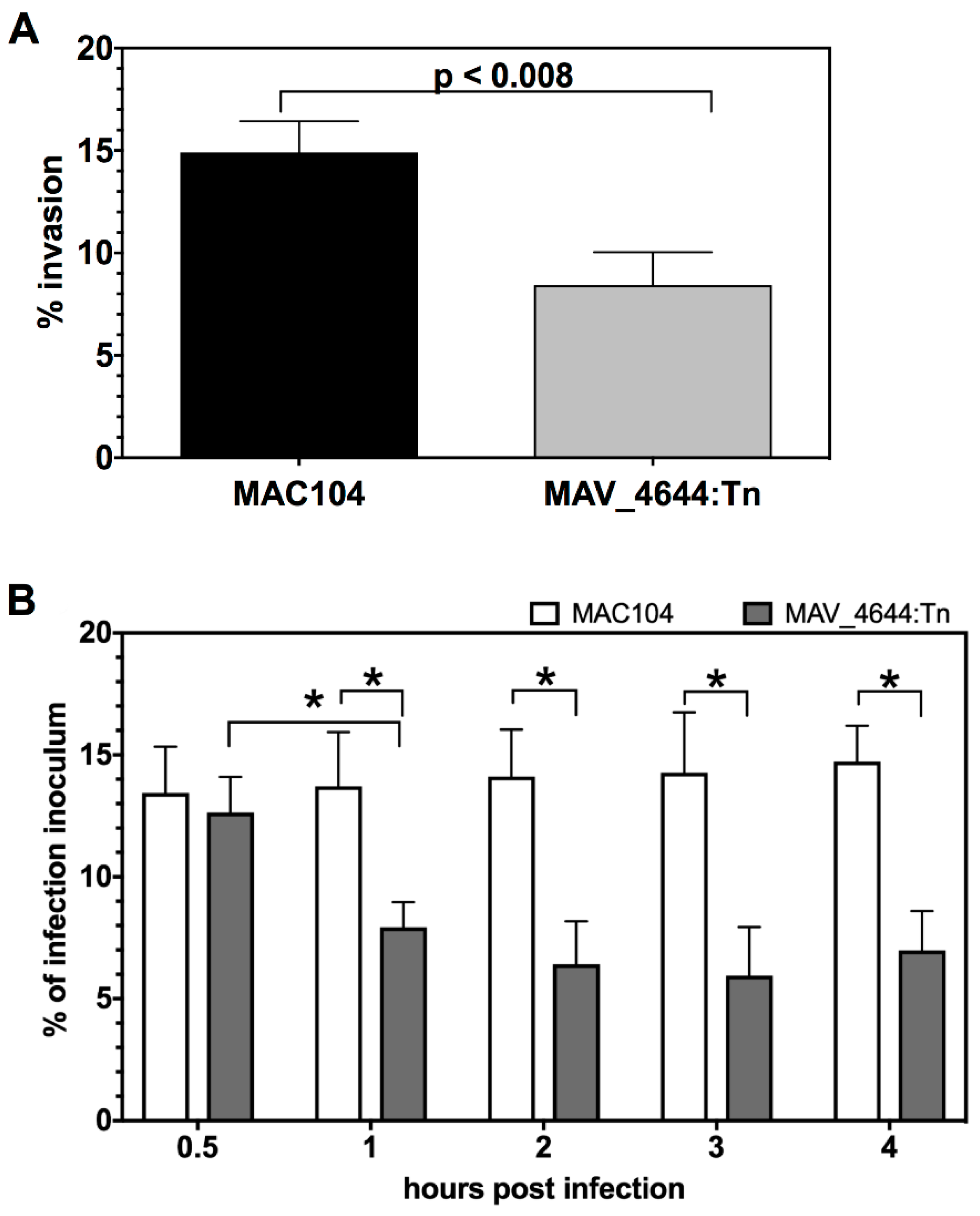
3.3. MAV_4644:Tn Is Attenuated in Murine Macrophages
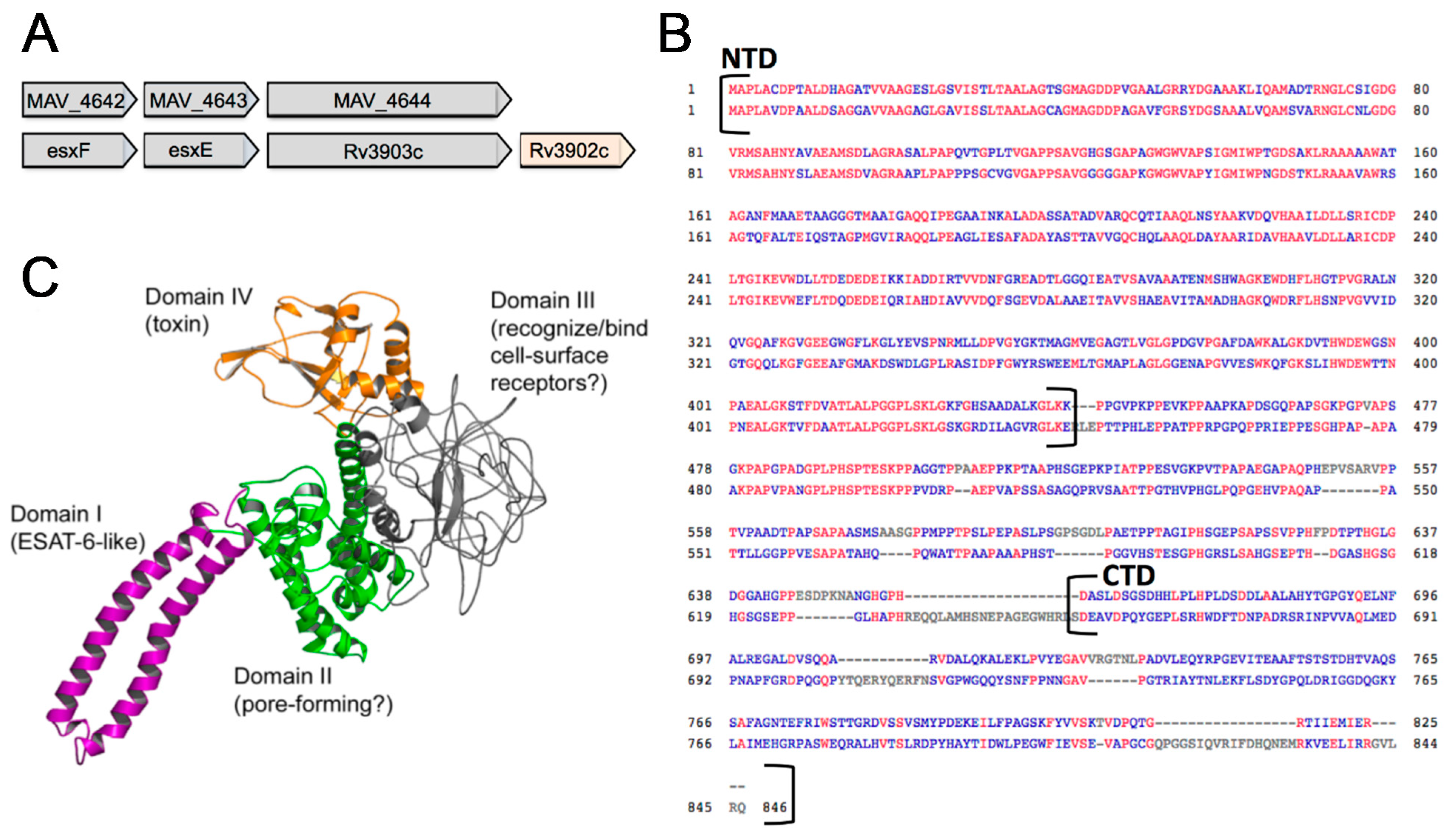
3.4. In Silico Analysis of MAV_4644
3.5. Complementation of the MAV_4644:Tn Mutant Restores the Bacterial Survivability Phenotype
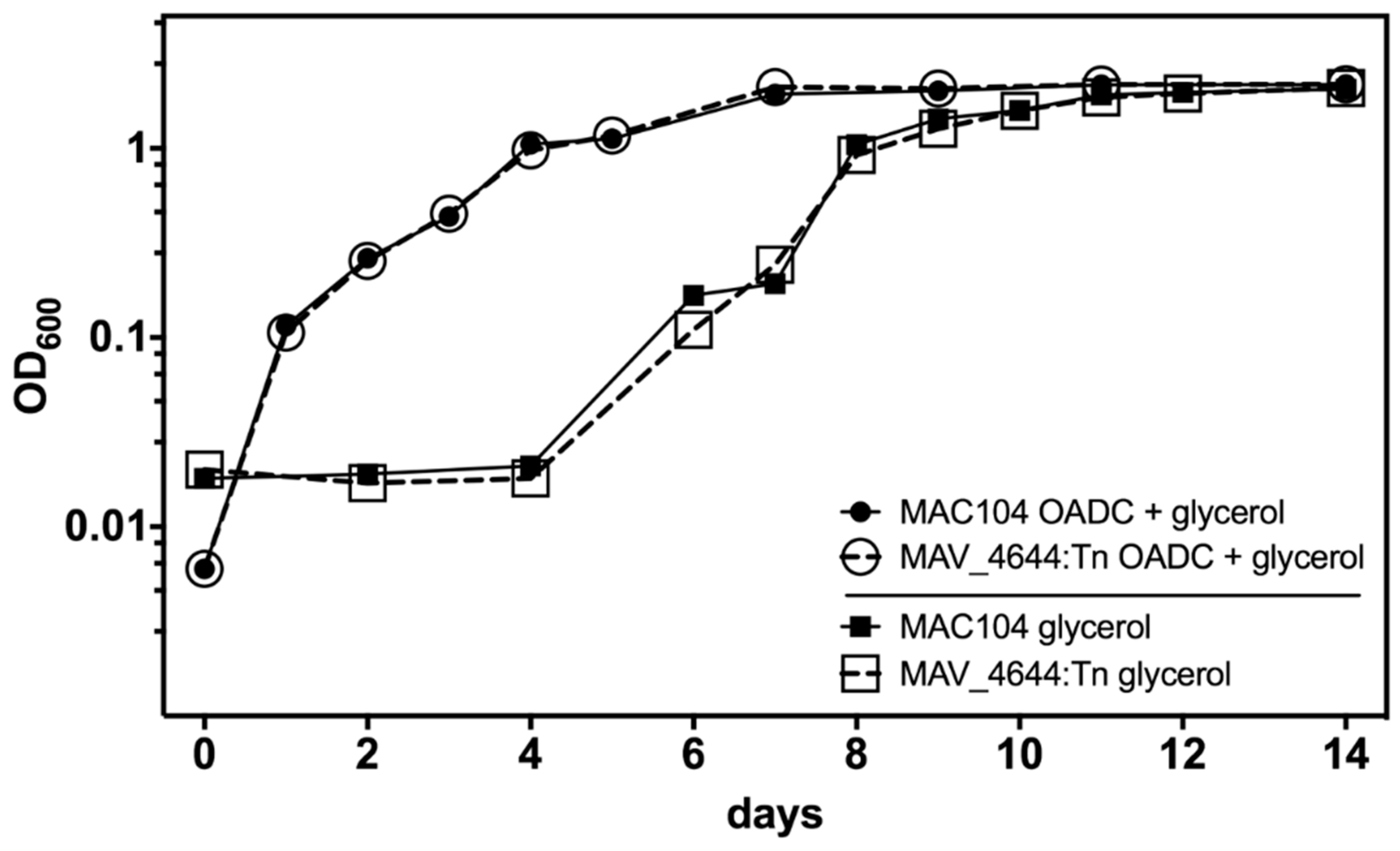
3.6. MAV_4644 Is Not Required for Growth in Defined Medium
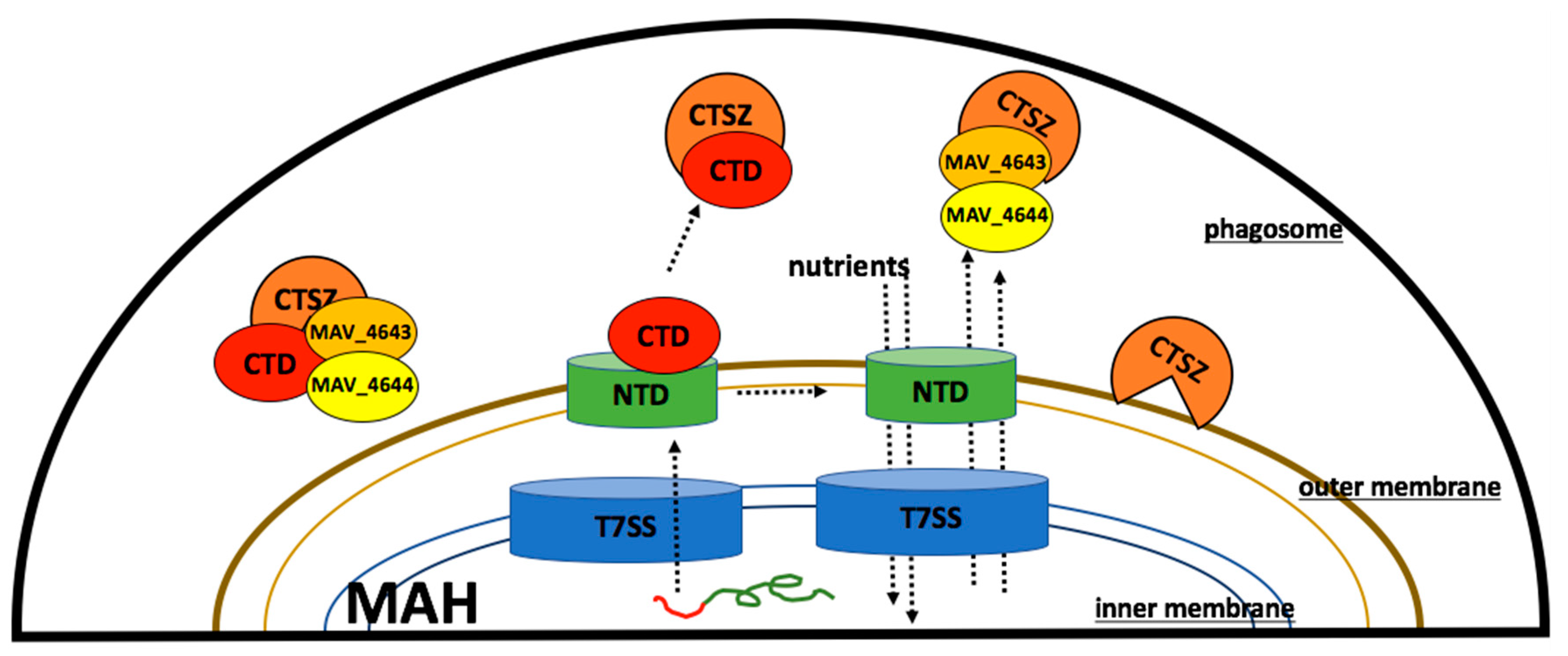
3.7. MAV_4644_CTD, MAV_4643, and MAV_4642 Recombinant Proteins Interact with the Cathepsin Z
3.8. MAV_4644:Tn Is Rapidly Killed in THP-1 Macrophages
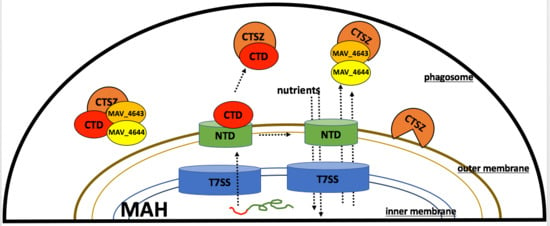
3.9. MAV_4644 Interferes with the Cathepsin Z Function
3.10. Effect of Cathepsin Z and NO on MAH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Inderlied, C.B.; Kemper, C.A.; Bermudez, L.E. The Mycobacterium avium complex. Clin. Microbiol. Rev. 1993, 6, 266–310. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.M.; Berney, M.; Gebhard, S.; Heinemann, M.; Cox, R.A.; Danilchanka, O.; Niederweis, M. Physiology of mycobacteria. Adv. Microb. Physiol. 2009, 55, 81–319. [Google Scholar] [PubMed]
- Gomez-Smith, C.K.; LaPara, T.M.; Hozalski, R.M. Sulfate Reducing Bacteria and Mycobacteria Dominate the Biofilm Communities in a Chloraminated Drinking Water Distribution System. Environ. Sci. Technol. 2015, 49, 8432–8440. [Google Scholar]
- Field, S.K.; Fisher, D.; Cowie, R.L. Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest 2004, 126, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Al Houqani, M.; Jamieson, F.; Chedore, P.; Mehta, M.; May, K.; Marras, T.K. Isolation prevalence of pulmonary nontuberculous mycobacteria in Ontario in 2007. Can. Respir. J. 2011, 18, 19–24. [Google Scholar] [CrossRef]
- Ishikane, M.; Tanuma, J. Mycobacterium avium complex enteritis in HIV-infected patient. IDCases 2014, 1, 22–23. [Google Scholar] [CrossRef]
- Lai, C.C.; Tan, C.K.; Chou, C.H.; Hsu, H.L.; Liao, C.H.; Huang, Y.T.; Yang, P.C.; Luh, K.T.; Hsueh, P.R. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000–2008. Emerg. Infect. Dis. 2010, 16, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Philley, J.V.; Griffith, D.E. Treatment of slowly growing mycobacteria. Clin. Chest Med. 2015, 36, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Marrakchi, H.; Laneelle, M.A.; Daffe, M. Mycolic acids: Structures, biosynthesis, and beyond. Chem. Biol. 2014, 21, 67–85. [Google Scholar] [CrossRef]
- Rocco, J.M.; Irani, V.R. Mycobacterium avium and modulation of the host macrophage immune mechanisms. Int. J. Tuberc. Lung Dis. 2011, 15, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Amer, A.O.; Swanson, M.S. A phagosome of one’s own: A microbial guide to life in the macrophage. Curr. Opin. Microbiol. 2002, 5, 56–61. [Google Scholar] [CrossRef]
- Sturgill-Koszycki, S.; Schaible, U.E.; Russell, D.G. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996, 15, 6960–6968. [Google Scholar] [CrossRef] [PubMed]
- Kelley, V.A.; Schorey, J.S. Mycobacterium’s arrest of phagosome maturation in macrophages requires Rab5 activity and accessibility to iron. Mol. Biol. Cell 2003, 14, 3366–3377. [Google Scholar] [CrossRef]
- Pires, D.; Marques, J.; Pombo, J.P.; Carmo, N.; Bettencourt, P.; Neyrolles, O.; Lugo-Villarino, G.; Anes, E. Role of Cathepsins in Mycobacterium tuberculosis Survival in Human Macrophages. Sci. Rep. 2016, 6, 32247. [Google Scholar] [CrossRef] [PubMed]
- Escuyer, V.; Haddad, N.; Frehel, C.; Berche, P. Molecular characterization of a surface-exposed superoxide dismutase of Mycobacterium avium. Microb. Pathog. 1996, 20, 41–55. [Google Scholar] [CrossRef]
- Liao, D.; Fan, Q.; Bao, L. The role of superoxide dismutase in the survival of Mycobacterium tuberculosis in macrophages. Jpn. J. Infect. Dis. 2013, 66, 480–488. [Google Scholar] [CrossRef] [PubMed]
- McNamara, M.; Tzeng, S.C.; Maier, C.; Wu, M.; Bermudez, L.E. Surface-exposed proteins of pathogenic mycobacteria and the role of cu-zn superoxide dismutase in macrophages and neutrophil survival. Proteome Sci. 2013, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.; Wolke, M.; Ernestus, K.; Plum, G. A mycobacterial gene involved in synthesis of an outer cell envelope lipid is a key factor in prevention of phagosome maturation. Infect. Immun. 2007, 75, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Houben, E.N.; Korotkov, K.V.; Bitter, W. Take five-Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta 2014, 1843, 1707–1716. [Google Scholar] [CrossRef]
- Groschel, M.I.; Sayes, F.; Simeone, R.; Majlessi, L.; Brosch, R. ESX secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016, 14, 677–691. [Google Scholar] [CrossRef]
- Simeone, R.; Bobard, A.; Lippmann, J.; Bitter, W.; Majlessi, L.; Brosch, R.; Enninga, J. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012, 8, e1002507. [Google Scholar] [CrossRef]
- Jeffrey, B.; Rose, S.J.; Gilbert, K.; Lewis, M.; Bermudez, L.E. Comparative analysis of the genomes of clinical isolates of Mycobacterium avium subsp. hominissuis regarding virulence-related genes. J. Med. Microbiol. 2017, 66, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- George, K.M.; Chatterjee, D.; Gunawardana, G.; Welty, D.; Hayman, J.; Lee, R.; Small, P.L. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science 1999, 283, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Fieldhouse, R.J.; Turgeon, Z.; White, D.; Merrill, A.R. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput. Biol. 2010, 6, e1001029. [Google Scholar] [CrossRef] [PubMed]
- Castagnini, M.; Picchianti, M.; Talluri, E.; Biagini, M.; Del Vecchio, M.; di Procolo, P.; Norais, N.; Nardi-Dei, V.; Balducci, E. Arginine-specific mono ADP-ribosylation in vitro of antimicrobial peptides by ADP-ribosylating toxins. PLoS ONE 2012, 7, e41417. [Google Scholar] [CrossRef] [PubMed]
- Park, E.K.; Jung, H.S.; Yang, H.I.; Yoo, M.C.; Kim, C.; Kim, K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007, 56, 45–50. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 2001, 98, 12712–12717. [Google Scholar] [CrossRef]
- Rose, S.J.; Bermudez, L.E. Identification of Bicarbonate as a Trigger and Genes Involved with Extracellular DNA Export in Mycobacterial Biofilms. mBio 2016, 7. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Mao, F.; Dam, P.; Chou, J.; Olman, V.; Xu, Y. DOOR: A database for prokaryotic operons. Nucleic Acids Res. 2009, 37, D459–D463. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.P.; St-Denis, N.A.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Bitter, W.; Houben, E.N.; Bottai, D.; Brodin, P.; Brown, E.J.; Cox, J.S.; Derbyshire, K.; Fortune, S.M.; Gao, L.Y.; Liu, J.; et al. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Siroy, A.; Lokareddy, R.K.; Speer, A.; Doornbos, K.S.; Cingolani, G.; Niederweis, M. The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol. 2015, 22, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Danilchanka, O.; Sun, J.; Pavlenok, M.; Maueroder, C.; Speer, A.; Siroy, A.; Marrero, J.; Trujillo, C.; Mayhew, D.L.; Doornbos, K.S.; et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc. Natl. Acad. Sci. USA 2014, 111, 6750–6755. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.S.; Florido, M.; Pais, T.F.; Appelberg, R. Improved clearance of Mycobacterium avium upon disruption of the inducible nitric oxide synthase gene. J. Immunol. 1999, 162, 6734–6739. [Google Scholar]
- Vazquez-Torres, A.; Stevanin, T.; Jones-Carson, J.; Castor, M.; Read, R.C.; Fang, F.C. Analysis of nitric oxide-dependent antimicrobial actions in macrophages and mice. Methods Enzymol. 2008, 437, 521–538. [Google Scholar]
- Tan, M.P.; Sequeira, P.; Lin, W.W.; Phong, W.Y.; Cliff, P.; Ng, S.H.; Lee, B.H.; Camacho, L.; Schnappinger, D.; Ehrt, S.; et al. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE 2010, 5, e13356. [Google Scholar] [CrossRef]
- Sohaskey, C.D. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 2008, 190, 2981–2986. [Google Scholar] [CrossRef]
- Knight, D.A.; Finck-Barbancon, V.; Kulich, S.M.; Barbieri, J.T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 1995, 63, 3182–3186. [Google Scholar]
- Rocha, C.L.; Coburn, J.; Rucks, E.A.; Olson, J.C. Characterization of Pseudomonas aeruginosa exoenzyme S as a bifunctional enzyme in J774A.1 macrophages. Infect. Immun. 2003, 71, 5296–5305. [Google Scholar] [CrossRef]
- Barbieri, A.M.; Sha, Q.; Bette-Bobillo, P.; Stahl, P.D.; Vidal, M. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect. Immun. 2001, 69, 5329–5334. [Google Scholar] [CrossRef] [PubMed]
- Rangel, S.M.; Diaz, M.H.; Knoten, C.A.; Zhang, A.; Hauser, A.R. The Role of ExoS in Dissemination of Pseudomonas aeruginosa during Pneumonia. PLoS Pathog. 2015, 11, e1004945. [Google Scholar] [CrossRef]
- Pallen, M.J.; Lam, A.C.; Loman, N.J.; McBride, A. An abundance of bacterial ADP-ribosyltransferases—Implications for the origin of exotoxins and their human homologues. Trends Microbiol. 2001, 9, 302–307. [Google Scholar] [CrossRef]
- Glowacki, G.; Braren, R.; Firner, K.; Nissen, M.; Kuhl, M.; Reche, P.; Bazan, F.; Cetkovic-Cvrlje, M.; Leiter, E.; Haag, F.; et al. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002, 11, 1657–1670. [Google Scholar] [CrossRef]
- Ohol, Y.M.; Goetz, D.H.; Chan, K.; Shiloh, M.U.; Craik, C.S.; Cox, J.S. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 2010, 7, 210–220. [Google Scholar] [CrossRef]
- Sreejit, G.; Ahmed, A.; Parveen, N.; Jha, V.; Valluri, V.L.; Ghosh, S.; Mukhopadhyay, S. The ESAT-6 protein of Mycobacterium tuberculosis interacts with beta-2-microglobulin (beta2M) affecting antigen presentation function of macrophage. PLoS Pathog. 2014, 10, e1004446. [Google Scholar] [CrossRef]
- Renshaw, P.S.; Panagiotidou, P.; Whelan, A.; Gordon, S.V.; Hewinson, R.G.; Williamson, R.A.; Carr, M.D. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 2002, 277, 21598–21603. [Google Scholar] [PubMed]
- Conus, S.; Simon, H.U. Cathepsins: Key modulators of cell death and inflammatory responses. Biochem. Pharmacol. 2008, 76, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Urk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar]
- Sendide, K.; Deghmane, A.E.; Pechkovsky, D.; Av-Gay, Y.; Talal, A.; Hmama, Z. Mycobacterium bovis BCG attenuates surface expression of mature class II molecules through IL-10-dependent inhibition of cathepsin S. J. Immunol. 2005, 175, 5324–5332. [Google Scholar] [CrossRef]
- Nepal, R.M.; Mampe, S.; Shaffer, B.; Erickson, A.H.; Bryant, P. Cathepsin L maturation and activity is impaired in macrophages harboring M. avium and M. tuberculosis. Int. Immunol. 2006, 18, 931–939. [Google Scholar] [CrossRef]
- Adams, L.A.; Moller, M.; Nebel, A.; Schreiber, S.; van der Merwe, L.; van Helden, P.D.; Hoal, E.G. Polymorphisms in MC3R promoter and CTSZ 3′UTR are associated with tuberculosis susceptibility. Eur. J. Hum. Genet. 2011, 19, 676–681. [Google Scholar] [CrossRef][Green Version]
- Baker, A.R.; Zalwango, S.; Malone, L.L.; Igo, R.P., Jr.; Qiu, F.; Nsereko, M.; Adams, M.D.; Supelak, P.; Mayanja-Kizza, H.; Boom, W.H.; et al. Genetic susceptibility to tuberculosis associated with cathepsin Z haplotype in a Ugandan household contact study. Hum. Immunol. 2011, 72, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, I.; Velasco, G.; Pendas, A.M.; Fueyo, A.; Lopez-Otin, C. Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J. Biol. Chem. 1998, 273, 16816–16823. [Google Scholar] [CrossRef]
- Garin, J.; Diez, R.; Kieffer, S.; Dermine, J.F.; Duclos, S.; Gagnon, E.; Sadoul, R.; Rondeau, C.; Desjardins, M. The phagosome proteome: Insight into phagosome functions. J. Cell. Biol. 2001, 152, 165–180. [Google Scholar] [CrossRef]
- Garg, R.; Tripathi, D.; Kant, S.; Chandra, H.; Bhatnagar, R.; Banerjee, N. The conserved hypothetical protein Rv0574c is required for cell wall integrity, stress tolerance, and virulence of Mycobacterium tuberculosis. Infect. Immun. 2015, 83, 120–129. [Google Scholar] [CrossRef]
- Bhutani, N.; Piccirillo, R.; Hourez, R.; Venkatraman, P.; Goldberg, A.L. Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes. J. Biol. Chem. 2012, 287, 17471–17482. [Google Scholar] [CrossRef]
- Tripathi, D.; Chandra, H.; Bhatnagar, R. Poly-L-glutamate/glutamine synthesis in the cell wall of Mycobacterium bovis is regulated in response to nitrogen availability. BMC Microbiol. 2013, 13, 226. [Google Scholar] [CrossRef]
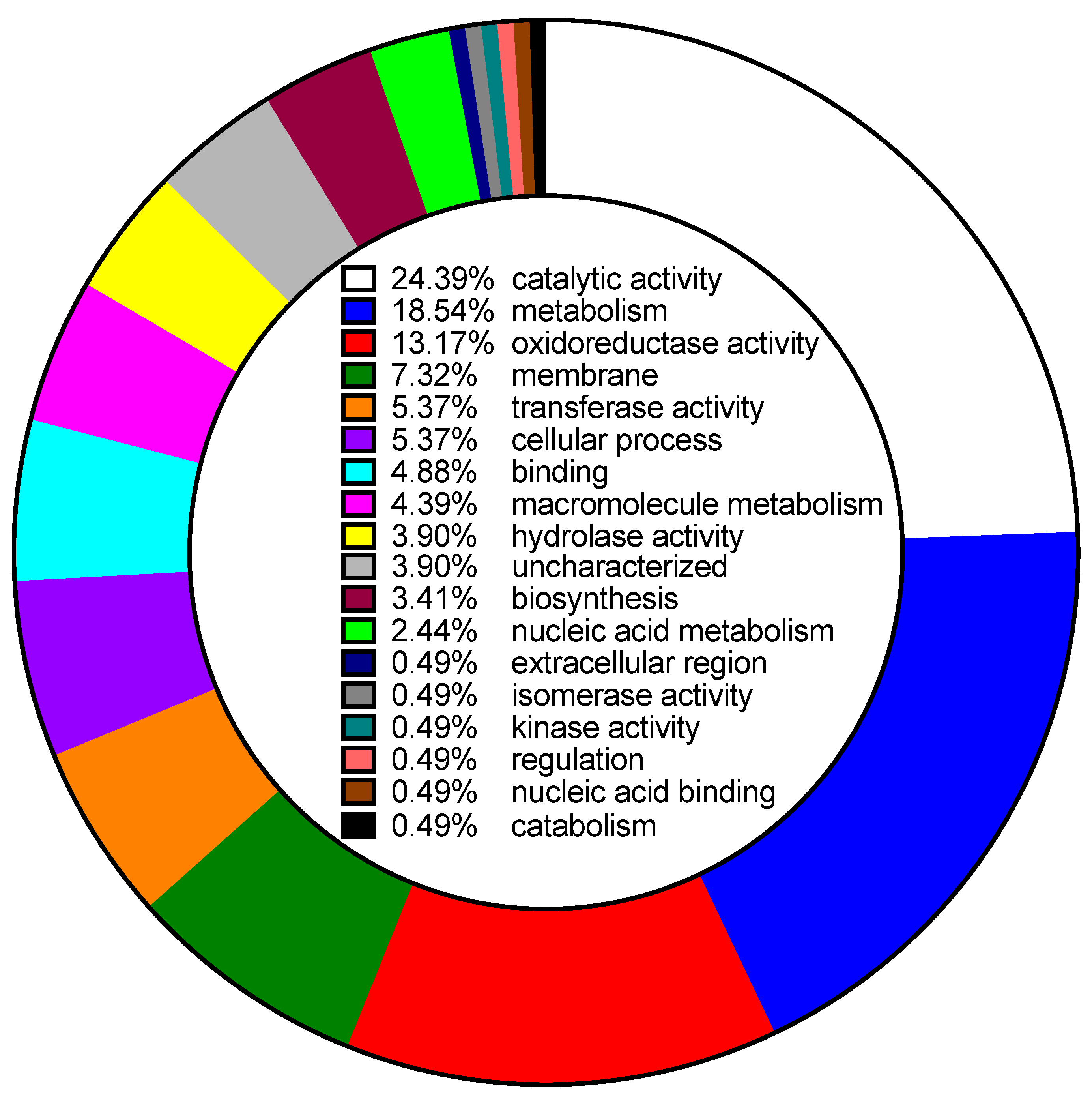
| Gene ID | Clone | % of WT | Description | Uniprot Accession | Locus Tag |
|---|---|---|---|---|---|
| MAV_0118 | 2B2 | 16.2 | Ppe family protein Subtilase family protein Monooxygenase, flavin-binding family protein Uncharacterized protein Uncharacterized protein Short chain dehydrogenase/reductase family protein Uncharacterized protein 3-Hydroxyacyl-CoA dehydrogenase type-2 Uncharacterized protein Gp108 protein RNA polymerase sigma factor Aldehyde dehydrogenase (NAD) family protein Putative acyl-CoA dehydrogenase Amidohydrolase 2 Uncharacterized protein Acyl-CoA dehydrogenase, C-domain protein Linear gramicidin synthetase subunit D Glycogen debranching enzyme GlgX Uncharacterized protein Uncharacterized protein Monooxygenase GTP pyrophosphokinase 2′-Hydroxybiphenyl-2-sulfinate desulfinase Ppe family protein Dehydrogenase 3-Ketosteroid dehydrogenase Isocitrate dehydrogenase, NADP-dependent Putative methyl transferase Putative NAD(+) arginine ADP-ribosyltransferase Cyclopropane-fatty-acyl-phospholipid synthase 2 Dehydrogenase Short chain dehydrogenase/reductase family protein NAD(P) transhydrogenase beta subunit Short chain dehydrogenase/reductase family protein | A0A0H2ZRQ5 | MAV_RS00580 |
| MAV_0158 | 1D3 | 40.1 | A0A0H2ZRQ5 | MAV_RS00770 | |
| MAV_0175 | 2D10 | 33.2 | A0A0H2ZWF6 | MAV_RS00840 | |
| MAV_0249 | 2F1 | 33.7 | A0A0H2ZY15 | MAV_RS01200 | |
| MAV_0273 | 1A9 | 39.7 | A0A0H2ZWS7 | MAV_RS01315 | |
| MAV_1264 | 1 E10 | 2.3 | A0A0H2ZTF8 | MAV_RS06065 | |
| MAV_1621 | 1G5 | 42.0 | A0A0H2ZZA4 | MAV_RS07760 | |
| MAV_1812 | 2F10 | 31.9 | A0A0H3A4S5 | MAV_RS08680 | |
| MAV_1816 | 1 E8 | 13.2 | A0A0H2ZUN3 | MAV_RS08700 | |
| MAV_2256 | 2D12 | 25.1 | A0A0H3A3J1 | MAV_RS10765 | |
| MAV_2426 | 1B12 | 1.5 | A0A0H2ZZS1 | MAV_RS11565 | |
| MAV_2564 | 1F10 | 6.8 | A0A0H2ZWS2 | MAV_RS12255 | |
| MAV_2590 | 2H4 | 33.9 | A0A0H3A2H6 | MAV_RS12365 | |
| MAV_2591 | 1H11 | 6.5 | A0A0H2ZVD5 | MAV_RS12360 | |
| MAV_2686 | 1B4 | 43.9 | A0A0H2ZUN8 | MAV_RS12825 | |
| MAV_2767 | 1 E3 | 40.1 | A0A0H3A0C8 | MAV_RS13195 | |
| MAV_3056 | 1B6 | 39.4 | A0A0H2ZTY5 | MAV_RS14575 | |
| MAV_3210 | 1H4 | 34.8 | A0A0H2ZZU9 | MAV_RS19660 | |
| MAV_3286 | 2F7 | 36.0 | A0A0H2ZX00 | MAV_RS15695 | |
| MAV_3296 | 2F3 | 43.5 | A0A0H2ZRF2 | MAV_RS15740 | |
| MAV_3337 | 1F1 | 11.1 | A0A0H2ZWN3 | MAV_RS15945 | |
| MAV_3464 | 2F6 | 29.9 | A0A0H3A3N2 | MAV_RS16570 | |
| MAV_3808 | 2 E11 | 38.2 | A0A0H3A3M7 | MAV_RS18245 | |
| MAV_4014 | 2A4 | 39.1 | A0A0H2ZTW6 | MAV_RS19230 | |
| MAV_4124 | 2F12 | 27.4 | A0A0H2ZX25 | MAV_RS19785 | |
| MAV_4249 | 2D11 | 41.6 | A0A0H3A167 | MAV_RS20390 | |
| MAV_4313 | 1 E12 | 0.9 | A0A0H2ZT90 | MAV_RS20695 | |
| MAV_4573 | 1G2 | 34.6 | A0QLB4 | MAV_RS21960 | |
| MAV_4644 | 1F2 | 29.0 | A0QLI5 | MAV_RS22305 | |
| MAV_4647 | 2G2 | 38.2 | A0A0H2ZZ17 | MAV_RS22320 | |
| MAV_4791 | 1D5 | 34.1 | A0A0H2ZUD0 | MAV_RS23020 | |
| MAV_4916 | 1A11 | 22.1 | A0A0H3A2P2 | MAV_RS23625 | |
| MAV_5140 | 1D2 | 40.9 | A0A0H2ZXD2 | MAV_RS24655 | |
| MAV_5205 | 2F8 | 38.4 | A0A0H3A044 | MAV_RS24970 |
| Name | Accession | Description | Amino Acids | E-Value |
|---|---|---|---|---|
| WXG100 | Pfam06013 | Proteins of 100 residues with WXG | 7–92 | 0.00371 |
| DedD | COG3147 | Cell division protein DedD | 455–527 | 0.00631 |
| PHA03247 | PHA03247 | Large tegument protein UL36 | 455–655 | 5.69 × 10−9 |
| Atrophin-1 | Pfam03154 | Atrophin-1 family | 459–634 | 2.74 × 10−6 |
| Rad23 | TIGR00601 | UV excision repair protein Rad23 | 475–543 | 0.00525 |
| ADPrib_exo_Tox | Pfam03496 | ADP-ribosyltransferase exoenzyme | 675–822 | 4.03 × 10−15 |
| VIP2 | Cd00233 | Family of actin-ADP-ribosylating toxin | 675–820 | 1.18 × 10−14 |
| Experimental Groups | Infection CFU/mL (Time 0) | Infection CFU/mL (Time 4 Days) |
|---|---|---|
| Wild-type MAH | 6.1 ± 0.5 × 105 | 7.8 ± 0.4 × 105 |
| MAV_4644:Tn | 2.6 ± 0.3 × 105 | 1.3 ± 0.3 × 104 * |
| Complemented MAV_4644:Tn | 5.8 ± 0.3 × 105 | 6.9 ± 0.5 × 105 |
| Cathepsin Z (mM/mL) | MAC104/MAV_4644:Tn CFU/mL | ||
|---|---|---|---|
| 103 | 104 | 105 | |
| 0 | 1.3 × 103/1.1 × 103 | 1.2 × 104/1.2 × 104 | 1.3 × 105/1.2 × 105 |
| 0 + NO | 1.4 × 103/9.1 × 102 | 1.4 × 104/8.3 × 103 | 1.1 × 105/8.1 × 104 |
| 0.5 | 1.3 × 103/ 1.4 × 103 | 1.5 × 104/ 1.2 × 104 | 1.4 × 105/1.3 × 105 |
| 0.5 + NO | 1.3 × 103/ 7.2 × 102 * | 1.3 × 104/6.4 × 103 * | 1.3 × 105/7.8 × 104 * |
| 1.0 | 1.4 × 103/1.3 × 103 | 1.4 × 104/1.2 × 104 | 1.4 × 105/1.2 × 105 |
| 1.0 + NO | 1.3 × 103/5.3 × 102 * | 1.5 × 104/4.2 × 103 * | 1.4 × 105/4.7 × 104 * |
| 2.0 | 1.4 × 103/1.2 × 103 | 1.3 × 104/1.2 × 104 | 1.2 × 105/1.1 × 105 |
| 2.0 + NO | 1.2 × 103/1.8 × 102 * | 1.3 × 104/1.4 × 103 * | 1.3 × 105/9.9 × 103 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewis, M.S.; Danelishvili, L.; Rose, S.J.; Bermudez, L.E. MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing. Microorganisms 2019, 7, 144. https://doi.org/10.3390/microorganisms7050144
Lewis MS, Danelishvili L, Rose SJ, Bermudez LE. MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing. Microorganisms. 2019; 7(5):144. https://doi.org/10.3390/microorganisms7050144
Chicago/Turabian StyleLewis, Matthew S., Lia Danelishvili, Sasha J. Rose, and Luiz E. Bermudez. 2019. "MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing" Microorganisms 7, no. 5: 144. https://doi.org/10.3390/microorganisms7050144
APA StyleLewis, M. S., Danelishvili, L., Rose, S. J., & Bermudez, L. E. (2019). MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing. Microorganisms, 7(5), 144. https://doi.org/10.3390/microorganisms7050144