Urbanization Altered Bacterial and Archaeal Composition in Tidal Freshwater Wetlands Near Washington DC, USA, and Buenos Aires, Argentina
Abstract
1. Introduction
2. Materials and Methods
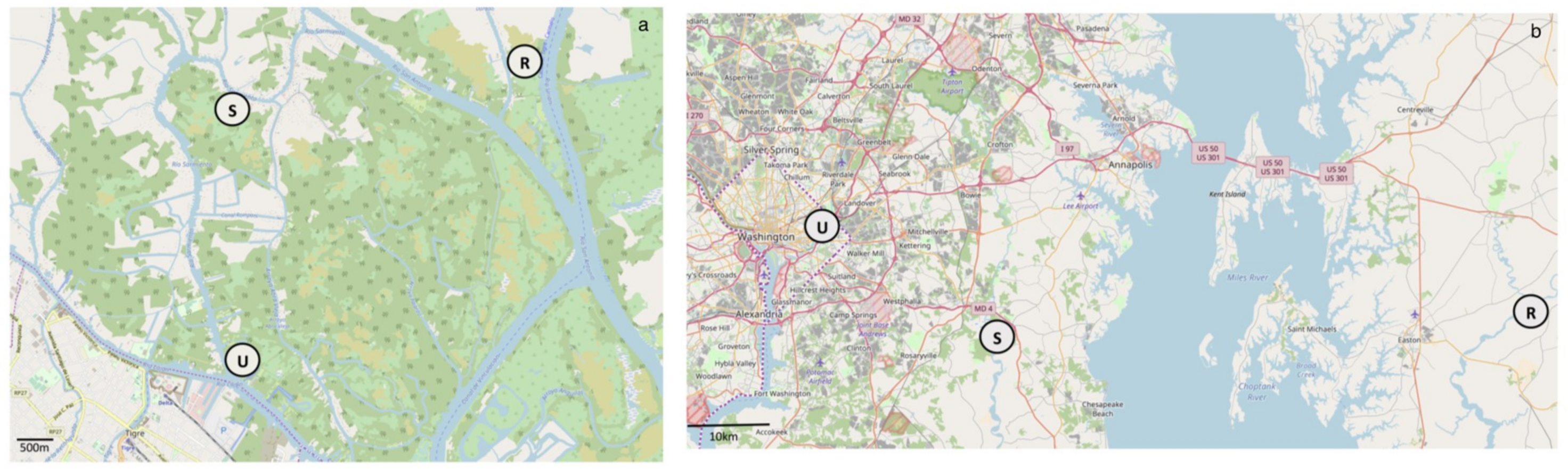
2.1. Site Description
2.2. Sample Collection
2.3. Soil Analysis
2.4. DNA Extraction and Illumina Library Preparation
2.5. Data Analysis
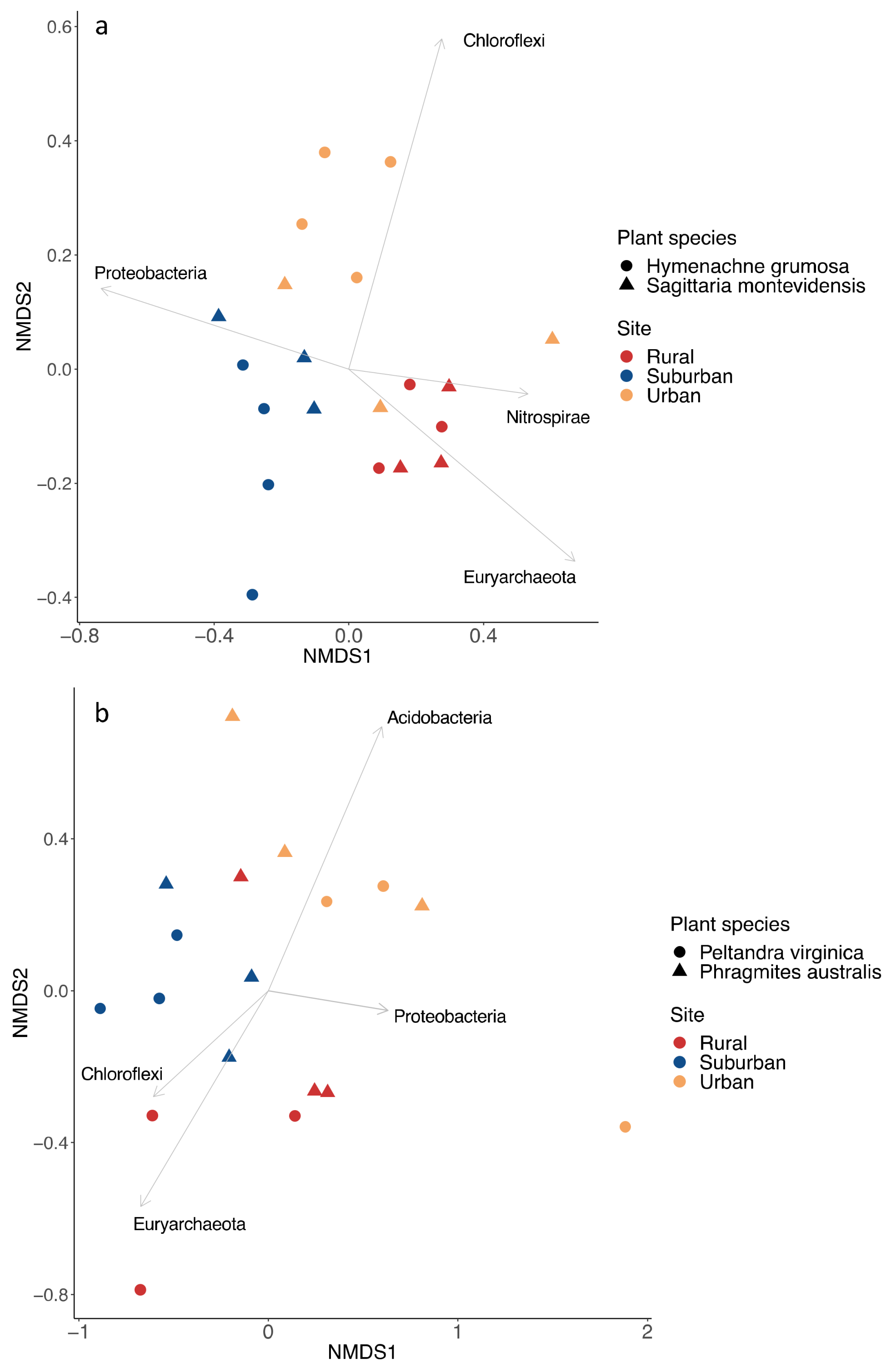
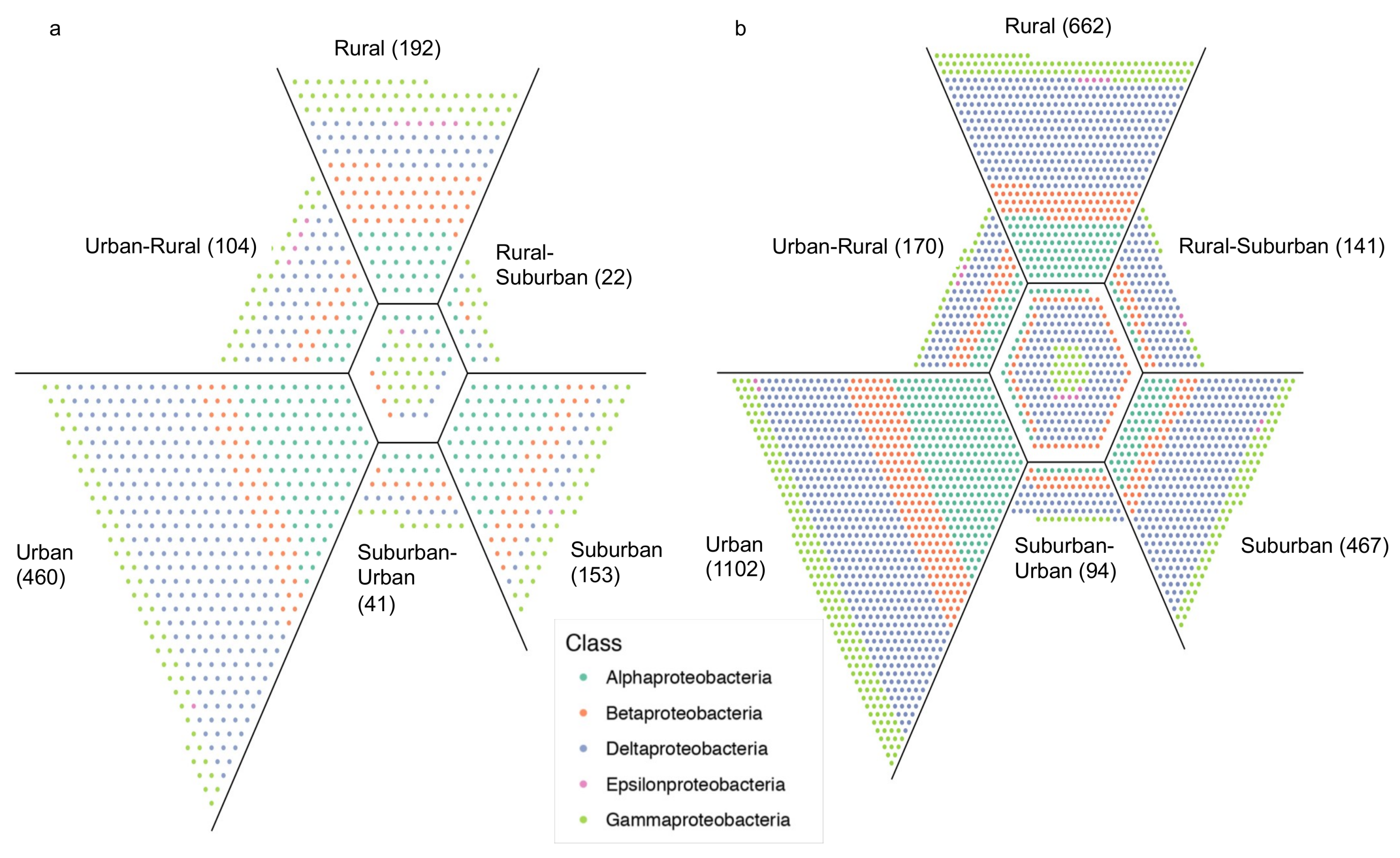
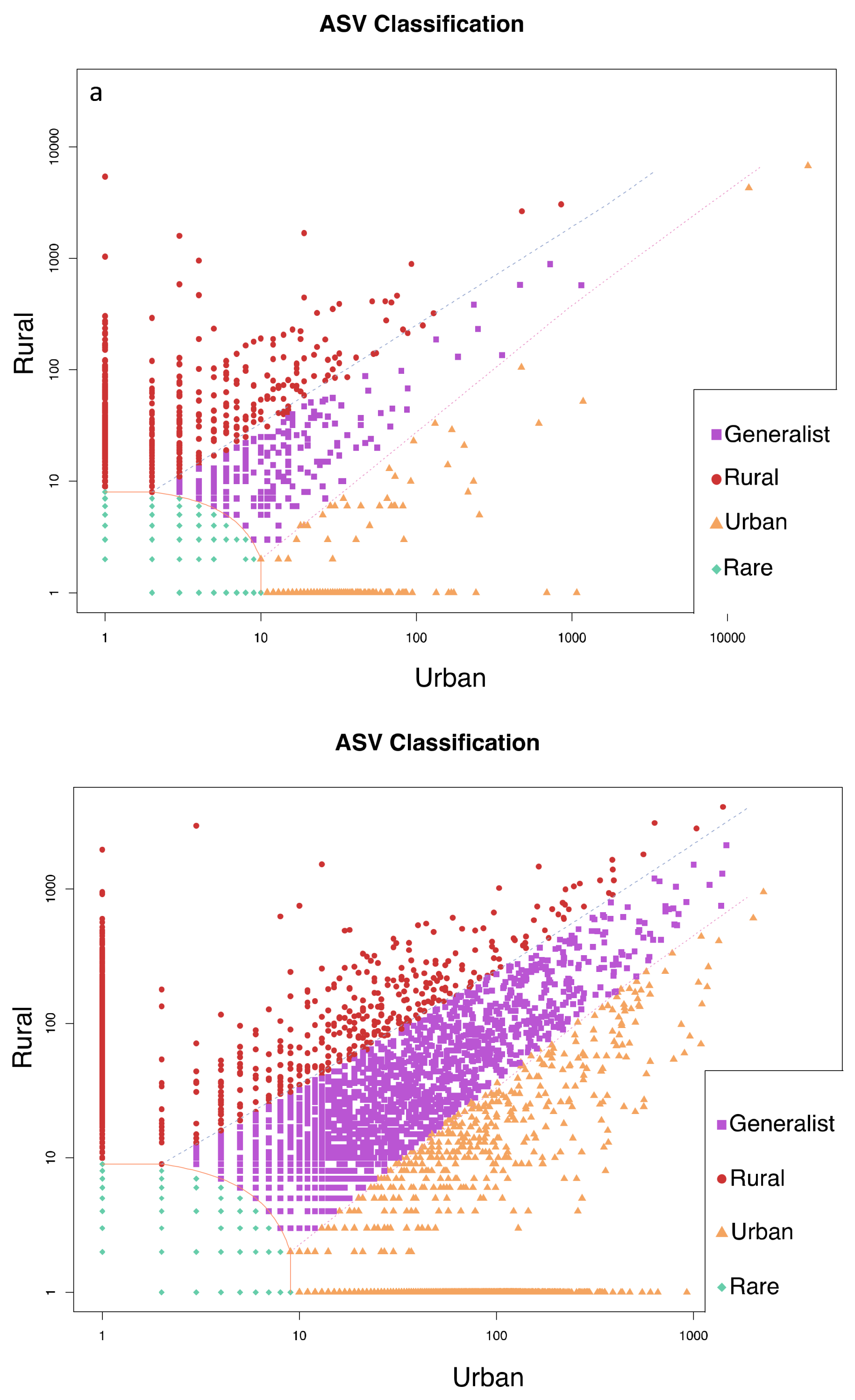
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division. World Urbanization Prospects: The 2014 Revision; ST/ESA/SER.A/366; United Nations: New York, NY, USA, 2015. [Google Scholar]
- Zedler, J.B.; Kercher, S. WETLAND RESOURCES: Status, Trends, Ecosystem Services, and Restorability. Annu. Rev. Environ. Resour. 2005, 30, 39–74. [Google Scholar] [CrossRef]
- Van Asselen, S.; Verburg, P.H.; Vermaat, J.E.; Janse, J.H. Drivers of Wetland Conversion: A Global Meta-Analysis. PLoS ONE 2013, 8, e81292. [Google Scholar] [CrossRef] [PubMed]
- Tibbetts, J. Coastal Cities: Living on the Edge. Environ. Health Perspect. 2002, 110, A674–A681. [Google Scholar] [CrossRef] [PubMed]
- Urbanization as a Major Cause of Biotic Homogenization. Biol. Conserv. 2006, 127, 247–260. [CrossRef]
- Millennium Ecosystem Assesment. Ecosystems and Human Well-Being: Wetlands and Water Synthesis. Millennium Ecosystem Assessment Report to the Ramsar Convention; World Resources Institute: Washington, DC, USA, 2005. [Google Scholar]
- Arnold, C.L.; Gibbons, C.J. Impervious Surface Coverage: The Emergence of a Key Environmental Indicator. J. Am. Plan. Assoc. 1996, 62, 243–258. [Google Scholar] [CrossRef]
- Schueler, T. Impacts of Impervious Cover on Aquatic Systems; Watershed Protection Research Monograph No 1; Center for Watershed Protection: Ellicot City, MD, USA, 2003. [Google Scholar]
- Hupp, C.R.; Woodside, M.D.; Yanosky, T.M. Sediment and Trace Element Trapping in a Forested Wetland, Chickahominy River, Virginia. Wetlands 1993, 13, 95–104. [Google Scholar] [CrossRef]
- Kimbrough, K.L.; Dickhut, R.M. Assessment of Polycyclic Aromatic Hydrocarbon Input to Urban Wetlands in Relation to Adjacent Land Use. Mar. Pollut. Bull. 2006, 52, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, J.G. Evaluating Wetlands within an Urban Context. Ecol. Eng. 2000, 15, 253–265. [Google Scholar] [CrossRef]
- Baldwin, A.H. Restoring Complex Vegetation in Urban Settings: The Case of Tidal Freshwater Marshes. Urban Ecosyst. 2004, 7, 125–137. [Google Scholar] [CrossRef]
- McKinney, M.L.; Lockwood, J.L. Biotic Homogenization: A Few Winners Replacing Many Losers in the next Mass Extinction. Trends Ecol. Evol. 1999, 14, 450–453. [Google Scholar] [CrossRef]
- Baiser, B.; Olden, J.D.; Record, S.; Lockwood, J.L.; McKinney, M.L. Pattern and Process of Biotic Homogenization in the New Pangaea. Proc. R. Soc. Lond. B Biol. Sci. 2012, 279, 4772–4777. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.T.; Hoverman, J.T.; McKenzie, V.J.; Blaustein, A.R.; Richgels, K.L.; Cadotte, M. Urbanization and wetland communities: Applying metacommunity theory to understand the local and landscape effects. J. Appl. Ecol. 2013, 50, 34–42. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Feng, X.; Zhai, L.; Zhu, G. Quantitative Analyses of Ammonia-Oxidizing Archaea and Bacteria in the Sediments of Four Nitrogen-Rich Wetlands in China. Appl. Microbiol. Biotechnol. 2011, 90, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.; Chapman, S.J.; Artz, R.R.E. Microbial Communities in Natural and Disturbed Peatlands: A Review. Soil Biol. Biochem. 2013, 57, 979–994. [Google Scholar] [CrossRef]
- Hartman, W.H.; Richardson, C.J.; Vilgalys, R.; Bruland, G.L. Environmental and Anthropogenic Controls over Bacterial Communities in Wetland Soils. Proc. Natl. Acad. Sci. USA 2008, 105, 17842–17847. [Google Scholar] [CrossRef] [PubMed]
- De Boer, W.; Kowalchuk, G.A.; van Veen, J.A. ‘Root-Food’ and the Rhizosphere Microbial Community Composition. New Phytol. 2006, 170, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Neori, A.; Agami, M. The Functioning of Rhizosphere Biota in Wetlands—A Review. Wetlands 2017, 37, 615–633. [Google Scholar] [CrossRef]
- Cadillo-Quiroz, H.; Yavitt, J.B.; Zinder, S.H.; Thies, J.E. Diversity and Community Structure of Archaea Inhabiting the Rhizoplane of Two Contrasting Plants from an Acidic Bog. Microb. Ecol. 2010, 59, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, S.A.; Baldwin, A.H.; Mateu, M.G.; Buyer, J.S. Archaeal Rhizosphere Communities Differ between the Native and Invasive Lineages of the Wetland Plant Phragmites australis (Common Reed) in a Chesapeake Bay Subestuary. Biol. Invas. 2016, 18, 2717–2728. [Google Scholar] [CrossRef]
- Bowen, J.L.; Kearns, P.J.; Byrnes, J.E.; Wigginton, S.; Allen, W.J.; Greenwood, M.; Tran, K.; Yu, J.; Cronin, J.T.; Meyerson, L.A. Lineage Overwhelms Environmental Conditions in Determining Rhizosphere Bacterial Community Structure in a Cosmopolitan Invasive Plant. Nat. Commun. 2017, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Prasse, C.E.; Baldwin, A.H.; Yarwood, S.A. Site History and Edaphic Features Override the Influence of Plant Species on Microbial Communities in Restored Tidal Freshwater Wetlands. Appl. Environ. Microbiol. 2015, 81, 3482–3491. [Google Scholar] [CrossRef] [PubMed]
- Cowardin, L.M.; Carter, V.; Golet, F.C.; LaRoe, E.T. Classification of Wetlands and Deepwater Habitats of the United States; U.S. Fish and Wildlife Service Report No. FWS/ OBS/-79/31; U.S. Fish and Wildlife Service: Washington, DC, USA, 1979.
- Whigham, D.F.; Baldwin, A.H.; Barendregt, A. Tidal Freshwater Wetlands” in Coastal Wetlands: An Integrated Ecosystem Approach; Elsevier: Amsterdam, the Netherlands, 2009. [Google Scholar]
- Simpson, R.L.; Good, R.E.; Leck, M.A.; Whigham, D.F. The Ecology of Freshwater Tidal Wetlands. BioScience 1983, 33, 255–259. [Google Scholar] [CrossRef]
- Barendregt, A.; Swarth, C.W. Tidal Freshwater Wetlands: Variation and Changes. Estuar. Coasts 2013, 36, 445–456. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Censos. Censo Nacional de Población, Hogares y Viviendas 2010. Available online: https://www.indec.gov.ar/nivel4_default.asp?id_tema_1=2&id_tema_2=41&id_tema_3=135 (accessed on 27 April 2018).
- DC Office of Planning. 2017 Annual Estimates of Population. Available online: https://planning.dc.gov/node/1296421 (accessed on 27 April 2018).
- Iriondo, M. The Littoral Complex at the Paraná Mouth. Q. Int. 2004, 114, 143–154. [Google Scholar] [CrossRef]
- Kandus, P.; Malvárez, A.I. Vegetation Patters and Change Analysis in the Lower Delta Islands of the Parana River (Argentina). Wetlands 2004, 24, 620–632. [Google Scholar] [CrossRef]
- Minotti, P.G.; Kandus, P. Actualización y Profundización del Mapa de Endicamientos y Terraplenes de la Región del Delta del Paraná. 2013. Available online: https://lac.wetlands.org/publicacion/actualizacion-y-profundizacion-del-mapa-de-endicamientos-y-terraplenes-de-la-region-del-delta-del-parana-2013/ (accessed on 27 April 2018).
- Fabricante, I.; Minotti, P.; Kandus, P. Urbanizaciones Cerradas en Humedales: Análisis espacial en el Delta del Paraná y en las Llanuras Aluviales de sus Principales Tributarios en Sector Continental de la Provincia de Buenos Aires. Urbanization in Wetlands. Fundación Humedales/Wetlands International; Laboratorio de Ecología Teledetección y Ecoinformática (LETyE) Instituto de Investigación e Ingeniería Ambiental (3iA) Universidad Nacional de General San Martín (UNSAM): San Martín, Argentina, 2012. [Google Scholar]
- Bisogno, M.C. Sustentabilidad de la Actividad Turística en la Primera Sección de Islas del Bajo Delta del Paraná (Tigre). Ph.D. Thesis, Universidad de Buenos Aires, Buenos Aires, Argentina, 2005. [Google Scholar]
- Fundación Metropolitana and Municipalidad de Tigre. Documento Base-Plan de Manejo Islas del Delta Tigre. 2012. Available online: http://metropolitana.org.ar/wp-content/uploads/downloads/2013/03/Plan-de-Manejo-Delta-Tigre-Documento-Base.pdf (accessed on 27 April 2018).
- Odum, W.E.; Smith, T.J.I.; Hoover, J.K.; McIvor, C.C. The Ecology of Tidal Freshwater Marshes of the United States East Coast: A Community Profile; FWS/OBS-83/17; U.S. Fish and Wildlife Service: Washington, DC, USA, 1984.
- Boesch, D.F.; Greer, J. Chesapeake Futures: Choices for the 21st Century; Chesapeake Research Consortium, Inc.: Edgewater, MD, USA, 2003. [Google Scholar]
- Dauer, D.M.; Ranasinghe, J.A.; Weisberg, S.B. Relationships between Benthic Community Condition, Water Quality, Sediment Quality, Nutrient Loads, and Land Use Patterns in Chesapeake Bay. Estuaries 2000, 23, 80–96. [Google Scholar] [CrossRef]
- Kemp, W.M.; Boynton, W.R.; Adolf, J.E.; Boesch, D.F.; Boicourt, W.C.; Brush, G.; Cornwell, J.C.; Fisher, T.R.; Glibert, P.M.; Hagy, J.D.; et al. Eutrophication of Chesapeake Bay: Historical Trends and Ecological Interactions. Mar. Ecol. Prog. Ser. 2005, 303, 1–29. [Google Scholar] [CrossRef]
- Ellis, J.B.; Wang, Y. Bacteriology of Urban Runoff: The Combined Sewer as a Bacterial Reactor and Generator. Water Sci. Technol. 1995, 31, 303. [Google Scholar] [CrossRef]
- Castañé, P.M.; Sánchez-Caro, A.; Salibián, A. Water Quality of the Luján River, a Lowland Watercourse near the Metropolitan Area of Buenos Aires (Argentina). Environ. Monit. Assess. 2015, 187, 645. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.R.; Hagy, J.D.; Boynton, W.R.; Williams, M.R. Cultural Eutrophication in the Choptank and Patuxent Estuaries of Chesapeake Bay. Limnol. Oceanogr. 2006, 51, 435–447. [Google Scholar] [CrossRef]
- Mozdzer, T.J.; Zieman, J.C. Ecophysiological Differences between Genetic Lineages Facilitate the Invasion of Non-native Phragmites Australis in North American Atlantic Coast Wetlands. J. Ecol. 2010, 98, 451–458. [Google Scholar] [CrossRef]
- League, M.T.; Colbert, E.P.; Seliskar, D.M.; Gallagher, J.L. Rhizome Growth Dynamics of Native and Exotic Haplotypes of Phragmites Australis (Common Reed). Estuar. Coasts 2006, 29, 269–276. [Google Scholar] [CrossRef]
- Gee, W.G.; Or, D. Particle-Size Analysis. In Methods of Soil Analysis: Part 4 Physical Methods; Soil Science Society of America, Inc.: Madison, WI, USA, 2002; pp. 255–293. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2006. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.4-6. 2018. Available online: https://CRAN.R-project.org/package=vegan (accessed on 27 April 2018).
- Hervé, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics, R package Version 0.9-69. Available online: https://CRAN.R-project.org/package=RVAideMemoire (accessed on 6 March 2019).
- Chazdon, R.L.; Chao, A.; Colwell, R.K.; Lin, S.Y.; Norden, N.; Letcher, S.G.; Clark, D.B.; Finegan, B.; Arroyo, J.P. A Novel Statistical Method for Classifying Habitat Generalists and Specialists. Ecology 2011, 92, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- De Caceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef] [PubMed]
- Woloszynek, S.; Mell, J.C.; Simpson, G.; Rosen, G.L. Exploring thematic structure in 16S rRNA marker gene surveys. bioRxiv 2017. [Google Scholar] [CrossRef]
- Wilkins, D.; Unioinplot. Github Repository. 2016. Available online: https://github.com/wilkox/unionplot (accessed on 1 February 2018).
- Hamann, C.; Hegemann, J.; Hildebrandt, A. Detection of polycyclic aromatic hydrocarbon degradation genes in different soil bacteria by polymerase chain reaction and DNA hybridization. FEMS Microbiol. Lett. 1999, 173, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Shuai, J.J.; Tian, Y.S.; Yao, Q.H.; Peng, R.H.; Xiong, F.; Xiong, A.S. Identification and Analysis of Polychlorinated Biphenyls (PCBs)-Biodegrading Bacterial Strains in Shanghai. Curr. Microbiol. 2010, 61, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 01369. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, M.; Legendre, P. Species Assemblages and Indicator Species:The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil PH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, P.; de Miera, L.E.S.; Ansola, G. Influence of Environmental Variables on the Structure and Composition of Soil Bacterial Communities in Natural and Constructed Wetlands. Sci. Total Environ. 2015, 506, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Li, H.; Yang, H.; Peng, C.; Peng, Z.; Lu, L. Shift in the Microbial Community Composition of Surface Water and Sediment along an Urban River. Sci. Total Environ. 2018, 627, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Jani, K.; Ghattargi, V.; Pawar, S.; Inamdar, M.; Shouche, Y.; Sharma, A. Anthropogenic Activities Induce Depletion in Microbial Communities at Urban Sites of the River Ganges. Curr. Microbiol. 2018, 75, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Hosen, J.D.; Febria, C.M.; Crump, B.C.; Palmer, M.A. Watershed Urbanization Linked to Differences in Stream Bacterial Community Composition. Front. Microbiol. 2017, 8, 01452. [Google Scholar] [CrossRef] [PubMed]
- Nold, S.C.; Zwart, G. Patterns and Governing Forces in Aquatic Microbial Communities. Aquat. Ecol. 1998, 32, 17–35. [Google Scholar] [CrossRef]
- Lv, X.; Yu, J.; Fu, Y.; Ma, B.; Qu, F.; Ning, K.; Wu, H. A Meta-Analysis of the Bacterial and Archaeal Diversity Observed in Wetland Soils. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Zhu, L.; Li, Y.; Xie, Y.; Zhao, M.; Luo, X. Composition and Variation of Sediment Bacterial and NirS-Harboring Bacterial Communities at Representative Sites of the Bohai Gulf Coastal Zone, China. World J. Microbiol. Biotechnol. 2014, 30, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, A.; Grönroos, M.; Siter, N.; Puhakka, R.; Vari, H.K.; Roslund, M.I.; Jumpponen, A.; Nurminen, N.; Laitinen, O.H.; Hyöty, H.; et al. Urbanization Reduces Transfer of Diverse Environmental Microbiota Indoors. Front. Microbiol. 2018, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.P.S. Water Microbiology. Bacterial Pathogens and Water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Lors, C.; Damidot, D.; Ponge, J.; Périé, F. Comparison of a Bioremediation Process of PAHs in a PAH-Contaminated Soil at Field and Laboratory Scales. Environ. Pollut. 2012, 165, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Selvakumaran, S.; Kapley, A.; Kashyap, S.M.; Daginawala, H.F.; Kalia, V.C.; Purohit, H.J. Diversity of Aromatic Ring-Hydroxylating Dioxygenase Gene in Citrobacter. Bioresour. Technol. 2011, 102, 4600–4609. [Google Scholar] [CrossRef] [PubMed]
- Foster, G.D.; Roberts, E.C.; Gruessner, B.; Velinsky, J. Hydrogeochemistry and transport of organic contaminants in an urban watershed of Chesapeake Bay (USA). Appl. Geochem. 2000, 15, 901–915. [Google Scholar] [CrossRef]
- Hwang, H.M.; Foster, G.D. Polychlorinated Biphenyls in Stormwater Runoff Entering the Tidal Anacostia River, Washington, DC, through Small Urban Catchments and Combined Sewer Outfalls. J. Environ. Sci. Health Part A 2008, 43, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Pinkney, A.E.; Harshbarger, J.C.; Rutter, M.A. Tumors in Brown Bullheads in the Chesapeake Bay Watershed: Analysis of Survey Data from 1992 through 2006. J. Aquat. Anim. Health 2009, 21, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Pinkney, A.E.; Harshbarger, J.C.; Karouna-Renier, N.K.; Jenko, K.; Balk, L.; Skarphéðinsdóttir, H.; Liewenborg, B.; Rutter, M.A. Tumor Prevalence and Biomarkers of Genotoxicity in Brown Bullhead (Ameiurus Nebulosus) in Chesapeake Bay Tributaries. Sci. Total Environ. 2011, 410, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Bodelier, P.L.E. Interactions between Nitrogenous Fertilizers and Methane Cycling in Wetland and Upland Soils. Curr. Opin. Environ. Sustain. 2011, 3, 379–388. [Google Scholar] [CrossRef]
- Liu, H.; Wu, X.; Li, Z.; Wang, Q.; Liu, D.; Liu, G. Responses of Soil Methanogens, Methanotrophs, Methane Fluxes to Land-Use Conversion and Fertilization in a Hilly Red Soil Region of Southern China. Environ. Sci. Pollut. Res. Int. 2017, 24, 8731–8743. [Google Scholar] [CrossRef] [PubMed]
- Höfferle, Š.; Nicol, G.W.; Pal, L.; Hacin, J.; Prosser, J.I.; Mandić-Mulec, I. Ammonium Supply Rate Influences Archaeal and Bacterial Ammonia Oxidizers in a Wetland Soil Vertical Profile. FEMS Microbiol. Ecol. 2010, 74, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, B.B.; Postgate, J.R. Ecology of the Bacteria of the Sulphur Cycle with Special Reference to Anoxic-Oxic Interface Environments. Philos. Trans. R. Soc. Lond. Ser. B 1982, 298, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Suárez, A.; López-López, A.; Tovar-Sánchez, A.; Yarza, P.; Orfila, A.; Terrados, J.; Arnds, J.; Marqués, S.; Niemann, H.; Schmitt-Kopplin, P.; et al. Response of Sulfate-reducing Bacteria to an Artificial Oil-spill in a Coastal Marine Sediment. Environ. Microbiol. 2011, 13, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Stams, A.J.M. The Ecology and Biotechnology of Sulphate-Reducing Bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Zhu, J.N.; Zhai, Z.H.; Zhang, Q. Endophytic Bacterial Diversity in Roots of Phragmites australis in Constructed Beijing Cuihu Wetland (China). FEMS Microbiol. Lett. 2010, 309, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Hsieh, Y.P. Sulfate Reduction in Freshwater Wetland Soils and the Effects of Sulfate and Substrate Loading. J. Environ. Qual. 1998, 27, 968–972. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Fu, X.; Yan, J.; Wu, J.; Tsang, Y.; Le, Y.; Sun, Y. Salinity and Nutrient Contents of Tidal Water Affects Soil Respiration and Carbon Sequestration of High and Low Tidal Flats of Jiuduansha Wetlands in Different Ways. Sci. Total Environ. 2016, 565, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, Y.; Wang, Y.; Chin, F.Y.L.; Zhang, T. Cellular Adhesiveness and Cellulolytic Capacity in Anaerolineae Revealed by Omics-Based Genome Interpretation. Biotechnol. Biofuels 2016, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Grube, M.; Erlacher, A.; Quehenberger, J.; Berg, G. The lettuce root microbiota. Environ. Microbiol. 2015, 17, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, S.A.; Högberg, M.N. Soil Bacteria and Archaea Change Rapidly in the First Century of Fennoscandian Boreal Forest Development. Soil Biol. Biochem. 2017, 114, 160–167. [Google Scholar] [CrossRef]
- Huang, S.; Vieira, S.; Bunk, B.; Riedel, T.; Spröer, C.; Overmann, J. First Complete Genome Sequence of a Subdivision 6 Acidobacterium Strain. Genome Announ. 2016, 4, e00469-16. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Velinsky, D.J.; Riedel, G.F.; Ashley, J.T.F.; Cornwell, J.C. Historical Contamination of the Anacostia River, Washington, D.C. Environ. Monit. Assess. 2011, 183, 307. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.; Groffman, P.M.; Findlay, S.E.G.; Arreola, A.E. Invasive Plant Species and Microbial Processes in a Tidal Freshwater Marsh. J. Environ. Qual. 1999, 28, 1252–1257. [Google Scholar] [CrossRef]
- Peralta, A.L.; Matthews, J.W.; Flanagan, D.N.; Kent, A.D. Environmental Factors at Dissimilar Spatial Scales Influence Plant and Microbial Communities in Restored Wetlands. Wetlands 2012, 32, 1125–1134. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete Nitrification by Nitrospira Bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Lücker, S.; Vejmelkova, D.; Kostrikina, N.A.; Kleerebezem, R.; Rijpstra, W.I.; Damsté, J.S.; Le Paslier, D.; Muyzer, G.; Wagner, M.; et al. Nitrification Expanded: Discovery, Physiology and Genomics of a Nitrite-Oxidizing Bacterium from the Phylum Chloroflexi. ISME J. 2012, 6, 2245–2256. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Buenos Aires | Maryland | ||||
|---|---|---|---|---|---|---|
| Rural | Suburban | Urban | Rural | Suburban | Urban | |
| pH | 6.4 ± 0.2 | 5.4 ± 0.08 | 5.8 ± 0.1 | 4 ± 0.3 | 4.7 ± 0.1 | 5.7 ± 0.1 |
| SOM (%) | 11.6 ± 0.5 | 6.6 ± 0.02 | 24.7 ± 2.2 | 35.6 ± 0.5 | 11.8 ± 1.2 | 6.5 ± 0.7 |
| C (%) | 2.7 ± 0.2 | 1.7 ± 0.1 | 3.54 ± 0.7 | 19 ± 0.2 | 5.6 ± 0.6 | 3 ± 0.4 |
| N (%) | 0.2 ± 0.02 | 0.2 ± 0.01 | 0.32 ± 0.1 | 0.12 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.03 |
| C/N | 13 | 11.67 | 11.77 | 15.63 | 14.1 | 15.7 |
| Sand (%) | 0.31 | 0.38 | 0.33 | 0.26 | 0.35 | 0.17 |
| Silt (%) | 0.43 | 0.4 | 0.42 | 0.34 | 0.53 | 0.58 |
| Clay (%) | 0.25 | 0.22 | 0.25 | 0.4 | 0.12 | 0.26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez Mateu, M.; Park, C.E.; McAskill, C.P.; Baldwin, A.H.; Yarwood, S.A. Urbanization Altered Bacterial and Archaeal Composition in Tidal Freshwater Wetlands Near Washington DC, USA, and Buenos Aires, Argentina. Microorganisms 2019, 7, 72. https://doi.org/10.3390/microorganisms7030072
Gonzalez Mateu M, Park CE, McAskill CP, Baldwin AH, Yarwood SA. Urbanization Altered Bacterial and Archaeal Composition in Tidal Freshwater Wetlands Near Washington DC, USA, and Buenos Aires, Argentina. Microorganisms. 2019; 7(3):72. https://doi.org/10.3390/microorganisms7030072
Chicago/Turabian StyleGonzalez Mateu, Martina, Cedric Evan Park, Cullen Patrick McAskill, Andrew H. Baldwin, and Stephanie A. Yarwood. 2019. "Urbanization Altered Bacterial and Archaeal Composition in Tidal Freshwater Wetlands Near Washington DC, USA, and Buenos Aires, Argentina" Microorganisms 7, no. 3: 72. https://doi.org/10.3390/microorganisms7030072
APA StyleGonzalez Mateu, M., Park, C. E., McAskill, C. P., Baldwin, A. H., & Yarwood, S. A. (2019). Urbanization Altered Bacterial and Archaeal Composition in Tidal Freshwater Wetlands Near Washington DC, USA, and Buenos Aires, Argentina. Microorganisms, 7(3), 72. https://doi.org/10.3390/microorganisms7030072





