Metagenomic Insights into the Effects of Seasonal Temperature Variation on the Activities of Activated Sludge
Abstract
1. Introduction
2. Materials and Methods
2.1. Wastewater and Seed Sludge
2.2. The Lab-Scale SBR and the Operating Conditions
2.3. Chemical Analysis
2.4. EPS Extraction and Characterization
2.5. DNA Extraction
2.6. Library Construction and High-Throughput Sequencing
2.7. Quality Control and Sequence Assembly
2.8. Gene Prediction and Gene Catalogue Construction
2.9. Taxonomic Assignment and Function Annotation
3. Results and Discussion
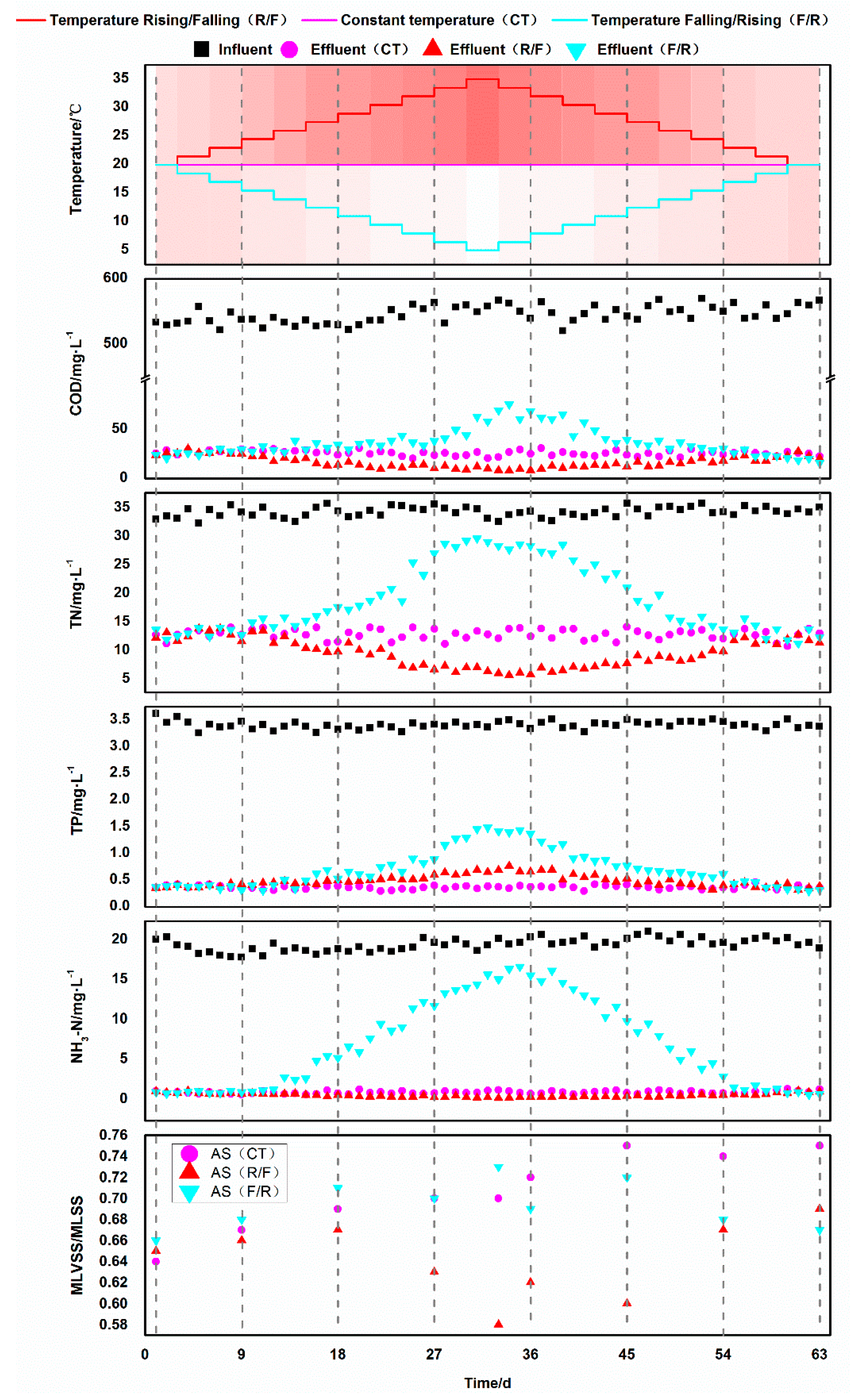
3.1. Degradation Effect of Activated Sludge at Different Temperatures
3.1.1. Chemical Oxygen Demand Removal
3.1.2. Nitrogen Removal
3.1.3. Phosphate Removal
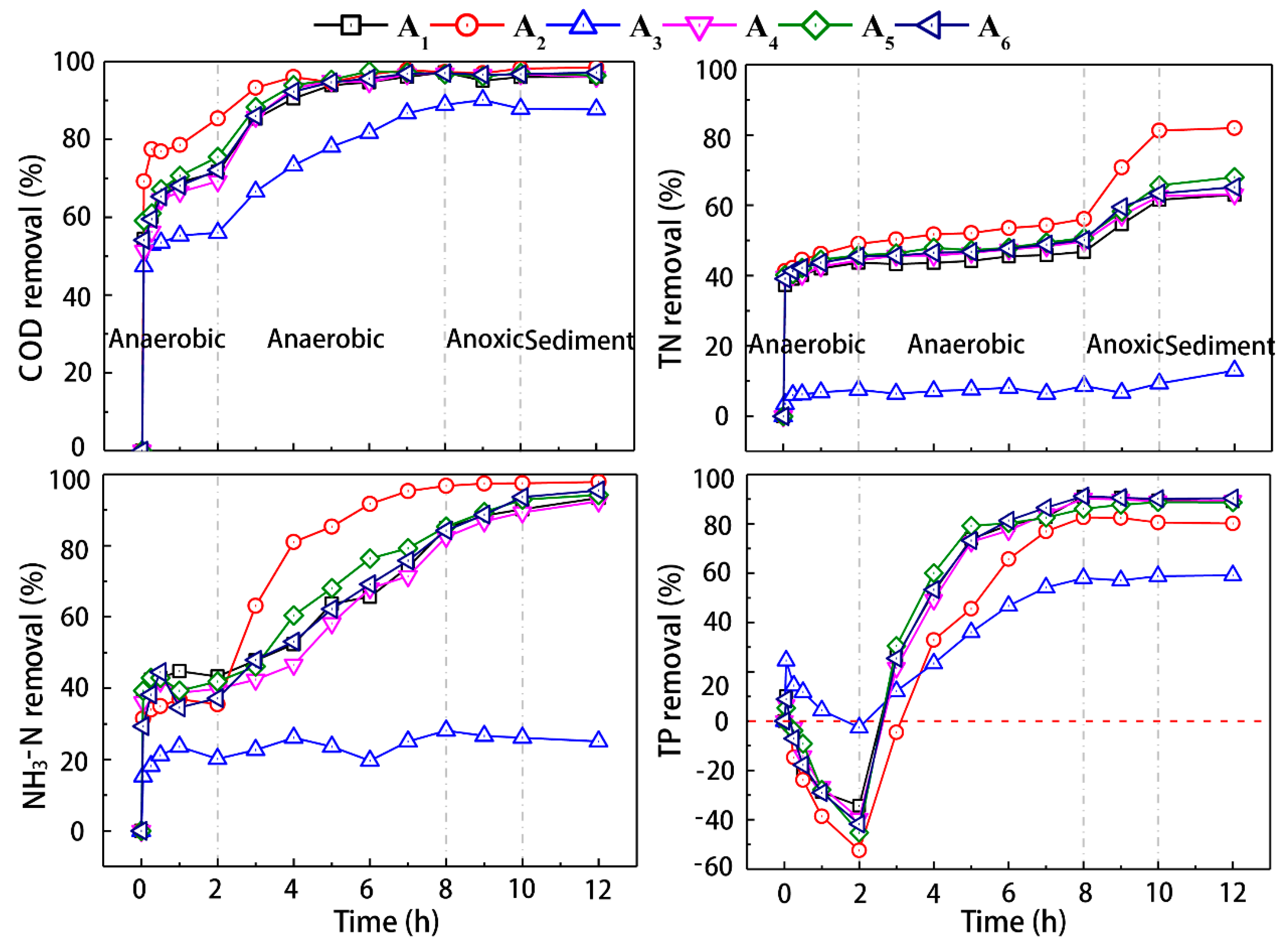
3.2. The Degradation Effect of the Activated Sludge under Typical Temperature in Single Cycle
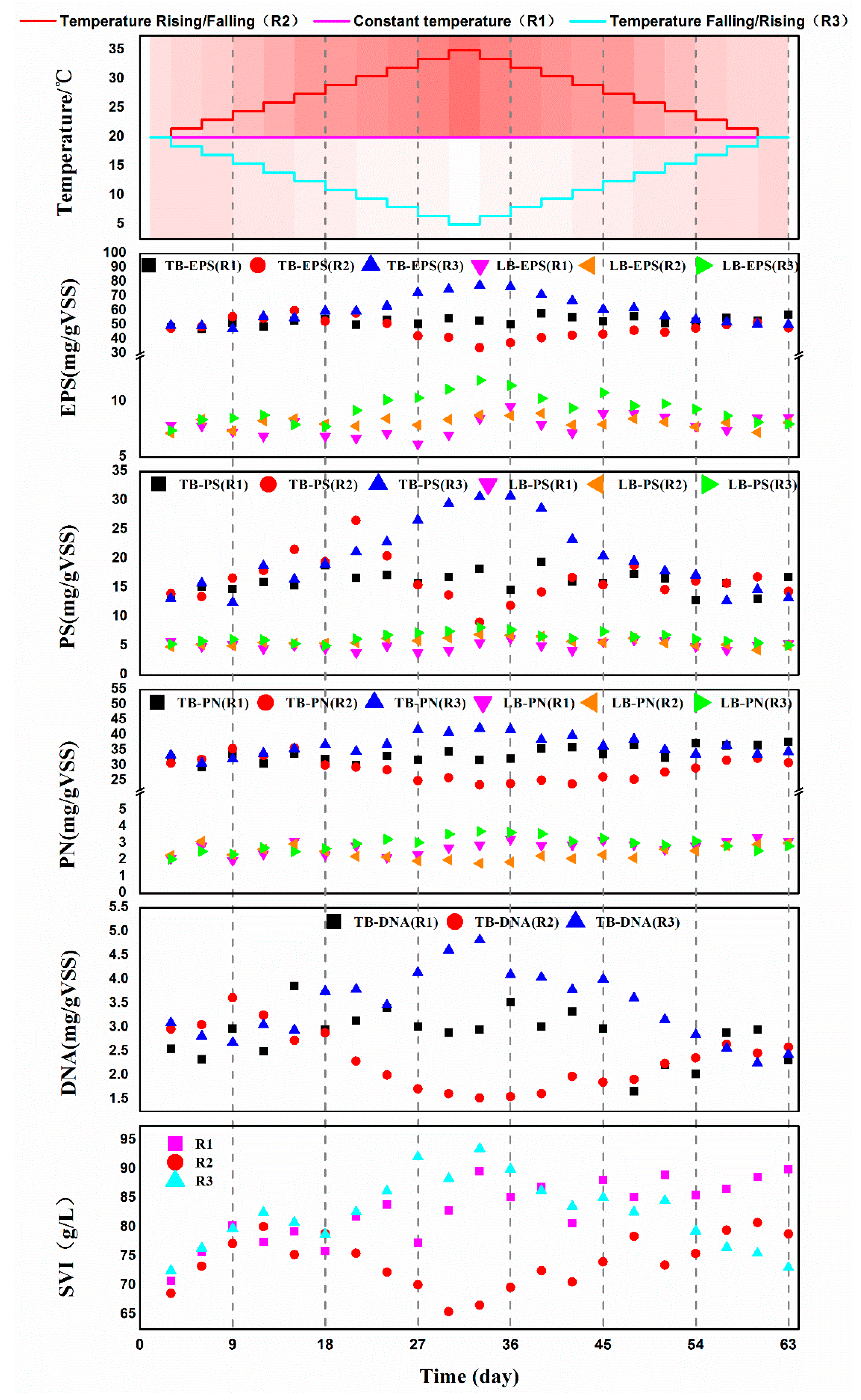
3.3. Changes of EPS Composition and the Sludge Index of Activated Sludge under Different Temperature Variation
3.4. Metagenomic Analysis
3.4.1. Overview of Metagenomic Sequence
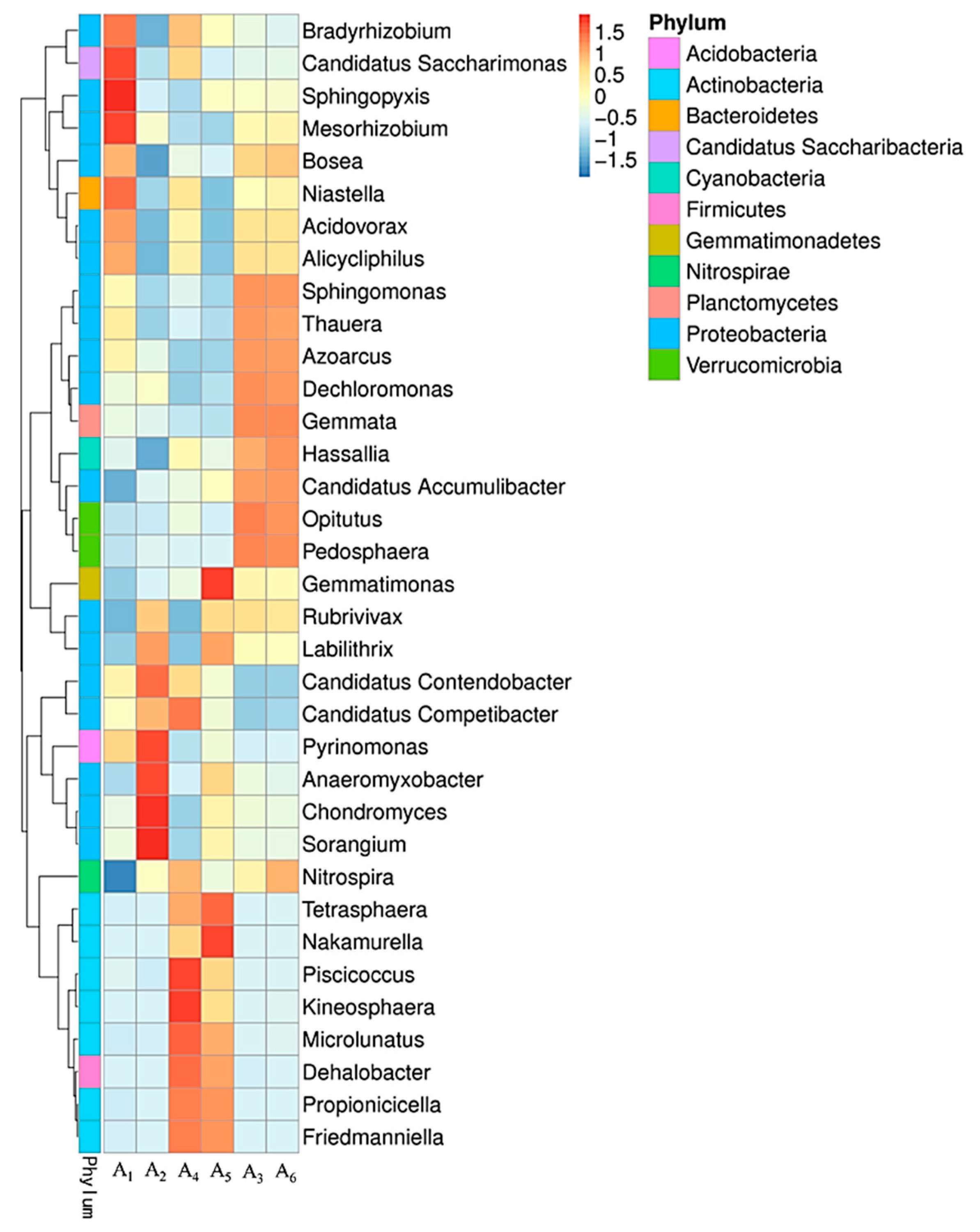
3.4.2. Variation of Microbial Community Structure in Activated Sludge
3.5. Activated Sludge Microbial Gene Function Annotation
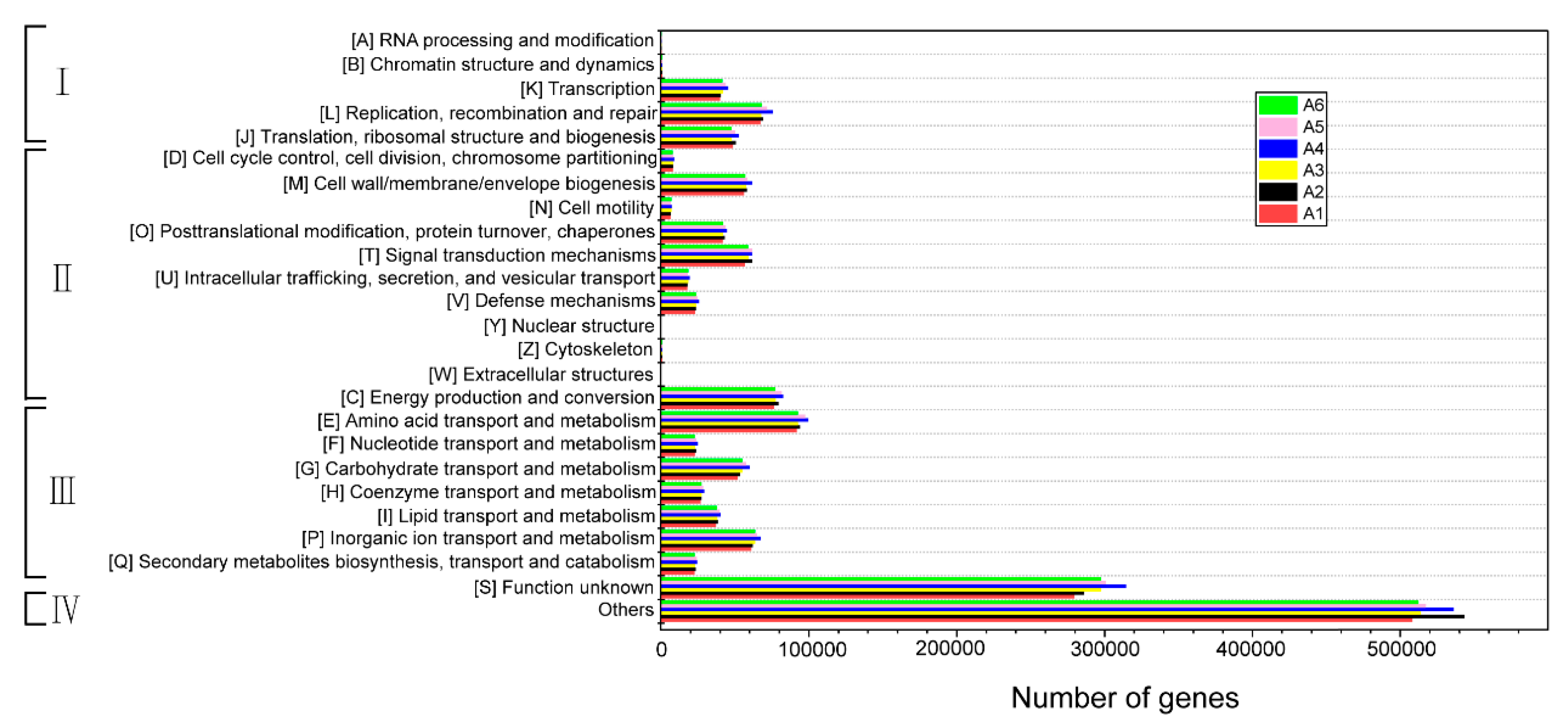
3.5.1. COG Functional Classification and Functional Potential Analysis
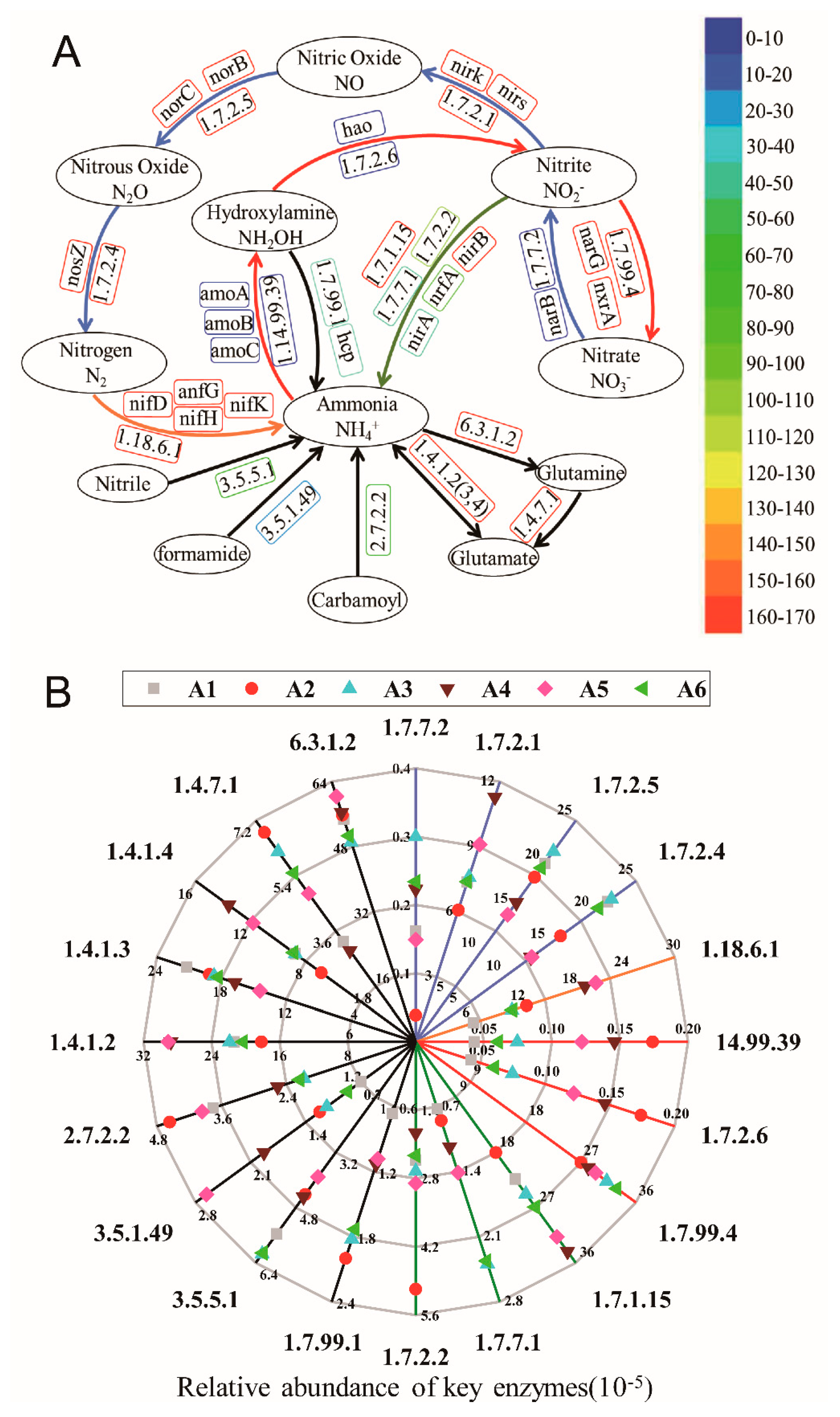
3.5.2. The Variation of Key Enzymes in Nitrogen Metabolic Pathways
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Papadimitriou, C.; Palaska, G.; Lazaridou, M.; Samaras, P.; Sakellaropoulos, G.P. The effects of toxic substances on the activated sludge microfauna. Desalination 2007, 211, 177–191. [Google Scholar] [CrossRef]
- Wagner, M.; Loy, A.; Nogueira, R.; Purkhold, U.; Lee, N.; Daims, H. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 2002, 81, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Loy, A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002, 13, 218–227. [Google Scholar] [CrossRef]
- Fang, H.; Cai, L.; Yu, Y.; Zhang, T. Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge. Bioresour. Technol. 2013, 129, 209–218. [Google Scholar] [CrossRef]
- Feng, J.; Feng, G.; Lin, Y.; Yu, X.; Tong, Z. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 2014, 6, 80–89. [Google Scholar]
- Guo, J.; Ni, B.J.; Han, X.; Chen, X.; Bond, P.; Peng, Y.; Yuan, Z. Unraveling microbial structure and diversity of activated sludge in a full-scale simultaneous nitrogen and phosphorus removal plant using metagenomic sequencing. Enzym. Microb. Technol. 2017, 102, 16–25. [Google Scholar] [CrossRef]
- Yi, J.C.; Mei, F.C.; Law, C.L. Biological treatment of anaerobically digested palm oil mill effluent (POME) using a lab-scale sequencing batch reactor (SBR). J. Environ. Manag. 2010, 91, 1738–1746. [Google Scholar]
- Li, X.Y.; Yang, S.F. Influence of loosely bound extracellular polymeric substances (EPS) on the flocculation, sedimentation and dewaterability of activated sludge. Water Res. 2007, 41, 1022–1030. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H. Measurement of protein using bicinchoninic acid. Anal. Chem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Q.; Zou, H.; Yu, Y.; Zhou, Z.; Mao, J.; Zhang, S. A metagenomic analysis of the relationship between microorganisms and flavor development in Shaoxing mechanized huangjiu fermentation mashes. Int. J. Food Microbiol. 2019, 303, 9–18. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Brdjanovic, D.; Logemann, S.; Loosdrecht, M.C.M.V.; Hooijmans, C.M.; Alaerts, G.J.; Heijnen, J.J. Influence of temperature on biological phosphorus removal: Process and molecular ecological studies. Water Res. 1998, 32, 1035–1048. [Google Scholar] [CrossRef]
- Chevalier, P.; Proulx, D.; Lessard, P.; Vincent, W.F.; Noüe, J.D.L. Nitrogen and phosphorus removal by high latitude mat-forming cyanobacteria for potential use in tertiary wastewater treatment. J. Appl. Phycol. 2000, 12, 105–112. [Google Scholar] [CrossRef]
- Kim, D.; Kim, K.Y.; Ryu, H.D.; Min, K.K.; Lee, S.I. Long term operation of pilot-scale biological nutrient removal process in treating municipal wastewater. Bioresour. Technol. 2009, 99, 3180–3184. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, L.; Chen, W.; Ma, F.; Liu, H.; Tian, Y. The regulation and control strategies of a sequencing batch reactor for simultaneous nitrification and denitrification at different temperatures. Bioresour. Technol. 2013, 133, 59–67. [Google Scholar] [CrossRef]
- Pollice, A.; Laera, G.; Saturno, D.; Giordano, C. Effects of sludge retention time on the performance of a membrane bioreactor treating municipal sewage. J. Membr. Sci. 2008, 317, 65–70. [Google Scholar] [CrossRef]
- Ducey, T.F.; Vanotti, M.B.; Shriner, A.D.; Szogi, A.A.; Ellison, A.Q. Characterization of a microbial community capable of nitrification at cold temperature. Bioresour. Technol. 2010, 101, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Brdjanovic, D.; Loosdrecht, M.C.M.V.; Versteeg, P.; Hooijmans, C.M.; Alaerts, G.J.; Heijnen, J.J. Modeling COD, N and P removal in a full-scale wwtp Haarlem Waarderpolder. Water Res. 2000, 34, 846–858. [Google Scholar] [CrossRef]
- Panswad, T.; Doungchai, A.; Jin, A. Temperature effect on microbial community of enhanced biological phosphorus removal system. Water Res. 2003, 37, 409–415. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Yang, F. Diameter control and stability maintenance of aerobic granular sludge in an A/O/A SBR. Sep. Purif. Technol. 2015, 149, 362–369. [Google Scholar] [CrossRef]
- Oehmen, A.; Lemos, P.C.; Carvalho, G.; Yuan, Z.; Keller, J.; Blackall, L.L.; Reis, M.A.M. Advances in enhanced biological phosphorus removal: From micro to macro scale. Water Res. 2007, 41, 2271–2300. [Google Scholar] [CrossRef]
- Sheng, G.P.; Yu, H.Q.; Li, X.Y. Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: A review. Biotechnol. Adv. 2010, 28, 882–894. [Google Scholar] [CrossRef]
- Dai, H.; Dai, Z.; Peng, L.; Wu, Y.; Zou, H.; Lu, X. Metagenomic and metabolomic analysis reveals the effects of chemical phosphorus recovery on biological nutrient removal system. Chem. Eng. J. 2017, 328, 1087–1097. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Z.; Li, H.; Park, H.D.; Wu, Z. Metagenomes reveal microbial structures, functional potentials, and biofouling-related genes in a membrane bioreactor. Appl. Microbiol. Biotechnol. 2016, 100, 5109–5121. [Google Scholar] [CrossRef]
- Wang, H.; Qun, S.; Jing, W.; Heng, Z.; Qiulai, H.; Wei, Z.; Jianyang, S.; Jinping, Z.; Hui, L. Simultaneous nitrification, denitrification and phosphorus removal in an aerobic granular sludge sequencing batch reactor with high dissolved oxygen: Effects of carbon to nitrogen ratios. Sci. Total Environ. 2018, 642, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Heylen, K.; Lebbe, L.; De Vos, P. Acidovorax caeni sp. nov., a denitrifying species with genetically diverse isolates from activated sludge. Int. J. Syst. Evol. Microbiol. 2008, 58, 73–77. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, F.; Feng, X.; Liu, Y.; Yan, X.; Zhang, X.; Wang, L.; Zhao, L. Thauera and Azoarcus as functionally important genera in a denitrifying quinoline-removal bioreactor as revealed by microbial community structure comparison. Fems Microbiol. Ecol. 2006, 55, 274–286. [Google Scholar] [CrossRef]
- Chen, D.Y.; Wang, Z.M.; Zhang, M.L.; Wang, X.H.; Lu, S. Effect of increasing salinity and low C/N ratio on the performance and microbial community of a sequencing batch reactor. Environ. Technol. 2019, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Hanada, S.; Marsh, T.L.; Kamagata, Y.; Nakamura, K. Kineosphaera limosa gen. nov., sp. nov., a novel Gram-positive polyhydroxyalkanoate-accumulating coccus isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2002, 52, 1845–1849. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Hiraishi, A.; Yoshimi, Y.; Kawaharasaki, M.; Masuda, K. Microlunatus phosphovorus gen. nov., sp. nov., a new gram-positive polyphosphate-accumulating bacterium isolated from activated sludge. Int. J. Syst. Bacteriol. 1995, 45, 17–22. [Google Scholar] [CrossRef]
- Hanada, S.; Liu, W.; Shintani, T.; Kamagata, Y. Tetrasphaera elongata sp. nov., a polyphosphate-accumulating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2002, 52, 883–887. [Google Scholar] [CrossRef]
- Huang, Z.; Gedalanga, P.B.; Asvapathanagul, P.; Olson, B.H. Influence of physicochemical and operational parameters on Nitrobacter and Nitrospira communities in an aerobic activated sludge bioreactor. Water Res. 2010, 44, 4351–4358. [Google Scholar] [CrossRef]
- Meng, F.; Gao, G.; Yang, T.-T.; Chen, X.; Chao, Y.; Na, G.; Ge, L.; Huang, L.-N. Effects of fluoroquinolone antibiotics on reactor performance and microbial community structure of a membrane bioreactor. Chem. Eng. J. 2015, 280, 448–458. [Google Scholar] [CrossRef]
- Terashima, M.; Yama, A.; Sato, M.; Yumoto, I.; Kamagata, Y.; Kato, S. Culture-dependent and ondependent identification of polyphosphate-accumulating Dechloromonas spp. predominating in a full-scale oxidation ditch wastewater treatment plant. Microbes Environ. 2016, 31, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Garcia Martin, H.; Ivanova, N.; Kunin, V.; Warnecke, F.; Barry, K.W.; McHardy, A.C.; Yeates, C.; He, S.; Salamov, A.A.; Szeto, E.; et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006, 24, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Hu, X.; He, Q.; Zhai, J.; Chen, Y.; Xiong, Q.; Vymazal, J. Comprehensive metagenomic analysis reveals the effects of silver nanoparticles on nitrogen transformation in constructed wetlands. Chem. Eng. J. 2019, 358, 1552–1560. [Google Scholar] [CrossRef]
- Xin, L.; Hu, H.Y.; Ke, G.; Sun, Y.X. Effects of different nitrogen and phosphorus concentrations on the growth, nutrient uptake, and lipid accumulation of a freshwater microalga Scenedesmus sp. Bioresour. Technol. 2010, 101, 5494–5500. [Google Scholar] [CrossRef] [PubMed]
- Asian, S.; Kapdan, I.K. Batch kinetics of nitrogen and phosphorus removal from synthetic wastewater by algae. Ecol. Eng. 2006, 28, 64–70. [Google Scholar]
- Bottomley, P.J.; Taylor, A.E.; Myrold, D.D. A consideration of the relative contributions of different microbial subpopulations to the soil N cycle. Front. Microbiol. 2012, 3, 373. [Google Scholar] [CrossRef]
- Dong, S.; Li, M.; Chen, Y. Inherent humic substance promotes microbial denitrification of landfill leachate via shifting bacterial community, improving enzyme activity and up-regulating gene. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Aslam, S.N.; Strauss, J.; Thomas, D.N.; Mock, T.; Underwood, G.J.C. Identifying metabolic pathways for production of extracellular polymeric substances by the diatom Fragilariopsis cylindrus inhabiting sea ice. Isme J. 2018, 12, 1237–1251. [Google Scholar] [CrossRef]
- Peng, T.; Zhou, D.; Liu, Y.; Yu, R.; Qiu, G.; Zeng, W. Effects of pH value on the expression of key iron/sulfur oxidation genes during bioleaching of chalcopyrite on thermophilic condition. Ann. Microbiol. 2019, 69, 627–635. [Google Scholar] [CrossRef]
- Ai, C.; Yan, Z.; Chai, H.; Gu, T.; Wang, J.; Chai, L.; Qiu, G.; Zeng, W. Increased chalcopyrite bioleaching capabilities of extremely thermoacidophilic Metallosphaera sedula inocula by mixotrophic propagation. J. Ind. Microbiol. Biotechnol. 2019, 46, 1113–1127. [Google Scholar] [CrossRef]
- Yu, K.; Zhang, T. Metagenomic and metatranscriptomic analys is of microbial community structure and gene expression of activated sludge. PLoS ONE 2012, 7, e38183. [Google Scholar] [CrossRef]
- Ai, C.; McCarthy, S.; Eckrich, V.; Rudrappa, D.; Qiu, G.; Blum, P. Increased acid resistance of the archaeon, Metallosphaera sedula by adaptive laboratory evolution. J. Ind. Microbiol. Biotechnol. 2016, 43, 1455–1465. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ai, C.; Yan, Z.; Zhou, H.; Hou, S.; Chai, L.; Qiu, G.; Zeng, W. Metagenomic Insights into the Effects of Seasonal Temperature Variation on the Activities of Activated Sludge. Microorganisms 2019, 7, 713. https://doi.org/10.3390/microorganisms7120713
Ai C, Yan Z, Zhou H, Hou S, Chai L, Qiu G, Zeng W. Metagenomic Insights into the Effects of Seasonal Temperature Variation on the Activities of Activated Sludge. Microorganisms. 2019; 7(12):713. https://doi.org/10.3390/microorganisms7120713
Chicago/Turabian StyleAi, Chenbing, Zhang Yan, Han Zhou, Shanshan Hou, Liyuan Chai, Guanzhou Qiu, and Weimin Zeng. 2019. "Metagenomic Insights into the Effects of Seasonal Temperature Variation on the Activities of Activated Sludge" Microorganisms 7, no. 12: 713. https://doi.org/10.3390/microorganisms7120713
APA StyleAi, C., Yan, Z., Zhou, H., Hou, S., Chai, L., Qiu, G., & Zeng, W. (2019). Metagenomic Insights into the Effects of Seasonal Temperature Variation on the Activities of Activated Sludge. Microorganisms, 7(12), 713. https://doi.org/10.3390/microorganisms7120713



