Activation of a Bovine Mammary Epithelial Cell Line by Ruminant-Associated Staphylococcus aureus is Lineage Dependent
Abstract
1. Introduction
2. Materials and Methods
2.1. PS Cell Culture Conditions
2.2. Bacteriological Culture
2.3. PS Cell Responses to S. aureus
2.4. IL-8 ELISA
2.5. Total RNA Extraction and Reverse Transcription
2.6. Statistical Analysis
3. Results
3.1. WTA and CP, but not Protein A, Modulate Activation of PS Cells
3.2. IL-8 Production by PS Cells Stimulated with S. aureus Isolates
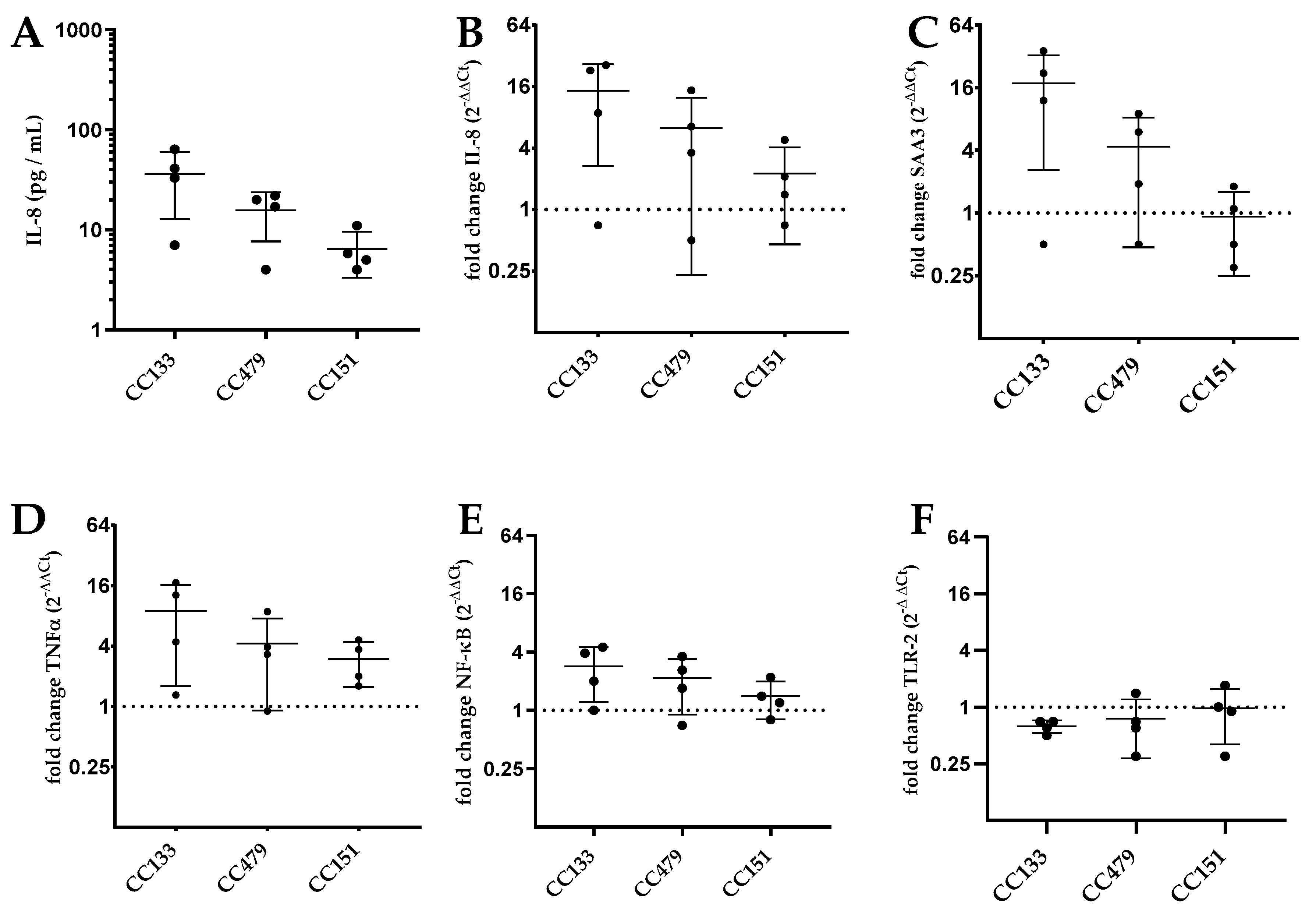
3.3. Transcription of Genes by PS Cells Following Stimulation with S. aureus Isolates
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aghamohammadi, M.; Haine, D.; Kelton, D.F.; Barkema, H.W.; Hogeveen, H.; Keefe, G.P.; Dufour, S. Herd-level mastitis-associated costs on Canadian dairy farms. Front. Vet. Sci. 2018, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Wellnitz, O.; Bruckmaier, R.M. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012, 192, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, D.; Worku, T.; Dad, R.; Rehman, Z.U.; Gong, X.; Zhang, S. Mechanism of pattern recognition receptors (PRRs) and host pathogen interplay in bovine mastitis. Microb. Pathog. 2018, 120, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Crispi, K.; Atalla, H.; Miglior, F.; Mallard, B.A. Bovine mastitis: Frontiers in immunogenetics. Front. Immunol. 2014, 5, 493. [Google Scholar] [CrossRef]
- Molenaar, A.J.; Harris, D.P.; Rajan, G.H.; Pearson, M.L.; Callaghan, M.R.; Sommer, L.; Farr, V.C.; Oden, K.E.; Miles, M.C.; Petrova, R.S.; et al. The acute-phase protein serum amyloid A3 is expressed in the bovine mammary gland and plays a role in host defence. Biomarkers 2009, 14, 26–37. [Google Scholar] [CrossRef]
- Cubeddu, T.; Cacciotto, C.; Pisanu, S.; Tedde, V.; Alberti, A.; Pittau, M.; Dore, S.; Cannas, A.; Uzzau, S.; Rocca, S.; et al. Cathelicidin production and release by mammary epithelial cells during infectious mastitis. Vet. Immunol. Immunopathol. 2017, 189, 66–70. [Google Scholar] [CrossRef]
- Gurao, A.; Kashyap, S.K.; Singh, R. β-defensins: An innate defense for bovine mastitis. Vet. World 2017, 10, 990–998. [Google Scholar] [CrossRef]
- Petzl, W.; Zerbe, H.; Günther, J.; Seyfert, H.-M.; Hussen, J.; Schuberth, H.-J. Pathogen-specific responses in the bovine udder. Models and immunoprophylactic concepts. Res. Vet. Sci. 2018, 116, 55–61. [Google Scholar] [CrossRef]
- Jensen, K.; Günther, J.; Talbot, R.; Petzl, W.; Zerbe, H.; Schuberth, H.-J.; Seyfert, H.-M.; Glass, E.J. Escherichia coli- and Staphylococcus aureus-induced mastitis differentially modulate transcriptional responses in neighbouring uninfected bovine mammary gland quarters. BMC Genomics 2013, 14, 36. [Google Scholar] [CrossRef]
- Fu, Y.; Zhou, E.; Liu, Z.; Li, F.; Liang, D.; Liu, B.; Song, X.; Zhao, F.; Fen, X.; Li, D.; et al. Staphylococcus aureus and Escherichia coli elicit different innate immune responses from bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 2013, 155, 245–252. [Google Scholar] [CrossRef]
- De Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Koymans, K.J.; Goldmann, O.; Karlsson, C.A.Q.; Sital, W.; Thänert, R.; Bisschop, A.; Vrieling, M.; Malmström, J.; van Kessel, K.P.M.; de Haas, C.J.C.; et al. The TLR2 Antagonist Staphylococcal Superantigen-Like Protein 3 Acts as a Virulence Factor to Promote Bacterial Pathogenicity in vivo. J. Innate Immun. 2017, 9, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Hilmi, D.; Parcina, M.; Stollewerk, D.; Ostrop, J.; Josten, M.; Meilaender, A.; Zaehringer, U.; Wichelhaus, T.A.; Bierbaum, G.; Heeg, K.; et al. Heterogeneity of Host TLR2 Stimulation by Staphylocoocus aureus Isolates. PLoS ONE 2014, 9, e96416. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, Y.; Shiratsuchi, A.; Kurokawa, K.; Gong, J.H.; Sekimizu, K.; Lee, B.L.; Nakanishi, Y. Inhibitory Role for D-Alanylation of Wall Teichoic Acid in Activation of Insect Toll Pathway by Peptidoglycan of Staphylococcus aureus. J. Immunol. 2010, 185, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Bar-Gal, G.K.; Blum, S.E.; Hadas, L.; Ehricht, R.; Monecke, S.; Leitner, G. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 2015, 176, 143–154. [Google Scholar] [CrossRef]
- Schlotter, K.; Ehricht, R.; Hotzel, H.; Monecke, S.; Pfeffer, M.; Donat, K. Leukocidin genes lukF-P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet. Res. 2012, 43, 42. [Google Scholar] [CrossRef]
- Budd, K.E.; Mitchell, J.; Keane, O.M.; Budd, K.E.; Mitchell, J.; Keane, O.M. Lineage associated expression of virulence traits in bovine-adapted Staphylococcus aureus. Vet. Microbiol. 2016, 189, 24–31. [Google Scholar] [CrossRef]
- Murphy, M.P.; Niedziela, D.A.; Leonard, F.C.; Keane, O.M. The in vitro host cell immune response to bovine-adapted Staphylococcus aureus varies according to bacterial lineage. Sci. Rep. 2019, 9, 6134. [Google Scholar] [CrossRef]
- Merz, A.; Stephan, R.; Johler, S. Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front. Microbiol. 2016, 7, 319. [Google Scholar] [CrossRef]
- Hoekstra, J.; Rutten, V.P.M.G.; van den Hout, M.; Spaninks, M.P.; Benedictus, L.; Koop, G. Differences between Staphylococcus aureus lineages isolated from ovine and caprine mastitis but not between isolates from clinical or subclinical mastitis. J. Dairy Sci. 2019, 102, 5430–5437. [Google Scholar] [CrossRef]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Guttman, D.S.; Fitzgerald, J.R. Population genomics of bacterial host adaptation. Nat. Rev. Genet. 2018, 19, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, J.; Rutten, V.; Sommeling, L.; van Werven, T.; Spaninks, M.; Duim, B.; Benedictus, L.; Koop, G. High production of LukMF’ in Staphylococcus aureus field strains is associated with clinical bovine mastitis. Toxins 2018, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Roussel, P.; Cunha, P.; Porcherie, A.; Petzl, W.; Gilbert, F.B.; Riollet, C.; Zerbe, H.; Rainard, P.; Germon, P. Investigating the contribution of IL-17A and IL-17F to the host response during Escherichia coli mastitis. Vet. Res. 2015, 46, 54–56. [Google Scholar] [CrossRef]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.E.; Peschel, A.; et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef]
- Watts, A.; Ke, D.; Wang, Q.; Pillay, A.; Nicholson-Weller, A.; Lee, J.C. Staphylococcus aureus strains that express serotype 5 or serotype 8 capsular polysaccharides differ in virulence. Infect. Immun. 2005, 73, 3502–3511. [Google Scholar] [CrossRef]
- Higgins, J.; Loughman, A.; van Kessel, K.P.M.; van Strijp, J.A.G.; Foster, T.J. Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol. Lett. 2006, 258, 290–296. [Google Scholar] [CrossRef]
- Deplanche, M.; Alekseeva, L.; Semenovskaya, K.; Fu, C.-L.; Dessauge, F.; Finot, L.; Petzl, W.; Zerbe, H.; Le Loir, Y.; Rainard, P.; et al. Staphylococcus aureus phenol-soluble modulins impair interleukin expression in bovine mammary epithelial cells. Infect. Immun. 2016, 84, 1682–1692. [Google Scholar] [CrossRef]
- Zbinden, C.; Stephan, R.; Johler, S.; Borel, N.; Bunter, J.; Bruckmaier, R.M.; Wellnitz, O. The inflammatory response of primary bovine mammary epithelial cells to Staphylococcus aureus strains is linked to the bacterial phenotype. PLoS ONE 2014, 9, e87374. [Google Scholar] [CrossRef][Green Version]
- Günther, J.; Petzl, W.; Zerbe, H.; Schuberth, H.-J.; Seyfert, H.-M. TLR ligands, but not modulators of histone modifiers, can induce the complex immune response pattern of endotoxin tolerance in mammary epithelial cells. Innate Immun. 2017, 23, 155–164. [Google Scholar] [CrossRef]
- Bulgari, O.; Dong, X.; Roca, A.L.; Caroli, A.M.; Loor, J.J. Innate immune responses induced by lipopolysaccharide and lipoteichoic acid in primary goat mammary epithelial cells. J. Anim. Sci. Biotechnol. 2017, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The Role of TLR2 in infection and immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef] [PubMed]
- Majcherczyk, P.A.; Rubli, E.; Heumann, D.; Glauser, M.P.; Moreillon, P. Teichoic acids are not required for Streptococcus pneumoniae and Staphylococcus aureus cell walls to trigger the release of tumor necrosis factor by peripheral blood monocytes. Infect. Immun. 2003, 71, 3707–3713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soell, M.; Diab, M.; Haan-Archipoff, G.; Beretz, A.; Herbelin, C.; Poutrel, B.; Klein, J.P. Capsular polysaccharide types 5 and 8 of Staphylococcus aureus bind specifically to human epithelial (KB) cells, endothelial cells, and monocytes and induce release of cytokines. Infect. Immun. 1995, 63, 1380–1386. [Google Scholar]
- Hilmi, D.; Parcina, M.; Bode, K.; Ostrop, J.; Schuett, S.; Heeg, K.; Ziebuhr, W.; Sommerburg, O.; Bekeredjian-Ding, I. Functional variation reflects intra-strain diversity of Staphylococcus aureus small colony variants in the host–pathogen interaction. Int. J. Med. Microbiol. 2013, 303, 61–69. [Google Scholar] [CrossRef]
- Fournier, B.; Philpott, D.J. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 2005, 18, 521–540. [Google Scholar] [CrossRef]
- Keinhörster, D.; George, S.E.; Weidenmaier, C.; Wolz, C. Function and regulation of Staphylococcus aureus wall teichoic acids and capsular polysaccharides. Int. J. Med. Microbiol. 2019, 309, 151333. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Joo, H.-S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins--critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Monecke, S.; Gavier-Widén, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus isolates in European wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- Moser, A.; Stephan, R.; Corti, S.; Johler, S. Comparison of genomic and antimicrobial resistance features of latex agglutination test-positive and latex agglutination test-negative Staphylococcus aureus isolates causing bovine mastitis. J. Dairy Sci. 2013, 96, 329–334. [Google Scholar] [CrossRef]
- Deb, R.; Kumar, A.; Chakraborty, S.; Verma, A.K.; Tiwari, R.; Dhama, K.; Singh, U.; Kumar, S. Trends in diagnosis and control of bovine mastitis: A review. Pakistan J. Biol. Sci. PJBS 2013, 16, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Raja, A.; Vignesh, A.R.; Mary, B.A.; Tirumurugaan, K.G.; Raj, G.D.; Kataria, R.; Mishra, B.P.; Kumanan, K. Sequence analysis of Toll-like receptor genes 1–10 of goat (Capra hircus). Vet. Immunol. Immunopathol. 2011, 140, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lacasse, P. Mammary tissue damage during bovine mastitis: Causes and control1. J. Anim. Sci. 2008, 86, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Foucras, G.; Fitzgerald, J.R.; Watts, J.L.; Koop, G.; Middleton, J.R. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg. Dis. 2017, 65, 149–165. [Google Scholar] [CrossRef] [PubMed]

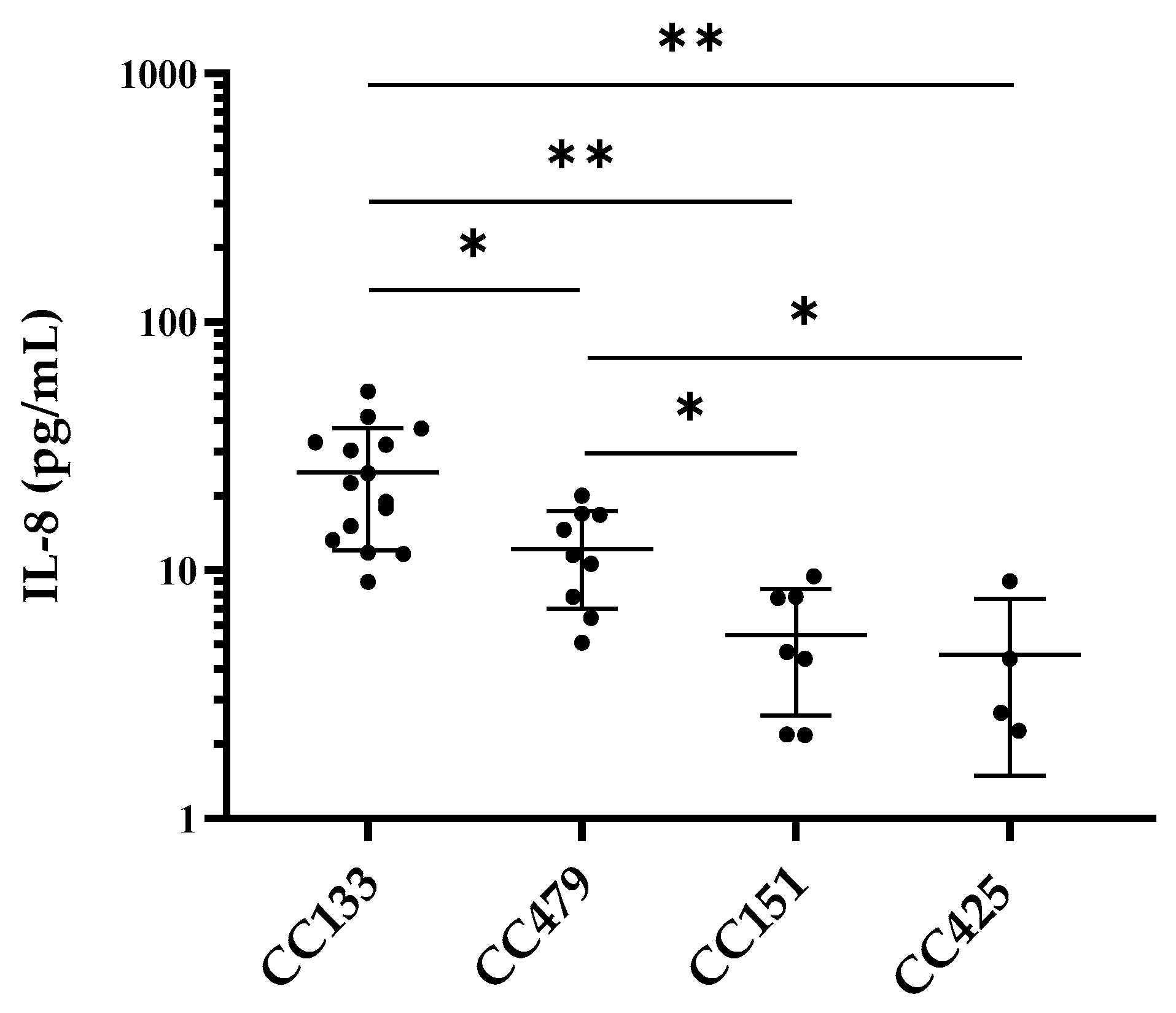
| Strain Name | Clonal Complex | Relevant Characteristics | Reference |
|---|---|---|---|
| Reynolds cap5 | 25 | wt Reynolds strain, expressing cap5 | [26] |
| Reynolds cap8 | 25 | Substitution of cap5 region with the cap8 region | [26] |
| Reynolds Δcap5 | 25 | Deletion of cap5, capsular polysaccharide negative strain | [26] |
| Newman wt | 8 | wt Newman strain | [27] |
| Newman Δspa | 8 | Deletion of spa gene | [27] |
| RN4220 wt | 8 | wt RN4220 strain | [25] |
| RN 4220 ΔtarO | 8 | Deletion of tarO, essential for wall teichoic acid (WTA) synthesis | [25] |
| RN 4220 ΔtarS/ΔtarM | 8 | Deletion of tarS and tarM, responsible for WTA glycosylation | [25] |
| Gene | Primer Sequence | Product Size (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| IL-8 | Fwd: ATGACTTCCAAGCTGGCTGTTG Rev: TTGATAAATTTGGGGTGGAAAG | 149 | 60 | [29] |
| TNF-α | Fwd: CCACGTTGTAGCCGACATC Rev: CCCTGAAGAGGACCTGTGAG | 155 | 60 | [29] |
| SAA3 | Fwd: CTTTCCACGGGCATCATTTT Rev: CTTCGGGCAGCGTCATAGTT | 188 | 60 | [30] |
| NF-κB | Fwd: CTGGAAGCACGAATGACAGA Rev: GCTGTAAACATGAGCCGTACC | 179 | 60 | [31] |
| TLR-2 | Fwd: CATTCCCTGGCAAGTGGATTATC Rev: GGAATGGCCTTCTTGTCAATGG | 201 | 62 | [29] |
| Ubiquitin | Fwd: AGATCCAGGATAAGGAAGGCAT Rev: GCTCCACCTCCAGGGTGAT | 198 | 62 | [29] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoekstra, J.; Rutten, V.P.M.G.; Lam, T.J.G.M.; Van Kessel, K.P.M.; Spaninks, M.P.; Stegeman, J.A.; Benedictus, L.; Koop, G. Activation of a Bovine Mammary Epithelial Cell Line by Ruminant-Associated Staphylococcus aureus is Lineage Dependent. Microorganisms 2019, 7, 688. https://doi.org/10.3390/microorganisms7120688
Hoekstra J, Rutten VPMG, Lam TJGM, Van Kessel KPM, Spaninks MP, Stegeman JA, Benedictus L, Koop G. Activation of a Bovine Mammary Epithelial Cell Line by Ruminant-Associated Staphylococcus aureus is Lineage Dependent. Microorganisms. 2019; 7(12):688. https://doi.org/10.3390/microorganisms7120688
Chicago/Turabian StyleHoekstra, Jurriaan, Victor P. M. G. Rutten, Theo J. G. M. Lam, Kok P. M. Van Kessel, Mirlin P. Spaninks, J. Arjan Stegeman, Lindert Benedictus, and Gerrit Koop. 2019. "Activation of a Bovine Mammary Epithelial Cell Line by Ruminant-Associated Staphylococcus aureus is Lineage Dependent" Microorganisms 7, no. 12: 688. https://doi.org/10.3390/microorganisms7120688
APA StyleHoekstra, J., Rutten, V. P. M. G., Lam, T. J. G. M., Van Kessel, K. P. M., Spaninks, M. P., Stegeman, J. A., Benedictus, L., & Koop, G. (2019). Activation of a Bovine Mammary Epithelial Cell Line by Ruminant-Associated Staphylococcus aureus is Lineage Dependent. Microorganisms, 7(12), 688. https://doi.org/10.3390/microorganisms7120688





