Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare
Abstract
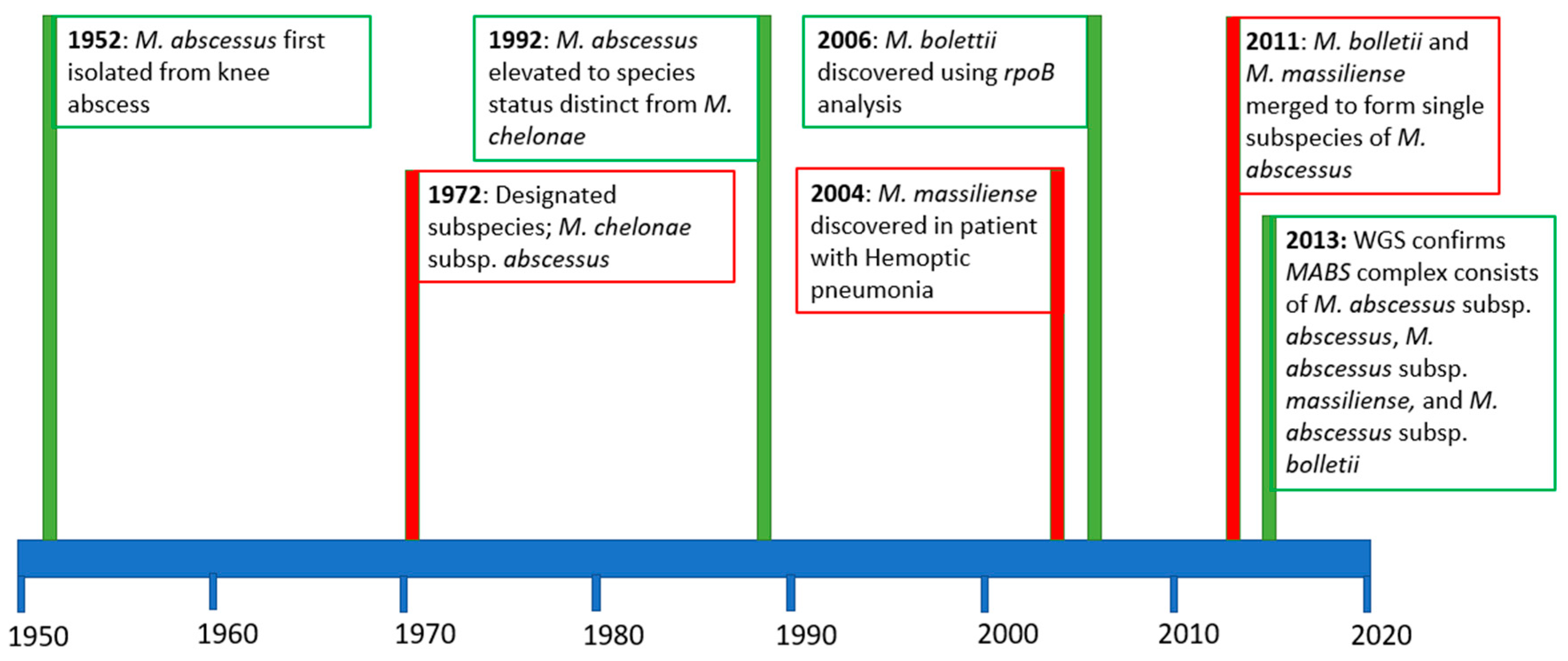
1. Introduction
2. M. abscessus and Cystic Fibrosis
3. M. abscessus Infection in Non-CF Populations
4. Environmental Reservoirs and Transmission
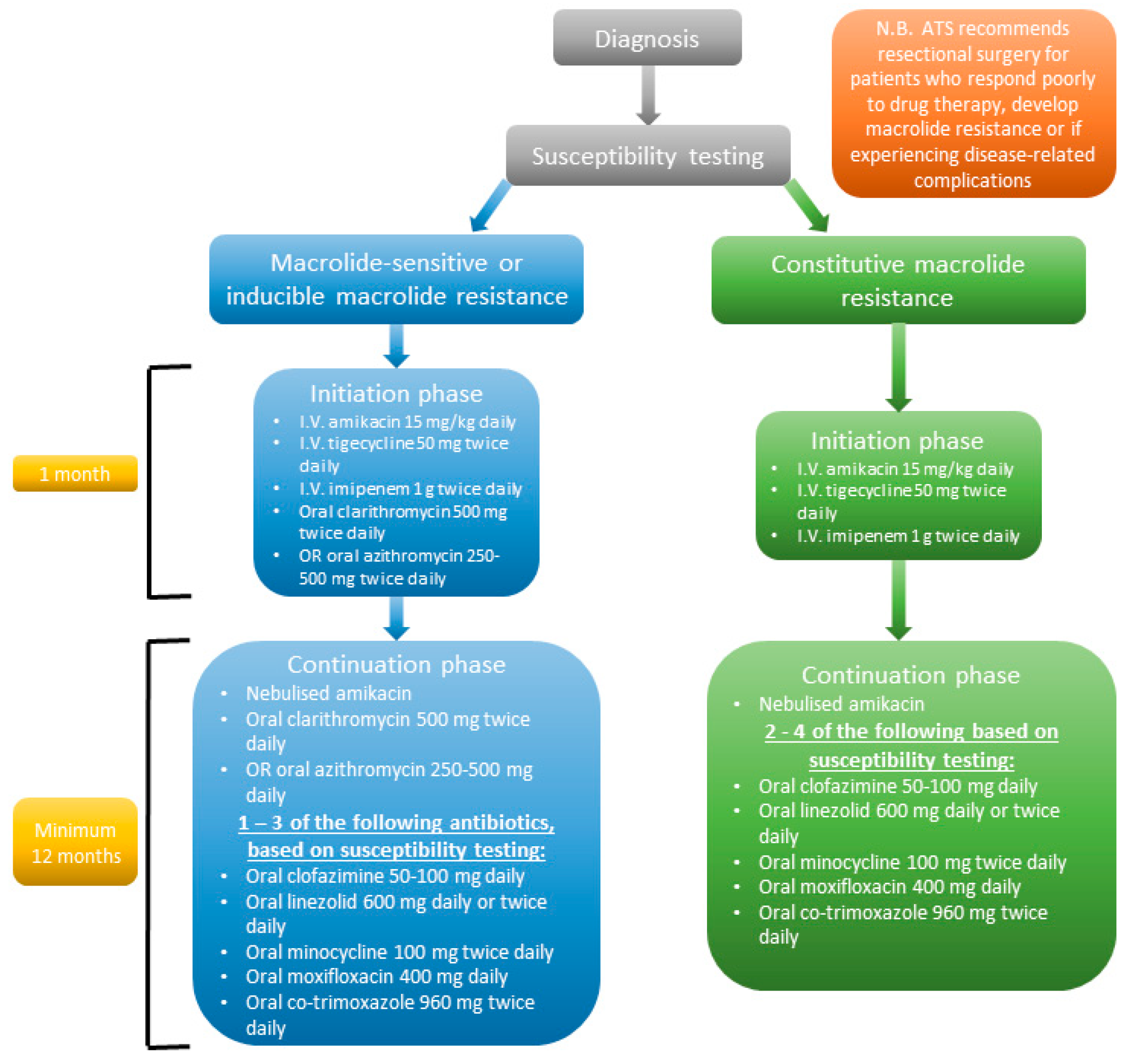
5. Diagnosis and Treatment:
6. Treatment
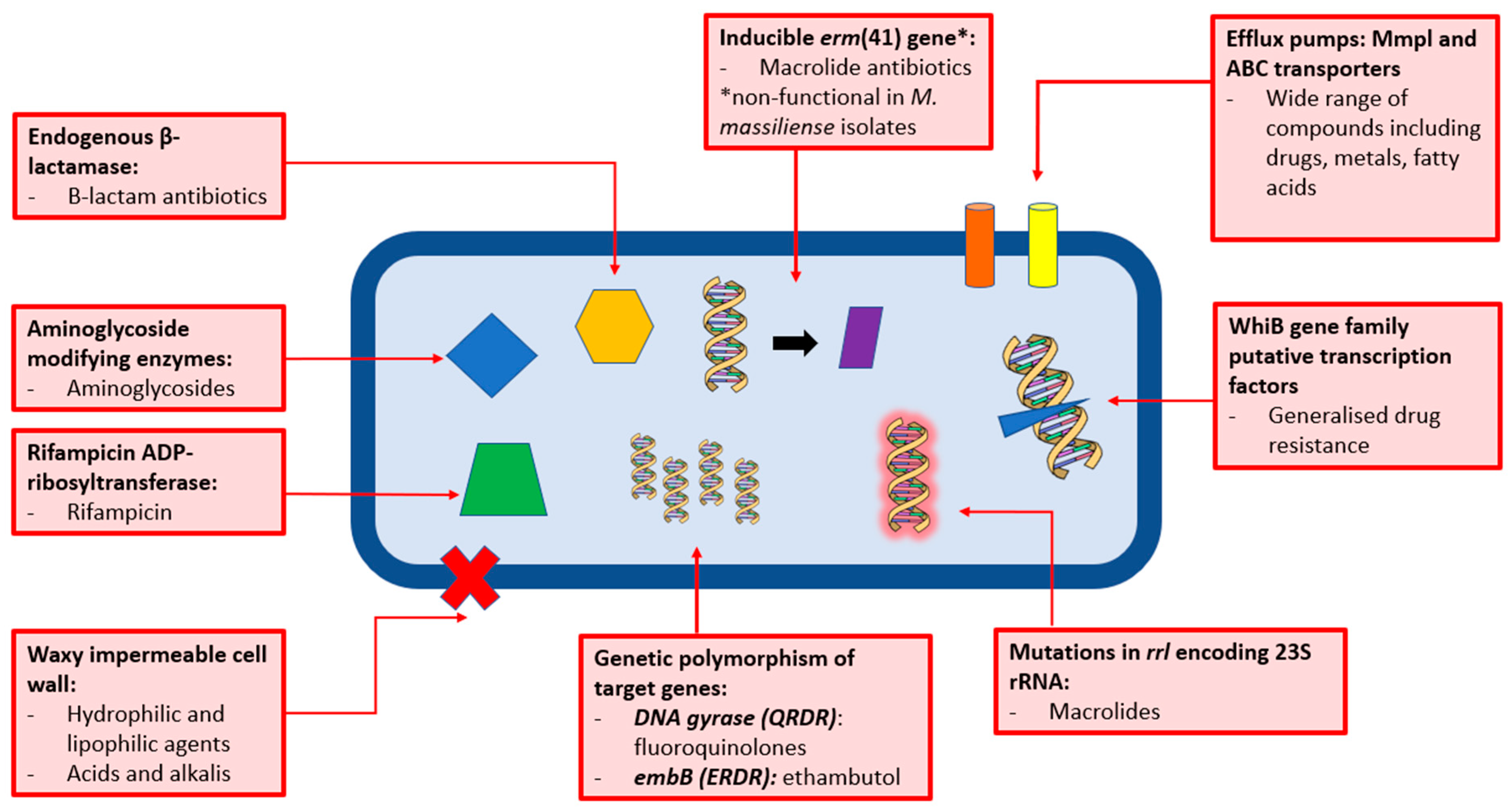
The Resistance Problem: Why the Drugs Don’t Work
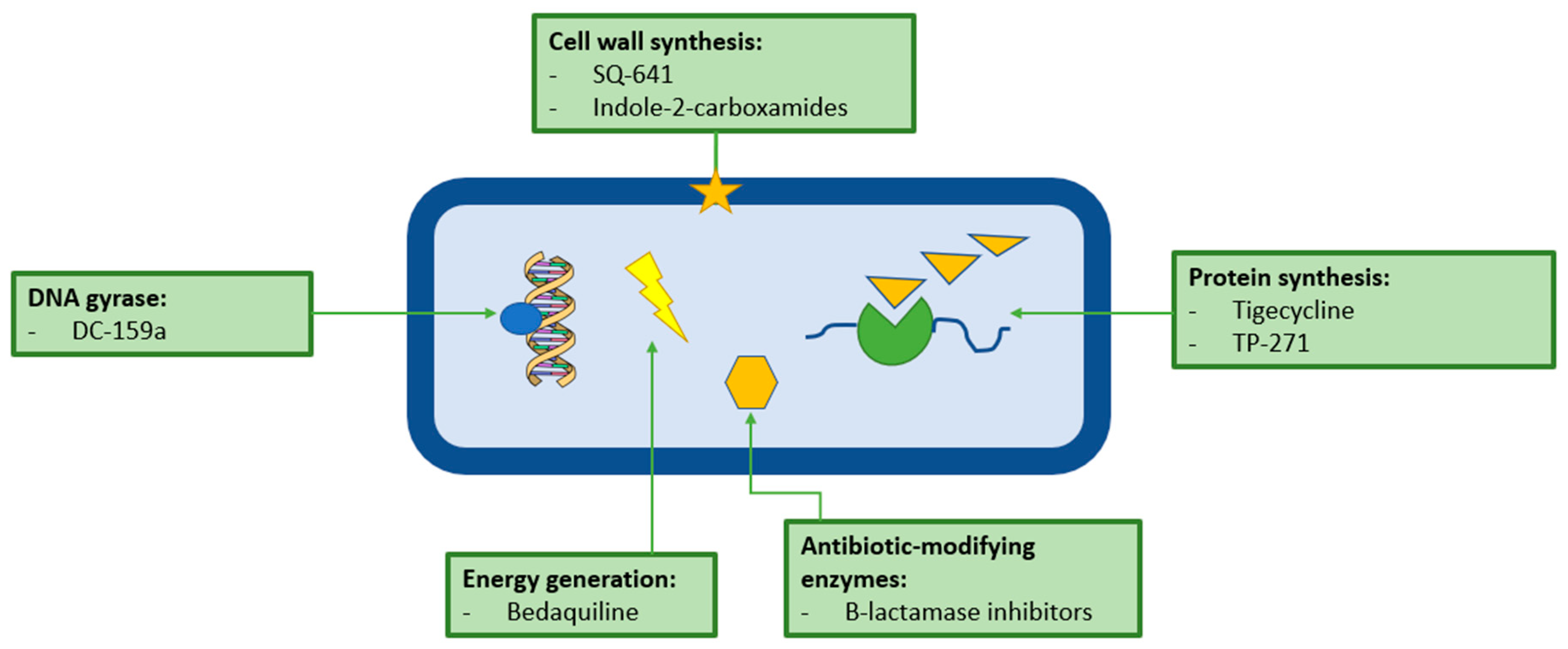
7. Future Perspectives for M. abscessus
Future Treatments
8. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moore, M.; Frerichs, J.B. An Unusual Acid-Fast Infection of the Knee with Subcutaneous, Abscess-Like Lesions of the Gluteal Region. J. Investig. Dermatol. 1953, 20, 133–169. [Google Scholar] [CrossRef]
- Griffith, D.E.; Girard, W.M.; Wallace, R.J. Clinical Features of Pulmonary Disease Caused by Rapidly Growing Mycobacteria: An Analysis of 154 Patients. Am. Rev. Respir. Dis. 1993, 147, 1271–1278. [Google Scholar] [CrossRef]
- Sopko, J.A.; Fieselmann, J.; Kasik, J.E. Pulmonary disease due to Mycobacterium chelonei subspecies abscessus: A report of four cases. Tubercle 1980, 61, 165–169. [Google Scholar] [CrossRef]
- Kubica, G.P.; Baess, I.; Gordon, R.E.; Jenkins, P.A.; Kwapinski, J.B.G.; McDurmont, C.; Pattyn, S.R.; Saito, H.; Silcox, V.; Stanford, J.L.; et al. A Co-operative Numberical Analysis of Rapidly Growing Mycobacteria. J. Gen. Microbiol. 1972, 73, 55–70. [Google Scholar] [CrossRef]
- Kusunoki, S.; Ezaki, T. Proposal of Mycobacterium peregrinum sp. Nov., nom. Rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 1992, 42, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Adekambi, T.; Reynaud-Gaubert, M.; Greub, G.; Gevaudan, M.J.; La Scola, B.; Raoult, D.; Drancourt, M. Amoebal Coculture of “Mycobacterium massiliense” sp. nov. from the Sputum of a Patient with Hemoptoic Pneumonia. J. Clin. Microbiol. 2004, 42, 5493–5501. [Google Scholar] [CrossRef] [PubMed]
- Adekambi, T.; Berger, P.; Raoult, D.; Drancourt, M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. Nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Conville, P.; Witebsky, F.G. Variables Affecting Results of Sodium Chloride Tolerance Test for Identification of Rapidly Growing Mycobacteria. J. Clin. Microbiol. 1998, 36, 1555–1559. [Google Scholar]
- Levy-Frebault, V.; Grimont, F.; Grimont, P.A.D.; David, L.D. Deoxyribonucleic Acid Relatedness Study of the Mycobacterium chelonae-Mycobacterium chelonae Complex. Int. J. Syst. Bacteriol. 1986, 36, 458–460. [Google Scholar] [CrossRef]
- Wallace, R.J.; Silcox, V.A.; Tsukamura, M.; Brown, B.A.; Kilburn, J.O.; Butler, W.R.; Onyi, G. Clinical Significance, Biochemical Features and Susceptibility Patterns of Sporadic Isolates of the Mycobacterium chelonae-Like Organism. J. Clin. Microbiol. 1993, 31, 3231–3239. [Google Scholar]
- Silcox, V.A.; Good, R.C.; Floyd, M.M. Identification of Clinically Significant Mycobacterium chelonae Complex Isolates. J. Clin. Microbiol. 1981, 14, 686–691. [Google Scholar] [PubMed]
- Wilson, R.W.; Steingrube, V.A.; Bottger, E.C.; Springer, B.; Brown-Elliot, B.A.; Vincent, V.; Jost, K.C., Jr.; Zhang, Y.; Garcia, M.J.; Chiu, S.H.; et al. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: An international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 2001, 51, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Leao, S.C.; Tortoli, E.; Euzeby, J.P.; Garcia, M.J. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacteri. Int. J. Syst. Evol. Microbiol. 2011, 61, 2311–2313. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.M.; Grogono, D.M.; Greaves, D.; Foweraker, J.; Roddick, I.; Inns, T.; Reacher, M.; Haworth, C.S.; Curran, M.D.; Harris, S.R.; et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: A retrospective cohort study. Lancet 2013, 381, 1551–1560. [Google Scholar] [CrossRef]
- Tortoli, E.; Kohl, T.A.; Brown-Elliot, B.A. Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. nov. Int. J. Syst. Evolut. Microbiol. 2016, 66, 4471–4479. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Trovato, A.; Baldan, R.; Costa, D.; Simonetti, T.M.; Cirillo, D.M.; Tortoli, E. Molecular typing of Mycobacteirum abscessus isolated from cystic fibrosis patients. Int. J. Mycobacteriol. 2017, 6, 138–141. [Google Scholar] [PubMed]
- Shaw, L.P.; Doyle, R.M.; Kavaliunaite, E.; Spencer, H.; Ballous, F.; Dixon, G.; Harris, K.A. Children with cystoc fibrosis are infected with multiple subpopulations of Mycobacterium abscessus with different antimicrobial resistance profiles. Clin. Infect. Dis. 2019, ciz069. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.R. Understanding nontuberculous mycobacterial lung disease: Its been a long time coming. F1000 Res. 2016, 5, 2797. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Assocaited with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Olivier, K.N. The natural history of nontuberculous mycobacteria in patients with cystic fibrosis. Paediatr. Respir. Rev. 2004, 5 (Suppl. A), S213–S216. [Google Scholar] [CrossRef]
- Roux, A.L.; Catherinot, E.; Ripoll, F.; Soismier, N.; Macheras, E.; Ravilly, S.; Bellis, G.; Vibet, M.A.; Le Roux, E.; Lemonnier, L.; et al. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J. Clin. Microbiol. 2009, 47, 4124–4128. [Google Scholar] [CrossRef] [PubMed]
- Seddon, P.; Fidler, K.; Raman, S.; Wyatt, H.; Ruiz, G.; Elston, C.; Perrin, F.; Gyi, K.; Bilton, D.; Drobniewski, F.; et al. Prevalence of nontuberculous mycobacteria in cystic fibrosis clinics, United Kingdom, 2009. Emerg. Infect. Dis. 2013, 19, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Adjemian, J.; Olivier, K.N.; Prevots, D.R. Nontuberculous Mycobacteria among Patients with Cystic Fibrosis in the United States. Screening Practices and Environmental Risk. Am. J. Crit. Care Med. 2014, 190, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Mussaffi, H.; Rivlin, J.; Shalit, I.; Ephros, M.; Blau, H. Nontuberculous mycobacteria in cystic fibrosis associated with allergic bronchopulmonary aspergillosis and steroid therapy. Eur. Respir. J. 2005, 25, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.E.; Aksamit, T.; Brown-Elliot, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Ats J. 2007, 175. [Google Scholar] [CrossRef]
- Viviani, L.; Harrison, M.J.; Zolin, A.; Haworth, C.S.; Floto, R.A. Epidemiology of nontuberculous myocbacteria (NTM) amongst individuals with cystic fibrosis (CF). J. Cyst. Fibros. 2016, 15, 619–623. [Google Scholar] [CrossRef]
- Gardner, A.I.; McClenaghan, E.; Saint, G.; McNamara, P.S.; Brodlie, M.; Thomas, M.F. Epidemiology of Non-tuberculous Mycobacteria infection in children and young people with cystic fibrosis: Analysis of UK Cystic Fibrosis Registry. Clin. Infect. Dis. 2018, 68, 731–737. [Google Scholar] [CrossRef]
- Levy, I.; Grisaru-Soen, G.; Lerner-Geva, L.; Kerem, E.; Blau, H.; Bentur, L.; Aviram, M.; Rivlin, J.; Picard, E.; Lavy, A.; et al. Multicenter Cross-Sectional Study of Nontuberculous Mycobacteria Infections among Cystic Fibrosis Patients, Israel. Emerg. Infect. Dis. 2008, 14, 378–384. [Google Scholar] [CrossRef]
- Osmani, M.; Sotello, D.; Alvarez, S.; Odell, J.A.; Thomas, M. Mycobacterium abscessus infections in lung transplants: 15 year experience from a single institution. Transpl. Infect. Dis. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Taylor, J.L.; Palmer, S.M. Mycobacterium abscessus chest wall and pulmonary infection in a cystic fibrosis lung transplant recipient. J. Heart Lung Transpl. 2006, 25, 985–988. [Google Scholar] [CrossRef] [PubMed]
- Lobo, L.J.; Chang, L.C.; Esther, C.R., Jr.; Gilligan, P.H.; Tulu, Z.; Noone, P.G. Lung transplant outcomes in cystic fibrosis patients with pre-operative Mycobacterium abscessus respiratory infections. Clin. Transplant. 2013, 27, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Gillijam, M.; Schersten, H.; Silverborn, M.; Jonsson, B.; Ericsson Hollsing, A. Lung transplantation in patients with cystic fibrosis and Mycobacterium abscessus infection. J. Cyst. Fibros. 2010, 9, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Lord, G.M.; Matarese, G.; Howard, J.K.; Baker, R.J.; Bloom, S.R.; Lechler, R.I. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature 1998, 394, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Brown-Elliott, B.A.; Wallace, R.J. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 2002, 15, 716–746. [Google Scholar] [CrossRef] [PubMed]
- Furuya, E.Y.; Paez, A.; Srinivasan, A.; Cooksey, R.; Augenbraun, M.; Baron, M.; Brudney, K.; Della-Latta, P.; Estivariz, C.; Fischer, S.; et al. Outbreak of Mycobacterium abscessus wound infections among “lipotourists” from the United States who underwent abdominoplasty in the Dominican Republic. Clin. Infect. Dis. 2008, 46, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Appelgren, P.; Farnebo, F.; Dotevall, L.; Studahl, M.; Jonsson, B.; Petrini, B. Late-Onset Posttraumatic Skin and Soft-Tissue Infections Caused by Rapid-Growing Mycobacteria in Tsunami Survivors. Clin. Infect. Dis. 2008, 47, 11–16. [Google Scholar] [CrossRef]
- Lamb, G.S.; Starke, J.R. Mycobacterium abscessus infection in children: A review of current literature. J. Pediatr. Infect. Dis. Soc. 2018, 7, e131–e144. [Google Scholar] [CrossRef]
- Johnson, L.R. Microcolony and biofilm formation as a survival strategy for bacteria. J. Theor. Biol. 2008, 251, 24–34. [Google Scholar] [CrossRef]
- McGrath, E.E.; Blades, Z.; McCabe, J.; Jarry, H.; Anderson, P.B. Nontuberculosis mycobacteria and the lung: From suspicion to treatment. Lung 2010, 188, 269–282. [Google Scholar] [CrossRef]
- Primm, T.; Lucero, C.A.; Falkinham, J.O. Health Impacts of Environmental Mycobacteria. Clin. Microbiol. Rev. 2004, 17, 98–106. [Google Scholar] [CrossRef]
- Brennan, P.; Nikaido, H. The Envelope of Mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef]
- Bendinger, B.; Rijnaarts, H.H.M.; Altendorf, K.; Zehnder, A.J.B. Physiochemical Cell Surface and Adhesive Properties of Coryneform Bacteria Related to the Presence and Chain Length of Mycolic Acids. Appl. Environ. Microbiol. 1993, 59, 3973–3977. [Google Scholar]
- Falkinham, J.O. Impact of human activities on the ecology of nontuberculosis mycobacteria. Future Microbiol. 2010, 5, 951–960. [Google Scholar] [CrossRef]
- Nessar, R.; Cambau, E.; Reyrat, J.M.; Murray, A.; Gicquel, B. Mycobacterium abscessus: A new antibiotic nightmare. J. Antimicrob. Chemother. 2012, 67, 810–818. [Google Scholar] [CrossRef]
- Jarlier, V.; Nikaido, H. Permeability Barrier to Hydrophilic Solutes in Mycobacterium chelonei. J. Bacteriol. 1990, 172, 1418–1423. [Google Scholar] [CrossRef]
- Grogono, D.; Bryant, J.; Rodriguez-Rincon, D.; Everall, I.; Brown, K.; Moreno, P.; Verma, D.; Hill, E.; Drijkoningen, J.; Haworth, C.; et al. Whole-Genome Sequencing Reveals Global Spread of Mycobacterium abscessus Clones Amongst Patients with Cystic Fibrosis; Non-Tuberculosis Mycobacteria: From Bench to Clinic; American Thoracic Society: Washington, DC, USA, 2017; Volume 195. [Google Scholar]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic Pathogens Enriched in Showerhead Biofilms. Proc. Natl. Acad. Sci. USA 2009, 106, 16393–16399. [Google Scholar] [CrossRef]
- Honda, J.R.; Hasan, N.A.; Davidson, R.M.; Williams, M.D.; Epperson, L.E.; Reynolds, P.R.; Smith, T.; Iakhiaeva, E.; Bankowski, M.J.; Wallace, R.J., Jr.; et al. Environmental Nontuberculous Mycobacteria in the Hawaiian Islands. PLoS Negl. Trop. Dis. 2016, 10, 1–17. [Google Scholar] [CrossRef]
- Carter, K.K.; Lundgren, I.; Correll, S.; Schmalz, T.; McCarter, T.; Stroud, J.; Bruesch, A.; Hahn, C.G. First United States Outbreak of Mycobacterium abscessus Hand and Foot Disease Among Children Associated with a Wading Pool. Pediatr. Infect. Dis. Soc. 2018, 1–6. [Google Scholar] [CrossRef]
- Faria, S.; Joao, I.; Jordao, L. General Overview on Nontuberculous Mycobacteria, Biofilms and Human Infection. J. Pathog. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Cystic Fibrosis Trust Mycobacterium abscessus Infection Control Working Group. Mycobacterium abscessus: Suggestions for Infection Prevention and Control (Interim Guidance—October 2013); Cystic Fibrosis Trust: Bromley, UK, 2013. [Google Scholar]
- Bange, F.C.; Brown, B.A.; Smaczny, C.; Wallace, R.J., Jr.; Bottger, E.C. Lack of Transmission of Mycobacterium abscessus among Patients with Cystic Fibrosis Attending a Single Clinic. Clin. Infect. Dis. 2001, 32, 1648–1650. [Google Scholar] [CrossRef]
- Harris, K.A.; Underwood, A.; Kenna, D.T.; Brooks, A.; Kavaliunaite, E.; Kapatai, G.; Tewolde, R.; Aurora, P.; Dixon, G. Whole-Genome Sequencing and Epidemiological Analysis Do Not Provide Evidence for Cross-transmission of Mycobacterium abscessus in a Cohort of Pediatric Cystic Fibrosis Patients. Clin. Infect. Dis. 2015, 60, 1007–1016. [Google Scholar] [CrossRef]
- Haworth, C.S.; Banks, J.; Capstick, T.; Fisher, A.J.; Gorsuch, T.; Laurenson, I.F.; Leitch, A.; Loebinger, M.R.; Milburn, H.J.; Nightingale, M.; et al. British Thoracic Society Guideline for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). BMJ Open Respir. Res. 2017, 4, 1–12. [Google Scholar] [CrossRef]
- Jeon, K.; Kwon, O.J.; Lee, N.Y.; Kim, B.J.; Kook, Y.H.; Lee, S.H.; Park, Y.K.; Kim, C.K.; Koh, W.J. Antibiotic Treatment of Mycobacterium abscessus Lung Disease. Am. J. Respir. Crit. Care Med. 2009, 180, 896–902. [Google Scholar] [CrossRef]
- Novosad, S.A.; Beekman, S.E.; Polgreen, P.M.; Mackey, K.; Winthrop, K.L. Treatment of Mycobacterium abscessus infection. Emerg. Infect. Dis. 2016, 22, 511–514. [Google Scholar] [CrossRef]
- Heffernan, C.B.; McKeon, M.G.; Molony, S.; Kawai, K.; Stiles, D.J.; Lachenauer, C.S.; Kenna, M.A.; Watters, K. Does Clarithromycin Cause Hearing Loss? A 12-Year Review of Clarithromycin Therapy for Nontuberculous Mycobacterial Lymphadenitis in Children. Ann. Otol. Rhinol. Laryngol. 2018, 127, 687–693. [Google Scholar] [CrossRef]
- Coulston, J.; Balaratnam, N. Irreversible sensorineural hearing loss due to clarithromycin. Postgrad. Med. J. 2005, 81, 58–59. [Google Scholar] [CrossRef]
- Guay, D.R.; Patterson, D.R.; Seipman, N.; Craft, J.C. Overview of the tolerability profile of clarithromycin in preclinical and clinical trials. Drug Saf. 1993, 8, 350–364. [Google Scholar] [CrossRef]
- Alcaide, F.; Pfyffer, G.E.; Telenti, A. Role of embB in Natural and Acquired Resistance to Ethambutol in Mycobacteria. Antimicrob. Agents Chemother. 1997, 41, 2270–2273. [Google Scholar] [CrossRef]
- Griffith, D.E. The talking Mycobacterium abscessus blues. Clin. Infect. Dis. 2011, 52, 572–574. [Google Scholar] [CrossRef]
- Jarlier, V.; Nikaido, H. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 1994, 123, 11–18. [Google Scholar] [CrossRef]
- Daffe, M.; Draper, P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 1998, 39, 131–203. [Google Scholar]
- Nguyen, L.; Thompson, C.J. Foundations of antibiotic resistance in bacterial physiology: The mycobacteria paradigm. Trends Microbiol. 2006, 14, 304–312. [Google Scholar] [CrossRef]
- De Rossi, E.; Ainsa, J.A.; Riccardi, G. Role of mycobacterial efflux transporters in drug resistance: An unresolved question. FEMS Microbiol. Rev. 2006, 30, 36–52. [Google Scholar] [CrossRef]
- Louw, G.E.; Warren, R.M.; Gey van Pittius, N.C.; McEvoy, C.R.; Van Helden, P.D.; Victor, T.C. A Balancing Act: Efflux/Influx in Mycobacterial Drug Resistance. Antimicrob. Agents Chemother. 2009, 53, 3181–3189. [Google Scholar] [CrossRef]
- Ripoll, F.; Pasek, S.; Schenowitz, C.; Dossat, C.; Barbe, V.; Rottman, M.; Macheras, E.; Heym, B.; Herrmann, J.L.; Daffe, M.; et al. Non Mycobacterial Virulence Genes in the Genome of the Emerging Pathogen Mycobacterium abscessus. PLoS ONE 2009, 4, e5660. [Google Scholar] [CrossRef]
- Tekaia, F.; Gordon, S.V.; Garnier, T.; Brosch, R.; Barrell, B.G.; Cole, S.T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 1999, 79, 329–342. [Google Scholar] [CrossRef]
- Pasca, M.R.; Guglierame, P.; De Rossi, E.; Zara, F.; Riccardi, G. mmpL7 Gene of Mycobacterium tuberculosis Is Responsible for Isoniazid Efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 2005, 49, 4775–4777. [Google Scholar] [CrossRef]
- Nash, K.A.; Brown-Elliot, B.A.; Wallce, R.J. A Novel Gene, erm(41), Confers Inducible Macrolide Resistance to Clinical Isolaes of Mycobacterium abscessus but is Absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 2009, 53, 1367–1376. [Google Scholar] [CrossRef]
- Liu, W.; Li, B.; Chu, H.; Zhang, Z.; Luo, L.; Ma, W.; Yang, S.; Guo, Q. Rapid detection of mutations in erm(41) and rrl associated with clarithromycin resistance in Mycobacterium abscessus complex by denaturing gradient gel electrophoresis. J. Microbiol. Methods 2017, 143, 87–93. [Google Scholar] [CrossRef]
- Brown-Elliot, B.A.; Vasireddy, S.; Vasireddy, R.; Iakhiaeva, E.; Howard, S.T.; Nash, K.; Parodi, N.; Strong, A.; Gee, M.; Smith, T.; et al. Utility of Seqencing the erm(41) Gene in Isolates of Mycobacterium abscessus subsp. abscessus with Low and Intermediate Clarithromycin MICs. J. Clin. Microbiol. 2015, 53, 1211–1215. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Kim, B.J.; Kook, Y.; Yun, Y.J.; Shin, J.H.; Kim, B.J.; Kook, Y.H. Mycobacterium massiliense in differentiated from Mycobacterium abscesssus and Mycobacterium bolletii by erthythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol. Immunol. 2010, 54, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Slechta, E.S.; Muir, H.; Barker, A.P. Rapid Molecular Detection of Inducible Macrolide Resistance in Mycobacterium chelonae and M. abscessus Strains: A Replacement for 14-Day Susceptibility Testing? J. Clin. Microbiol. 2014, 52, 1705–1707. [Google Scholar] [CrossRef] [PubMed]
- Bastian, S.; Veziris, N.; Roux, A.L.; Brossier, F.; Gaillard, J.L.; Jarlier, V.; Cambau, E. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob. Agents Chemother. 2011, 55, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Sander, P.; Prammananan, T.; Meier, A.; Frischkorn, K.; Bottger, E.C. The role of ribosomal RNAs in macrolide resistance. Mol. Microbiol. 1997, 26, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, I.; Jarlier, V.; Cambau, E. Correlation between Quinolone Susceptibility Patterns and Sequences in the A and B Subunits of DNA Gyrase in Mycobacteria. Antimicrob. Agents Chemother. 1998, 42, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Morais Cabral, J.H.; Jackson, A.P.; Smith, C.V.; Shikotra, N.; Maxwell, A.; Liddington, R.C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 1997, 388, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Soroka, D.; Dubee, V.; Soulier-Eschrihuela, O.; Cuinet, G.; Hugonnet, J.E.; Gutmann, L.; Mainardi, J.L.; Arthur, M. Characterization of broad-spectrum Mycobacterium abscessus class A Beta-lactamase. J. Antimicrob. Chemother. 2014, 69, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Rominski, A.; Roditscheff, A.; Selchow, P.; Bottger, E.C.; Sander, P. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J. Antimicrob. Chemother. 2016, 72, 376–384. [Google Scholar] [CrossRef]
- Burian, J.; Ramon-Garcia, S.; Howes, C.G.; Thompson, C.J. WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti-Infect. Ther. 2012, 10, 1037–1047. [Google Scholar] [CrossRef]
- Hurst-Hess, K.; Rudra, P.; Ghosh, P. Mycobacterium abscessus WhiB7 Regulates a Species-Specific Repertoire of Genes to Confer Extreme Antibiotic Resistance. Antimicrob. Agents Chemother. 2017, 61, e01347-17. [Google Scholar] [CrossRef]
- Tanaka, E.; Kimoto, T.; Tsuyuguchi, K.; Suzuki, K.; Amitani, R. Successful treatment with faropenem and clarithroymcin of pulmonary Mycobacterium abscessus infection. J. Infect. Chemother. 2002, 8, 252–255. [Google Scholar] [CrossRef]
- Okazaki, A.; Takato, H.; Ohkura, N.; Katayama, N.; Kasahara, K.; Fujimura, M. Successful treatment with chemotherapy and corticosteroids of pulmonary Mycobacterium abscessus infection accompanied by pleural effusion. J. Infect. Chemother. 2013, 19, 964–968. [Google Scholar] [CrossRef]
- Dubee, V.; Bernut, A.; Cortes, M.; Lesne, T.; Dorchene, D.; Lefebvre, A.L.; Hugonnet, J.E.; Gutmann, L.; Mainardi, J.L.; Herrmann, J.L.; et al. Beta-lactamase inhibition by avibactam in Mycobacterium abscessus. J. Antimicrob. Chemother. 2015, 70, 1051–1058. [Google Scholar]
- Lefebvre, A.L.; Le Moigne, V.; Bernut, A.; Veckerle, C.; Compain, F.; Herrmann, J.L.; Kremer, L.; Arthur, M.; Mainardi, J.L. Inhibition of the Beta-lactamase BlaMab by Avibactam Improves the In Vitro and In Vivo Efficacy of Imipenem against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2017, 61, e02440-16. [Google Scholar] [CrossRef]
- Disratthakit, A.; Doi, N. In Vitro Activities of DC-159a, a Novel Fluoroquinolone, against Mycobacterium Species. Antimicrob. Agents Chemother. 2010, 54, 2684–2686. [Google Scholar] [CrossRef]
- Abrahams, K.A.; Besra, G.S. Mycobacterial cell wall biosynthesis: A multifaceted antibiotic target. Parasitology 2018, 145, 116–133. [Google Scholar] [CrossRef]
- Dubuisson, T.; Bogatcheva, E.; Krishnan, M.Y.; Collins, M.T.; Einck, L.; Nacy, C.A.; Reddy, V.M. In vitro antimicrobial activities of capuramycin analogues against non-tuberculous mycobacteria. J. Antimicrob. Chemother. 2010, 65, 2590–2597. [Google Scholar] [CrossRef]
- Reddy, V.M.; Einck, L.; Nacy, C.A. In vitro antimycobacterial activities of capuramycin analogues. Antimicrob. Agents Chemother. 2008, 52, 719–721. [Google Scholar] [CrossRef]
- Franz, N.D.; Belardinella, J.M.; Kaminski, M.A.; Dunn, L.C.; Calado Nogueira de Moura, V.; Blaha, M.A.; Truong, D.D.; Li, W.; Jackson, M.; North, E.J. Design, synthesis and evaluation of indole-2-carboxamides with pan anti-mycobacterial activity. Bioorg. Med. Chem. 2017, 25, 3746–3755. [Google Scholar] [CrossRef]
- Pandya, A.N.; Prathipati, P.K.; Hegde, P.; Li, W.; Graham, K.F.; Mandal, S.; Drescher, K.M.; Destache, C.J.; Ordway, D.; Jackson, M.; et al. Indole-2-carboxamides are Active Against an Actute Mycobacterium abscessus Infected Mouse Model. Antimicrob. Agents Chemother. 2019, 1–16. [Google Scholar] [CrossRef]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, 1–14. [Google Scholar] [CrossRef]
- Wallace, R.J.; Dukart, G.; Brown-Elliot, B.A.; Griffith, D.E.; Scerpella, E.G.; Marshall, B. Clinical experience in 52 patients with tigecycline-containing regimens for salvage treatment of Mycobacterium abscessus and Mycobacterium chelonae infections. J. Antimicrob. Chemother. 2014, 69, 1945–1953. [Google Scholar] [CrossRef]
- Cynamon, M.; Jureller, J.; Desai, B.; Ramachandran, K.; Sklaney, M.; Grossman, T.H. In Vitro Activity of TP-271 against Mycobacterium abscessus, Mycobacterium chelonae, and Nocardia Species. Antimicrob. Agents Chemother. 2012, 56, 3986–3988. [Google Scholar] [CrossRef]
- Obregon-Henao, A.; Arnett, K.A.; Henao-Tamayo, M.; Massoudi, L.; Creissen, E.; Andries, K.; Lenaerts, A.J.; Ordway, D.J. Susceptibility of Mycobacterium abscessus to Antimycobacterial Drugs in Preclinical Models. Antimicrob. Agents Chemother. 2015, 59, 6904–6912. [Google Scholar] [CrossRef]
- Vesenbeckh, S.; Schonfeld, N.; Roth, A.; Bettermann, G.; Krieger, D.; Bauer, T.T.; Russmann, H.; Mauch, H. Bedaquiline as a potential agent in the treatment of Mycobacterium abscessus infections. Eur. Respir. J. 2017, 49, 1700083. [Google Scholar] [CrossRef]
- Brown-Elliott, B.A.; Wallace, R.J., Jr. In vitro susceptibility testing of tedizolid against nontuberculous mycobacteria. J. Clin. Microbiol. 2017, 55, 1747–1754. [Google Scholar] [CrossRef]
- Ruth, M.M.; Sangen, J.J.N.; Remmers, K.; Pennings, L.J.; Svensson, E.; Aarnoutse, R.E.; Zweijpfenning, S.M.H.; Hoefsloot, W.; Kuipers, S.; Magis-Escurra, C.; et al. A bedaquiline/clofazimine combination regimen might add activity to the treatment of clinically relevant non-tuberculous mycobacteria. J. Antimicrob. Chemother. 2019, dky526. [Google Scholar] [CrossRef]
| Study | Location | Sample Size | NTM Prevalence in CF |
|---|---|---|---|
| Oliver, KN (2004) [21] | USA | 750 | 13% (majority M. avium complex) |
| Roux, AL, et al. (2009) [22] | France | 1582 | 6.6% (M. abs most common) |
| Seddon, P, et al. (2013) [23] | UK | 3805 adults 3317 children | 5% adults 3.3% children |
| Adjemian, J, et al. (2014) [24] | USA | 18,003 | 10–20%; depending on area |
| Mussaffi, H, et al. (2005) [25] | Israel | 139 | 8.6% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopeman, R.C.; Harrison, J.; Desai, M.; Cox, J.A.G. Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare. Microorganisms 2019, 7, 90. https://doi.org/10.3390/microorganisms7030090
Lopeman RC, Harrison J, Desai M, Cox JAG. Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare. Microorganisms. 2019; 7(3):90. https://doi.org/10.3390/microorganisms7030090
Chicago/Turabian StyleLopeman, Rose C., James Harrison, Maya Desai, and Jonathan A. G. Cox. 2019. "Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare" Microorganisms 7, no. 3: 90. https://doi.org/10.3390/microorganisms7030090
APA StyleLopeman, R. C., Harrison, J., Desai, M., & Cox, J. A. G. (2019). Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare. Microorganisms, 7(3), 90. https://doi.org/10.3390/microorganisms7030090






