Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria
Abstract
1. Introduction
2. Cells of the Intestine and Their Interactions with Microbes
2.1. Enterocytes
2.2. Goblet Cells
2.3. Paneth Cells
2.4. Enteroendocrine Cells
2.5. M Cells
2.6. Tuft Cells
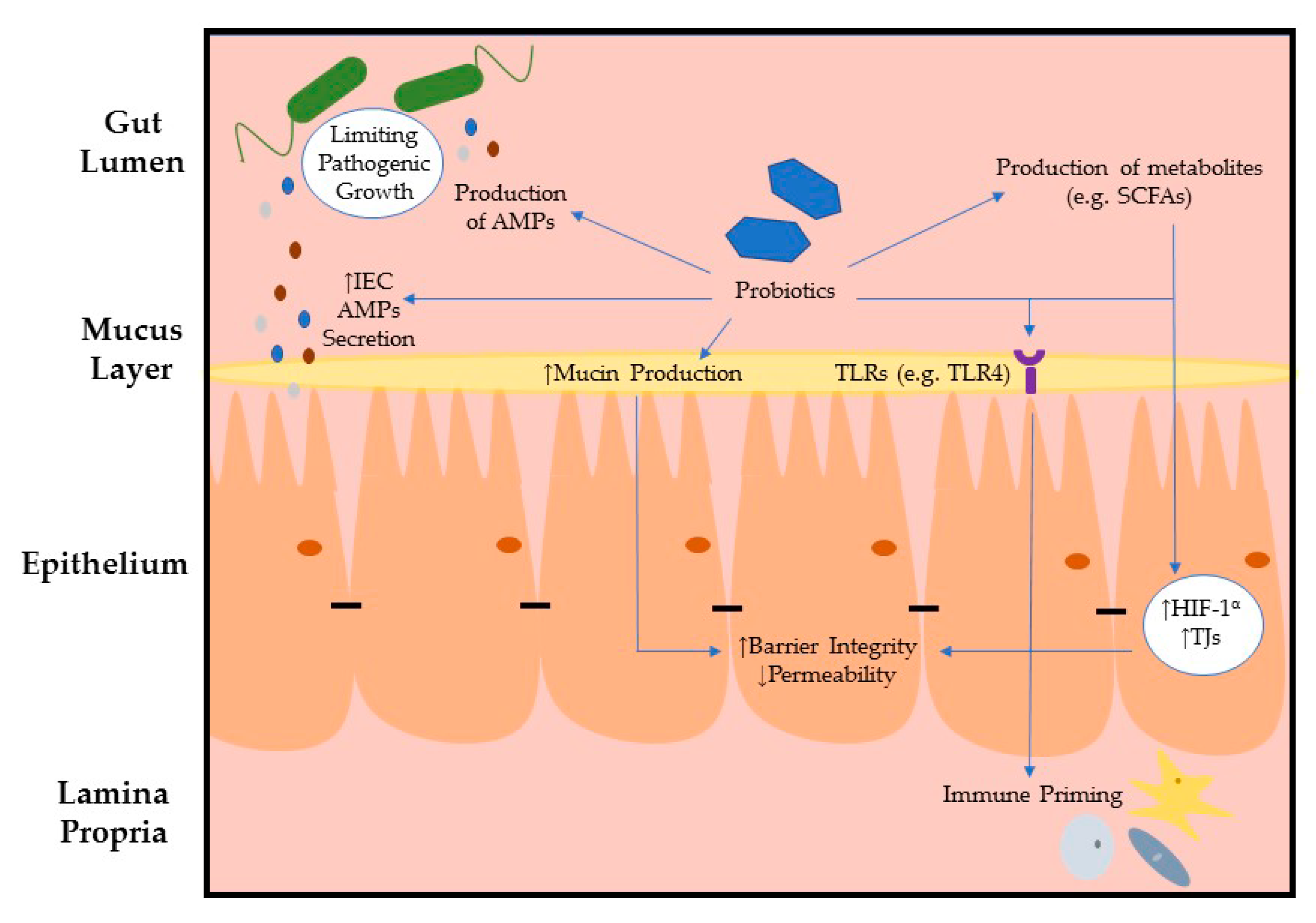
3. Intestinal Epithelial Barrier Integrity Influenced by Microbes
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moog, F. The lining of the small intestine. Sci. Am. 1981, 245, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Brookes, S.J.; Hennig, G.W. Anatomy and physiology of the enteric nervous system. Gut 2000, 47 (Suppl. S4), iv15–iv19. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; de Vos, W.M.; Belzer, C. Glycobiome: Bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 25–38. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1. [Google Scholar] [CrossRef]
- Nava, G.M.; Friedrichsen, H.J.; Stappenbeck, T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011, 5, 627–638. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694.e3. [Google Scholar] [CrossRef]
- Mooseker, M.S. Organization, chemistry, and assembly of the cytoskeletal apparatus of the intestinal brush border. Annu. Rev. Cell Biol. 1985, 1, 209–241. [Google Scholar] [CrossRef]
- Maury, J.; Nicoletti, C.; Guzzo-Chambraud, L.; Maroux, S. The filamentous brush border glycocalyx, a mucin-like marker of enterocyte hyper-polarization. Eur J. Biochem. 1995, 228, 323–331. [Google Scholar] [CrossRef]
- Frey, A.; Giannasca, K.T.; Weltzin, R.; Giannasca, P.J.; Reggio, H.; Lencer, W.I.; Neutra, M.R. Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: Implications for microbial attachment and oral vaccine targeting. J. Exp. Med. 1996, 184, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Hershberg, R.M.; Mayer, L.F. Antigen processing and presentation by intestinal epithelial cells - polarity and complexity. Immunol. Today 2000, 21, 123–128. [Google Scholar] [CrossRef]
- Hershberg, R.M.; Cho, D.H.; Youakim, A.; Bradley, M.B.; Lee, J.S.; Framson, P.E.; Nepom, G.T. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J. Clin. Investig. 1998, 102, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Atuma, C.; Strugala, V.; Allen, A.; Holm, L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G922–G929. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Szentkuti, L.; Riedesel, H.; Enss, M.L.; Gaertner, K.; Von Engelhardt, W. Pre-epithelial mucus layer in the colon of conventional and germ-free rats. Histochem. J. 1990, 22, 491–497. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef]
- Smirnova, M.G.; Guo, L.; Birchall, J.P.; Pearson, J.P. LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell Immunol. 2003, 221, 42–49. [Google Scholar] [CrossRef]
- Wlodarska, M.; Thaiss, C.A.; Nowarski, R.; Henao-Mejia, J.; Zhang, J.P.; Brown, E.M.; Frankel, G.; Levy, M.; Katz, M.N.; Philbrick, W.M.; et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014, 156, 1045–1059. [Google Scholar] [CrossRef]
- Shimotoyodome, A.; Meguro, S.; Hase, T.; Tokimitsu, I.; Sakata, T. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2000, 125, 525–531. [Google Scholar] [CrossRef]
- Bergström, A.; Kristensen, M.B.; Bahl, M.I.; Metzdorff, S.B.; Fink, L.N.; Frøkiaer, H.; Licht, T.R. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res. Notes 2012, 5, 402. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Velcich, A.; Yang, W.; Heyer, J.; Fragale, A.; Nicholas, C.; Viani, S.; Kucherlapati, R.; Lipkin, M.; Yang, K.; Augenlicht, L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002, 295, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Cunliffe, R.N.; Mahida, Y.R. Expression and regulation of antimicrobial peptides in the gastrointestinal tract. J. Leukoc. Biol. 2004, 75, 49–58. [Google Scholar] [CrossRef]
- Yokoi, Y.; Nakamura, K.; Yoneda, T.; Kikuchi, M.; Sugimoto, R.; Shimizu, Y.; Ayabe, T. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci. Rep. 2019, 9, 2710. [Google Scholar] [CrossRef]
- Gong, J.; Xu, J.; Zhu, W.; Gao, X.; Li, N.; Li, J. Epithelial-specific blockade of MyD88-dependent pathway causes spontaneous small intestinal inflammation. Clin. Immunol. 2010, 136, 245–256. [Google Scholar] [CrossRef]
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118. [Google Scholar] [CrossRef]
- Farin, H.F.; Karthaus, W.R.; Kujala, P.; Rakhshandehroo, M.; Schwank, G.; Vries, R.G.; Kalkhoven, E.; Nieuwenhuis, E.E.; Clevers, H. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J. Exp. Med. 2014, 211, 1393–1405. [Google Scholar] [CrossRef]
- Burger, E.; Araujo, A.; López-Yglesias, A.; Rajala, M.W.; Geng, L.; Levine, B.; Hooper, L.V.; Burstein, E.; Yarovinsky, F. Loss of Paneth Cell Autophagy Causes Acute Susceptibility to Toxoplasma gondii-Mediated Inflammation. Cell Host Microbe. 2018, 23, 177–190.e4. [Google Scholar] [CrossRef]
- Schoenborn, A.A.; von Furstenberg, R.J.; Valsaraj, S.; Hussain, F.S.; Stein, M.; Shanahan, M.T.; Henning, S.J.; Gulati, A.S. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes 2019, 10, 45–58. [Google Scholar] [CrossRef]
- Cazorla, S.I.; Maldonado-Galdeano, C.; Weill, R.; De Paula, J.; Perdigón, G.D.V. Oral Administration of Probiotics Increases Paneth Cells and Intestinal Antimicrobial Activity. Front. Microbiol. 2018, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Eriguchi, Y.; Takashima, S.; Oka, H.; Shimoji, S.; Nakamura, K.; Uryu, H.; Shimoda, S.; Iwasaki, H.; Shimono, N.; Ayabe, T.; et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood 2012, 120, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Eriguchi, Y.; Nakamura, K.; Hashimoto, D.; Shimoda, S.; Shimono, N.; Akashi, K.; Ayabe, T.; Teshima, T. Decreased secretion of Paneth cell α-defensins in graft-versus-host disease. Transpl. Infect. Dis. 2015, 17, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Sternini, C.; Anselmi, L.; Rozengurt, E. Enteroendocrine cells: A site of ‘taste’ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008, 15, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.L.; Egerod, K.L.; Engelstoft, M.S.; Dmytriyeva, O.; Theodorsson, E.; Patel, B.A.; Schwartz, T.W. Enterochromaffin 5-HT cells—A major target for GLP-1 and gut microbial metabolites. Mol. Metab. 2018, 11, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, L.J.; Lenaerts, K.; Kiers, D.; Pais de Barros, J.P.; Le Guern, N.; Plesnik, J.; Thomas, C.; Bourgeois, T.; Dejong, C.H.C.; Kox, M.; et al. Enteroendocrine L Cells Sense LPS after Gut Barrier Injury to Enhance GLP-1 Secretion. Cell Rep. 2017, 21, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Kidd, M.; Gustafsson, B.I.; Drozdov, I.; Modlin, I.M. IL1beta- and LPS-induced serotonin secretion is increased in EC cells derived from Crohn’s disease. Neurogastroenterol. Motil. 2009, 21, 439–450. [Google Scholar] [CrossRef]
- Larraufie, P.; Doré, J.; Lapaque, N.; Blottière, H.M. TLR ligands and butyrate increase Pyy expression through two distinct but inter-regulated pathways. Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef]
- Grøndahl, M.F.; Keating, D.J.; Vilsbøll, T.; Knop, F.K. Current Therapies That Modify Glucagon Secretion: What Is the Therapeutic Effect of Such Modifications? Curr. Diab. Rep. 2017, 17, 128. [Google Scholar] [CrossRef]
- Loh, K.; Herzog, H.; Shi, Y.C. Regulation of energy homeostasis by the NPY system. Trends Endocrinol. Metab. 2015, 26, 125–135. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed]
- Tankou, S.K.; Regev, K.; Healy, B.C.; Cox, L.M.; Tjon, E.; Kivisakk, P.; Vanande, I.P.; Cook, S.; Gandhi, R.; Glanz, B.; et al. Investigation of probiotics in multiple sclerosis. Mult. Scler. 2018, 24, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Nøhr, M.K.; Pedersen, M.H.; Gille, A.; Egerod, K.L.; Engelstoft, M.S.; Husted, A.S.; Sichlau, R.M.; Grunddal, K.V.; Poulsen, S.S.; Han, S.; et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs. FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 2013, 154, 3552–3564. [Google Scholar] [CrossRef]
- Larraufie, P.; Martin-Gallausiaux, C.; Lapaque, N.; Dore, J.; Gribble, F.M.; Reimann, F.; Blottiere, H.M. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci. Rep. 2018, 8, 74. [Google Scholar] [CrossRef]
- Greiner, T.U.; Bäckhed, F. Microbial regulation of GLP-1 and L-cell biology. Mol. Metab. 2016, 5, 753–758. [Google Scholar] [CrossRef]
- Neutra, M.R.; Frey, A.; Kraehenbuhl, J.P. Epithelial M cells: Gateways for mucosal infection and immunization. Cell 1996, 86, 345–348. [Google Scholar] [CrossRef]
- Hase, K.; Kawano, K.; Nochi, T.; Pontes, G.S.; Fukuda, S.; Ebisawa, M.; Kadokura, K.; Tobe, T.; Fujimura, Y.; Kawano, S.; et al. Uptake through glycoprotein 2 of FimH+ bacteria by M cells initiates mucosal immune response. Nature 2009, 462, 226–230. [Google Scholar] [CrossRef]
- Nakato, G.; Fukuda, S.; Hase, K.; Goitsuka, R.; Cooper, M.D.; Ohno, H. New approach for m-cell-specific molecules screening by comprehensive transcriptome analysis. DNA Res. 2009, 16, 227–235. [Google Scholar] [CrossRef]
- Kaufmann, S.H. Heat shock proteins and the immune response. Immunol Today 1990, 11, 129–136. [Google Scholar] [CrossRef]
- Rios, D.; Wood, M.B.; Li, J.; Chassaing, B.; Gewirtz, A.T.; Williams, I.R. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal. Immunol. 2016, 9, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Sato, A. Tuft cells. Anat. Sci. Int. 2007, 82, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Voss, U.; Ekblad, E. A novel serotonin-containing tuft cell subpopulation in mouse intestine. Cell Tissue Res. 2019, 376, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; O’Leary, C.E.; von Moltke, J.; Liang, H.E.; Ang, Q.Y.; Turnbaugh, P.J.; Radhakrishnan, S.; Pellizzon, M.; Ma, A.; Locksley, R.M. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 2018, 174, 271–284.e14. [Google Scholar] [CrossRef]
- Nadjsombati, M.S.; McGinty, J.W.; Lyons-Cohen, M.R.; Jaffe, J.B.; DiPeso, L.; Schneider, C.; Miller, C.N.; Pollack, J.L.; Nagana Gowda, G.A.; Fontana, M.F.; et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 2018, 49, 33–41.e7. [Google Scholar] [CrossRef]
- Farquhar, M.G.; Palade, G.E. Junctional complexes in various epithelia. J. Cell Biol 1963, 17, 375–412. [Google Scholar] [CrossRef]
- Viswanathan, V.K.; Koutsouris, A.; Lukic, S.; Pilkinton, M.; Simonovic, I.; Simonovic, M.; Hecht, G. Comparative analysis of EspF from enteropathogenic and enterohemorrhagic Escherichia coli in alteration of epithelial barrier function. Infect. Immun. 2004, 72, 3218–3227. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Kuwae, A.; Abe, A. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 alter epithelial paracellular permeability. Infect. Immun. 2005, 73, 6283–6289. [Google Scholar] [CrossRef]
- Dean, P.; Kenny, B. Intestinal barrier dysfunction by enteropathogenic Escherichia coli is mediated by two effector molecules and a bacterial surface protein. Mol. Microbiol. 2004, 54, 665–675. [Google Scholar] [CrossRef]
- Peralta-Ramírez, J.; Hernandez, J.M.; Manning-Cela, R.; Luna-Muñoz, J.; Garcia-Tovar, C.; Nougayréde, J.P.; Oswald, E.; Navarro-Garcia, F. EspF Interacts with nucleation-promoting factors to recruit junctional proteins into pedestals for pedestal maturation and disruption of paracellular permeability. Infect. Immun. 2008, 76, 3854–3868. [Google Scholar] [CrossRef]
- Hanajima-Ozawa, M.; Matsuzawa, T.; Fukui, A.; Kamitani, S.; Ohnishi, H.; Abe, A.; Horiguchi, Y.; Miyake, M. Enteropathogenic Escherichia coli, Shigella flexneri, and Listeria monocytogenes recruit a junctional protein, zonula occludens-1, to actin tails and pedestals. Infect. Immun. 2007, 75, 565–573. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boyle, E.C.; Brown, N.F.; Finlay, B.B. Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell Microbiol. 2006, 8, 1946–1957. [Google Scholar] [CrossRef]
- Köhler, H.; Sakaguchi, T.; Hurley, B.P.; Kase, B.A.; Kase, B.J.; Reinecker, H.C.; McCormick, B.A. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G178–G187. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, T.; Köhler, H.; Gu, X.; McCormick, B.A.; Reinecker, H.C. Shigella flexneri regulates tight junction-associated proteins in human intestinal epithelial cells. Cell Microbiol. 2002, 4, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Rajabian, T.; Gavicherla, B.; Heisig, M.; Müller-Altrock, S.; Goebel, W.; Gray-Owen, S.D.; Ireton, K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 2009, 11, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Clyne, M.; Tegtmeyer, N. Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun. Signal. 2011, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Araya, M.; Morelli, L.; Reid, G.; Sanders, M.E.; Stanton, C. Guidelines for the Evaluation of Probiotics in Food. In Proceedings of the Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food, London, ON, Canada, 30 April–1 May 2002. [Google Scholar]
- Madsen, K.; Cornish, A.; Soper, P.; McKaigney, C.; Jijon, H.; Yachimec, C.; Doyle, J.; Jewell, L.; De Simone, C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 2001, 121, 580–591. [Google Scholar] [CrossRef]
- Gupta, P.; Andrew, H.; Kirschner, B.S.; Guandalini, S. Is lactobacillus GG helpful in children with Crohn’s disease? Results of a preliminary, open-label study. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 453–457. [Google Scholar] [CrossRef]
- Galdeano, C.M.; Perdigón, G. Role of viability of probiotic strains in their persistence in the gut and in mucosal immune stimulation. J. Appl. Microbiol. 2004, 97, 673–681. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.; Young, L.H.; Caplan, M.J. AMP-activated protein kinase regulates the assembly of epithelial tight junctions. Proc. Natl. Acad. Sci. USA 2006, 103, 17272–17277. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Colgan, S.P.; Taylor, C.T. Hypoxia: An alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, R.C.; Samborska, B.; Faubert, B.; Ma, E.H.; Gravel, S.P.; Andrzejewski, S.; Raissi, T.C.; Pause, A.; St-Pierre, J.; Jones, R.G. AMPK Maintains Cellular Metabolic Homeostasis through Regulation of Mitochondrial Reactive Oxygen Species. Cell Rep. 2017, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Li, Z.R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef]
- Yu, H.; Qin, L.; Hu, H.; Wang, Z. Alteration of the Gut Microbiota and Its Effect on AMPK/NADPH Oxidase Signaling Pathway in 2K1C Rats. BioMed Res. Int. 2019, 2019, 8250619. [Google Scholar] [CrossRef]
- Olivier, S.; Leclerc, J.; Grenier, A.; Foretz, M.; Tamburini, J.; Viollet, B. AMPK Activation Promotes Tight Junction Assembly in Intestinal Epithelial Caco-2 Cells. Int J. Mol. Sci. 2019, 20, 5171. [Google Scholar] [CrossRef]
- Blackwood, B.P.; Yuan, C.Y.; Wood, D.R.; Nicolas, J.D.; Grothaus, J.S.; Hunter, C.J. Probiotic Lactobacillus Species Strengthen Intestinal Barrier Function and Tight Junction Integrity in Experimental Necrotizing Enterocolitis. J. Probiotics Health 2017, 5. [Google Scholar] [CrossRef]
- Yi, H.; Wang, L.; Xiong, Y.; Wang, Z.; Qiu, Y.; Wen, X.; Jiang, Z.; Yang, X.; Ma, X. LR1 Improved Expression of Genes of Tight Junction Proteins via the MLCK Pathway in IPEC-1 Cells during Infection with Enterotoxigenic. Mediators Inflamm. 2018, 2018, 6434910. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, L.; Dou, X.; Wang, C.; Zhang, W.; Gao, K.; Liu, J.; Wang, H. ZJ617 maintains intestinal integrity via regulating tight junction, autophagy and apoptosis in mice challenged with lipopolysaccharide. Oncotarget 2017, 8, 77489–77499. [Google Scholar] [CrossRef]
- Rhayat, L.; Maresca, M.; Nicoletti, C.; Perrier, J.; Brinch, K.S.; Christian, S.; Devillard, E.; Eckhardt, E. Effect of Bacillus subtilis Strains on Intestinal Barrier Function and Inflammatory Response. Front. Immunol. 2019, 10, 564. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, X.; Zhu, Y.; Ma, J.; Ma, H.; Zhang, H. Probiotic Mixture Protects Dextran Sulfate Sodium-Induced Colitis by Altering Tight Junction Protein Expressions and Increasing Tregs. Mediators Inflamm. 2018, 2018, 9416391. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Mechanism of Invasion | Effects on TJs | Reference |
|---|---|---|---|
| Escherichia coli AEEC | Type-three secretion system (TSS3) of effector proteins; EspF [57], EspG [58], and MAP [59] |
| |
| [60] | ||
| [60,61] | ||
| Salmonella typhimurium | Salmonella pathogenicity island (SPI1) injected into host cell using T3SS SPI1 effectors; SopB, SopE, SopE2, and SipA [62] |
| [63] |
| Shigella flexneria | T3SS injection of effector proteins; SepA [63] |
| [64] |
| Listeria monocytogenes | T3SS injection of effector protein; InlC [65] |
| |
| Helicobacter pylori | T4SS delivery of effector protein; CagA [66] |
|
| Probiotic Bacteria | Intestinal Disease | Effects on TJs | Reference |
|---|---|---|---|
| L. plantarum (ATCC 10241) & L. rhamnosus (ATCC 53103) | Necrotizing enterocolitis (NEC) |
| [78] |
| L. reuteri (LR1) | Enteric pathogen infection using ETEC K88 |
| [79] |
| L. rhamnosus (GG) & L. reuteri (ZJ617) | LPS-induced barrier dysfunction |
| [80] |
| Bacillus subtilis (29784) | Pro-inflammatory cytokine induced intestinal inflammation |
| [81] |
| Bifidobacterium, L. acidophilus & Enterococcus | Dextran sodium sulfate (DSS)- induced colitis |
| [82] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Callaghan, A.A.; Corr, S.C. Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria. Microorganisms 2019, 7, 663. https://doi.org/10.3390/microorganisms7120663
O’Callaghan AA, Corr SC. Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria. Microorganisms. 2019; 7(12):663. https://doi.org/10.3390/microorganisms7120663
Chicago/Turabian StyleO’Callaghan, Amy A., and Sinéad C. Corr. 2019. "Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria" Microorganisms 7, no. 12: 663. https://doi.org/10.3390/microorganisms7120663
APA StyleO’Callaghan, A. A., & Corr, S. C. (2019). Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria. Microorganisms, 7(12), 663. https://doi.org/10.3390/microorganisms7120663




