Paenibacillus lutrae sp. nov., A Chitinolytic Species Isolated from A River Otter in Castril Natural Park, Granada, Spain
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Phenotypic Characterization
2.3. Chemotaxonomic Analysis
2.4. Phylogenetic 16S rRNA Gene Analysis
2.5. G+C Content
2.6. Genome Sequencing and Assembly
2.7. In Silico ANI and DDH
2.8. Analysis of Multilocus Sequences and Core Orthologous Genes
3. Results and Discussion
3.1. Phenotypic Characterization
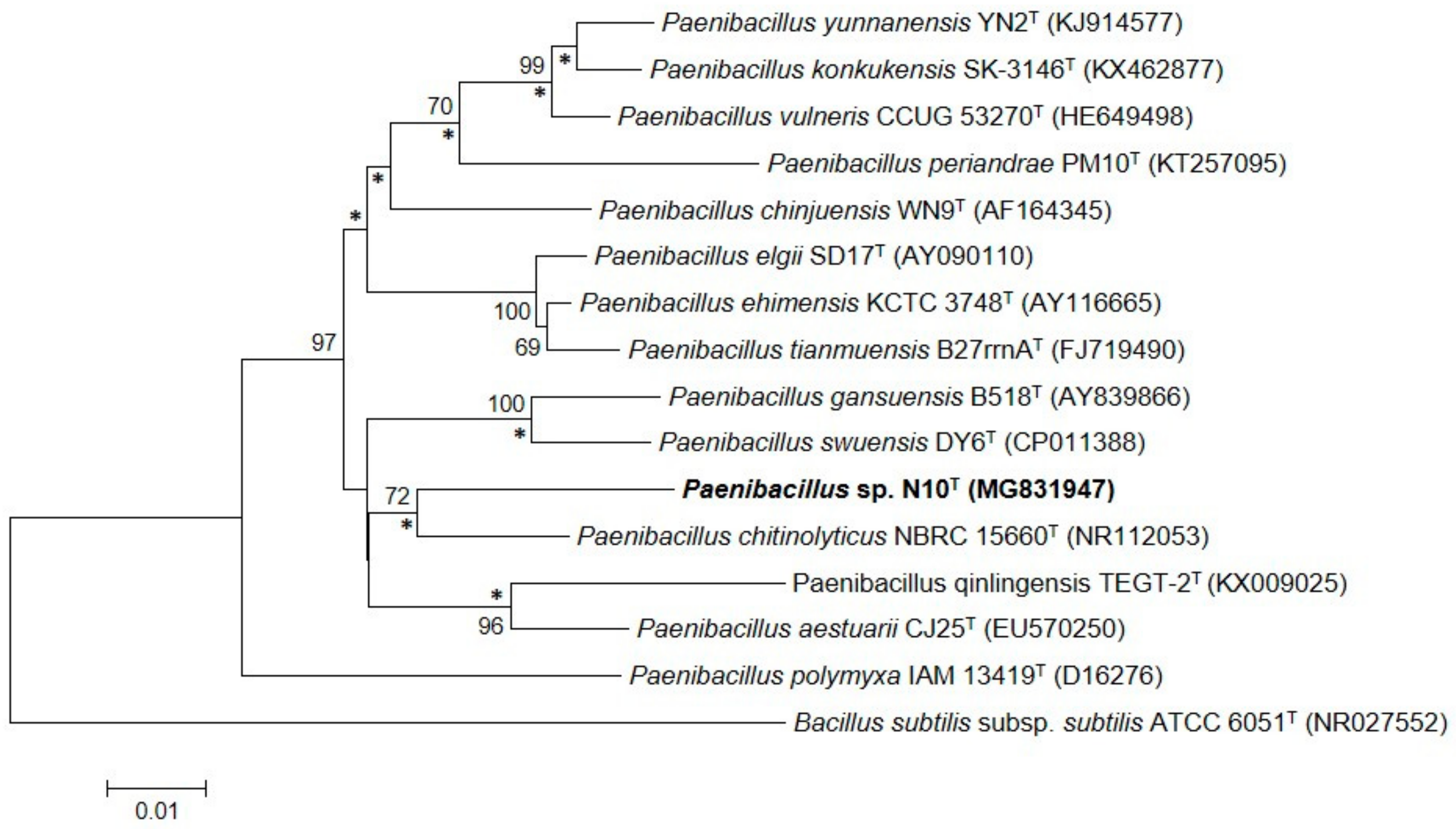
3.2. Phylogenetic Analysis Based on the 16S rRNA Gene Sequence
3.3. Analysis of Cellular Fatty Acids, Polar Lipids, Isoprenoid Quinones and Peptidoglycan Structure
3.4. G+C Content
3.5. Whole-Genome Sequencing and Assembly
3.6. ANI and dDDH Calculations
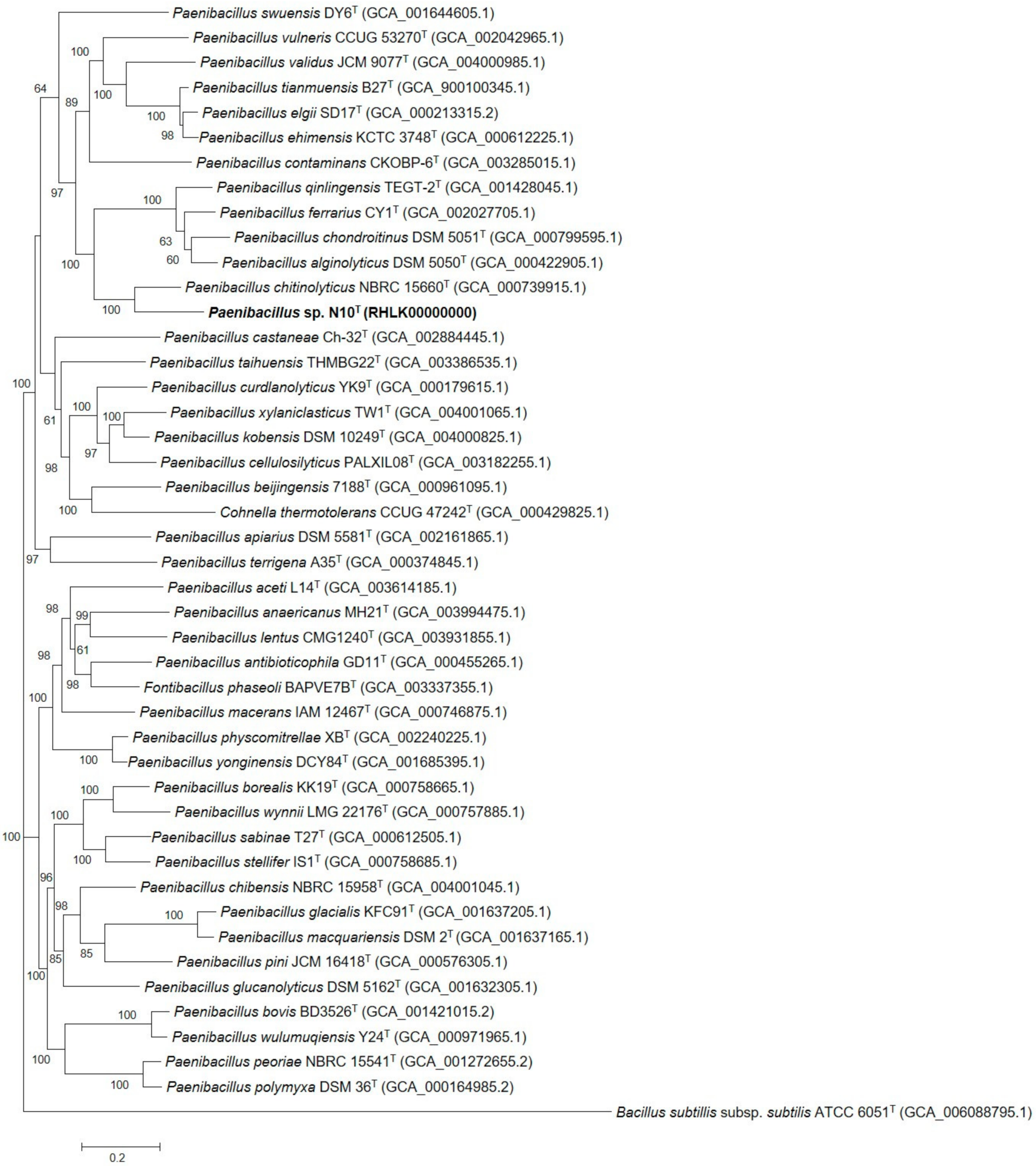
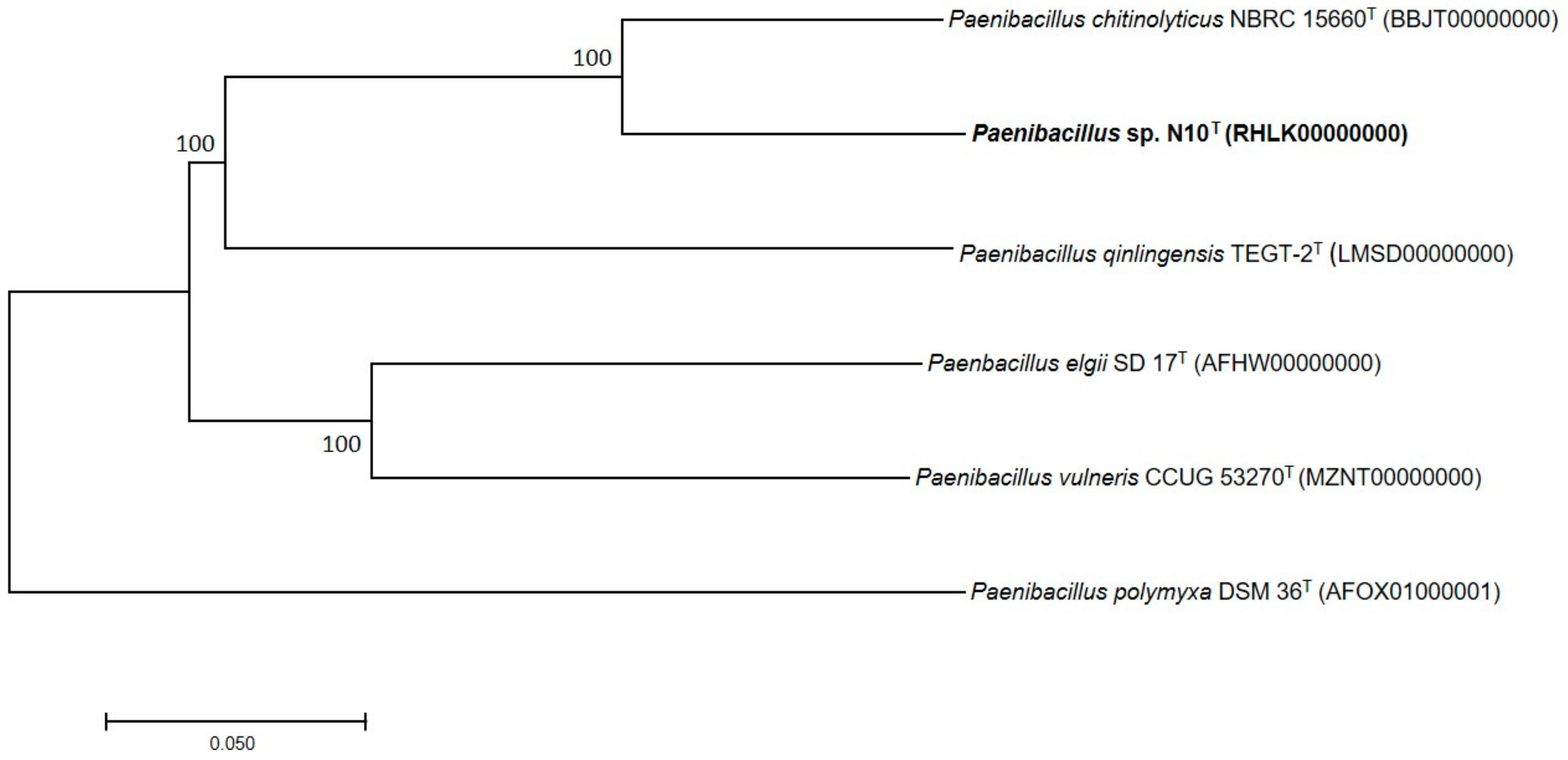
3.7. Phylogenetic Analysis of Multilocus Sequences and Core Orthologous Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Description of Paenibacillus lutrae sp. nov
References
- Flach, J.; Pilet, P.-E.; Jollès, P. What’s new in chitinase research? Experientia 1992, 48, 701–716. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.J.; Muthukrishnan, S. Insect chitinases: Molecular biology and potential use as biopesticides. Insect Biochem. Mol. Biol. 1997, 27, 887–900. [Google Scholar] [CrossRef]
- Gooday, G.W. Diversity of roles for chitinases in nature. In Chitin and Chitosan—The Versatile Environmentally Friendly Modern Materials; Zakaria, M.B., Muda, W.M.W., Abdullah, M.P., Eds.; Penerbit Universiti Kebangsaan Malasya: Bangi, Malasia, 1995; Volume 3, pp. 191–202. [Google Scholar]
- Cottrell, M.T.; Kirchman, D.L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 2000, 66, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Bertilsson, S. Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol. Oceanogr. 2011, 56, 1179–1188. [Google Scholar] [CrossRef]
- Reyes-Ramírez, A.; Escudero-Abarca, B.; Aguilar-Uscanga, G.; Hayward-Jones, P.; Barboza-Corona, J.E. Antifungal activity of Bacillus thuringiensis chitinase and its potential for the biocontrol of phytopathogenic fungi in soybean seeds. J. Food Sci. 2004, 69, M131–M134. [Google Scholar] [CrossRef]
- Huang, C.J.; Wang, T.K.; Chung, S.C.; Chen, C.Y. Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus 28-9. J. Biochem. Mol. Biol. 2005, 38, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Aktuganov, G.E.; Melent’ev, A.I.; Galimzyanova, N.F.; Shirokov, A.V. The study of mycolytic properties of aerobic spore-forming bacteria producing extracellular chitinases. Microbiology 2008, 77, 700–709. [Google Scholar] [CrossRef]
- Singh, A.K.; Ghodke, I.; Chhatpar, H.S. Pesticide tolerance of Paenibacillus sp. D1 and its chitinase. J. Environ. Manag. 2009, 91, 358–362. [Google Scholar] [CrossRef]
- Terahara, T.; Ikeda, S.; Noritake, C.; Minamisawa, K.; Ando, K.; Tsuneda, S.; Harayama, S. Molecular diversity of bacterial chitinases in arable soils and the effects of environmental factors on the chitinolytic bacterial community. Soil Biol. Biochem. 2009, 41, 473–480. [Google Scholar] [CrossRef]
- Brzezinska, M.S.; Donderski, W. Chitinolytic bacteria in two lakes of different trophic status. Pol. J. Ecol. 2006, 54, 295–301. [Google Scholar]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Someya, N.; Ikeda, S.; Morohoshi, T.; Tsujimoto, M.N.; Yoshida, T.; Sawada, H.; Ikeda, T.; Tsuchiya, K. Diversity of culturable chitinolytic bacteria from rhizospheres of agronomic plants in Japan. Microbes Environ. 2009, 26, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, Y.; Chet, I.; Cohn, E. Use of chitin for controlling plant plant-parasitic nematodes. Plant Soil 1987, 98, 337–345. [Google Scholar] [CrossRef]
- Vionis, A.P.; Niemeyer, F.; Karagouni, A.D.; Schrempf, H. Production and processing of a 59-kilodalton exochitinase during growth of Streptomyces lividans carrying pCHIO12 in soil microcosms amended with crab or fungal chitin. Appl. Environ. Microbiol. 1996, 62, 1774–1780. [Google Scholar]
- Williamson, N.; Brian, P.; Wellington, E. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek 2000, 78, 315–321. [Google Scholar] [CrossRef]
- Gomes, R.C.; Semêdo, L.T.; Soares, R.M.; Alviano, C.S.; Linhares, L.F.; Coelho, R.R. Chitinolytic activity of actinomycetes from a cerrado soil and their potential in biocontrol. Lett. Appl. Microbiol. 2000, 30, 146–150. [Google Scholar] [CrossRef]
- Gomes, R.; Semêdo, L.; Soares, R.; Linhares, L.; Ulhoa, C.; Alviano, C.; Coelho, R. Purification of a thermostable endochitinase from Streptomyces RC1071 isolated from a cerrado soil and its antagonism against phytopathogenic fungi. J. Appl. Microbiol. 2001, 90, 653–661. [Google Scholar] [CrossRef]
- Yasir, M.; Aslam, Z.; Kim, S.W.; Lee, S.W.; Jeon, C.O.; Chung, Y.R. Bacterial community composition and chitinase gene diversity of vermicompost with antifungal activity. Bioresour. Technol. 2009, 100, 4396–4403. [Google Scholar] [CrossRef]
- Cottrell, M.T.; Moore, J.A.; Kirchman, D.L. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 1999, 65, 2553–2557. [Google Scholar]
- Beier, S.; Jones, C.M.; Mohit, V.; Hallin, S.; Bertilsson, S. Global phylogeography of chitinase genes in aquatic metagenomes. Appl. Environ. Microbiol. 2011, 77, 1101–1106. [Google Scholar] [CrossRef]
- Eski, A.; Demir, I.; Gullu, M.; Demirbag, Z. Biodiversity and pathogenicity of bacteria associated with the gut microbiota of beet armyworm, Spodoptera exigua Hübner (Lepidoptera: Noctuidae). Microb. Pathog. 2018, 121, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Pasari, N.; Gupta, M.; Eqbal, D.; Yazdani, S.S. Genome analysis of Paenibacillus polymyxa A18 gives insights into the features associated with its adaptation to the termite gut environment. Sci. Rep. 2019, 9, 6091. [Google Scholar] [CrossRef] [PubMed]
- Beygmoradi, A.; Homaei, A. Marine chitinolytic enzymes, a biotechnological treasure hidden in the ocean? Appl. Microbiol. Biotechnol. 2018, 102, 9937–9948. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wei, Y.; Shi, M.; Li, Z.; Zhang, S.-H. An archaeal chitinase with a secondary capacity for catalyzing cellulose and its biotechnological applications in shell and straw degradation. Front. Microbiol. 2019, 10, 1253. [Google Scholar] [CrossRef]
- Chen, Q.; Peng, D. Nematode chitin and application. Adv. Exp. Med. Biol. 2019, 1142, 209–219. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Xu, N.; Zhang, A.; Chen, K.; Xin, F.; Zhang, W.; Ma, J.; Fang, Y.; Jiang, M.; et al. The broad-specificity chitinases: Their origin, characterization, and potential application. Appl. Microbiol. Biotechnol. 2019, 103, 3289–3295. [Google Scholar] [CrossRef]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef]
- Djukic, M.; Becker, D.; Poehlein, A.; Voget, S.; Daniel, R. Genome sequence of Paenibacillus alvei DSM 29, a secondary invader during European foulbrood outbreaks. J. Bacteriol. 2012, 194, 6365. [Google Scholar] [CrossRef]
- Kuroshima, K.-I.; Sakane, T.; Takata, R.; Yokota, A. Bacillus ehimensis sp. nov. and Bacillus chitinolyticus sp. nov., new chitinolytic members of the genus Bacillus. Int. J. Syst. Evol. Microbiol. 1996, 46, 76–80. [Google Scholar] [CrossRef]
- Lee, J.S.; Pyun, Y.R.; Bae, K.S. Transfer of Bacillus ehimensis and Bacillus chitinolyticus to the genus Paenibacillus with emended descriptions of Paenibacillus ehimensis comb. nov. and Paenibacillus chitinolyticus comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 929–933. [Google Scholar] [CrossRef]
- Sun, H.; Mao, X. Discovery and characterization of a novel chitosanase from Paenibacillus dendritiformis by phylogeny-based enzymatic product specificity prediction. J. Agric. Food Chem. 2018, 66, 4645–4651. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Park, S.K.; Hur, J.Y.; Kim, Y.C. Purification and characterization of a major extracellular chitinase from a biocontrol bacterium, Paenibacillus elgii HOA73. Plant. Pathol. J. 2017, 33, 318–328. [Google Scholar] [CrossRef]
- García-González, E.; Poppinga, L.; Fünfhaus, A.; Hertlein, G.; Hedtke, K.; Jakubowska, A.; Genersch, E. Paenibacillus larvae chitin-degrading protein PlCBP49 is a key virulence factor in American Foulbrood of honey bees. PLoS Pathog. 2014, 10, e1004284. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.R.; Kim, C.H.; Hwang, I.; Chun, J. Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Microbiol. 2000, 50, 1495–1500. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B. Reclamation of marine chitinous materials for chitosanase production via microbial conversion by Paenibacillus macerans. Mar. Drugs 2018, 16, 429. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B. Production of a thermostable chitosanase from shrimp heads via Paenibacillus mucilaginosus TKU032 conversion and its application in the preparation of bioactive chitosan oligosaccharides. Mar. Drugs 2019, 17, 217. [Google Scholar] [CrossRef] [PubMed]
- Passera, A.; Marcolungo, L.; Casati, P. Hybrid genome assembly and annotation of Paenibacillus pasadenensis strain R16 reveals insights on endophytic life style and antifungal activity. PLoS ONE 2018, 13, e0189993. [Google Scholar] [CrossRef]
- Cody, R.M. Distribution of chitinase and chitobiase in Bacillus. Curr. Microbiol. 1989, 19, 201–205. [Google Scholar] [CrossRef]
- Yahiaoui, M.; Laribi-Habchi, H.; Bouacem, K.; Asmani, K.L.; Mechri, S.; Harir, M.; Bendif, H.; Aissani-El Fertas, R.; Jaouadi, B. Purification and biochemical characterization of a new organic solvent-tolerant chitinase from Paenibacillus timonensis strain LK-DZ15 isolated from the Djurdjura Mountains in Kabylia, Algeria. Carbohydr. Res. 2019, 483, 107747. [Google Scholar] [CrossRef]
- Liao, W.; Liu, P.; Liao, W.; Miao, L. Complete genome of the chitin-degrading bacterium, Paenibacillus xylanilyticus W4. Genome Biol. Evol. 2019, 11, 3252–3255. [Google Scholar] [CrossRef]
- An, C.; Okamoto, Y.; Xu, S.; Eo, K.Y.; Kimura, J.; Yamamoto, N. Comparison of fecal microbiota of three captive carnivore species inhabiting Korea. J. Vet. Med. Sci. 2017, 79, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; Berge, O.; Bishop, A.H.; Busse, H.-J.; De Vos, P.; Fritze, D.; Heyndrickx, M.; Kämpfer, P.; Rabinovitch, L.; Salkinoja-Salonen, M.S.; et al. Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 1956, 178, 703. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.Y.; Lin, H.P.; Li, L.; Brown, R.; Goodfellow, M.; Deng, Z.; Hong, K. Verrucosispora wenchangensis sp. nov., isolated from mangrove soil. Antonie Van Leeuwenhoek 2012, 102, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.H. Hydrogen sulphide production by bacteria. J. Gen. Microbiol. 1953, 8, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Skerman, V.B.D. A Guide to the Identification of the Genera of Bacteria; The Williams & Wilkins, Co.: Baltimore, MD, USA, 1961; p. 251. [Google Scholar]
- Kovacs, N. Eine vereinfachte methode zum nachweis der indolbildung durch bakterien. Immunforsch 1928, 55, 311–315. [Google Scholar]
- Barritt, M.M. The intensification of the Voges-Proskauer reaction by the addition of α-naphthol. J. Pathol. Bacteriol. 1936, 42, 441–454. [Google Scholar] [CrossRef]
- Uttley, A.; Collins, C. Theory and practice of bacterial identification. In Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd ed.; Barrow, G.I., Feltham, R.K.A., Eds.; Cambridge University Press: Cambridge, UK, 1993; pp. 46–49. [Google Scholar]
- Matthews, S.; Suhaimi, M. Selection of suitable growth medium for free-living diazotrophs isolated from compost. J. Trop. Agric. Food Sci. 2010, 38, 211–219. [Google Scholar]
- Poonguzhali, S.; Madhaiyan, M.; Sa, T. Cultivation-dependent characterization of rhizobacterial communities from field grown Chinese cabbage Brassica campestris ssp. pekinensis and screening of traits for potential plant growth promotion. Plant Soil 2006, 286, 167–180. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fert. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Sikorski, J.; Smibert, R.A.; Krieg, N.R. Phenotypic characterization and the principles of comparative systematics. In Methods for General and Molecular Microbiology, 3rd ed.; Reddy, C., Beveridge, T., Breznak, J., Marzluf, G., Schmidt, T., Snyder, L.A., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar]
- Tindall, B.J. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 1990, 13, 128–130. [Google Scholar] [CrossRef]
- Tindall, B.J. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 1990, 66, 199–202. [Google Scholar] [CrossRef]
- Schumann, P. Peptidoglycan structure. Method. Microbiol. 2011, 38, 101–129. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Owen, R.; Pitcher, D. Current methods for estimating DNA base composition and levels of DNA-DNA hybridization. In Chemicals Methods in Bacterial Systematics; Goodfellow, M., Minnikin, D.E., Eds.; Academic Press: London, UK, 1985; Volume 20, pp. 67–93. [Google Scholar]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2015, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Kainer, D.; Lanfear, R. The effects of partitioning on phylogenetic inference. Mol. Biol. Evol. 2015, 32, 1611–1627. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Yao, R.; Wang, R.; Wang, D.; Su, J.; Zheng, S.; Wang, G. Paenibacillus selenitireducens sp. nov., a selenite-reducing bacterium isolated from a selenium mineral soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 805–811. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Pérez-Cataluña, A.; Salas-Massó, N.; Diéguez, A.L.; Balboa, S.; Lema, A.; Romalde, J.L.; Figueras, M.J. Revisiting the taxonomy of the genus Arcobacter: Getting order from the chaos. Front. Microbiol. 2018, 9, 2077. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
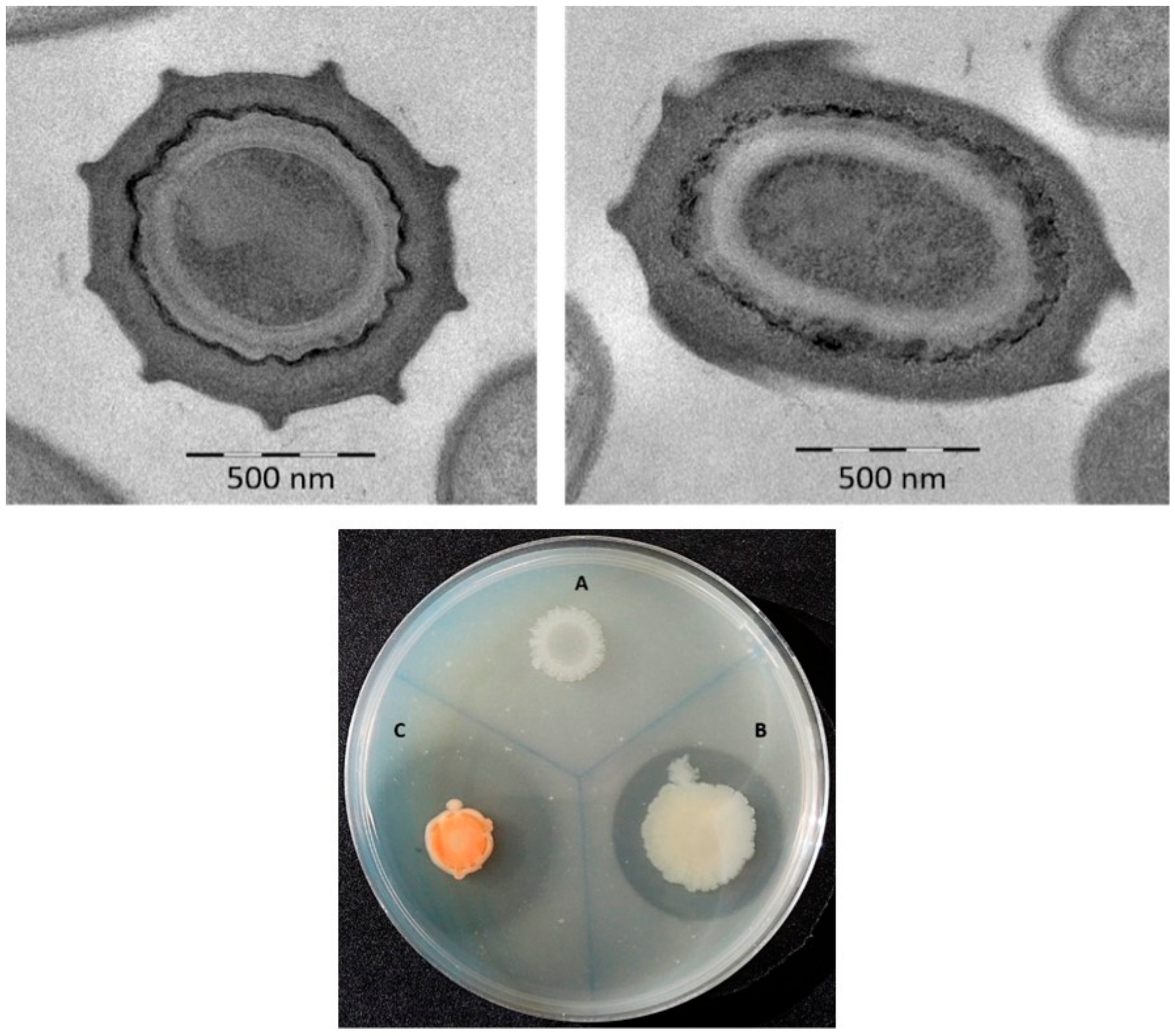
| Characteristic | 1 | 2 | 3 |
|---|---|---|---|
| Colony Pigmentation | Pink | Cream | Cream-White |
| NaCl range (%) (w/v) | 0–3 | 0–4 | 0–5 |
| NaCl optimum (%) (w/v) | 1 | 0 | 0 |
| pH range | 7–10 | 6–9 | 6–9 |
| pH optimum | 8 | 7 | 7 |
| Oxidase | + | v | - |
| Methyl red | - | - | + |
| Voges-Proskauer | - | - | + |
| Aerobic nitrate reduction | - | + | + |
| Hydrolysis of: | |||
| Gelatine | - | v | - |
| Chitin | + | + | - |
| Starch | - | - | + |
| Casein | - | v | + |
| Tween 80 | - | - | + |
| ACC deaminase | + | + | - |
| Assimilation of: | |||
| l-arabinose | - | - | + |
| d-mannose | - | + | + |
| d-mannitol | - | - | + |
| N-acetyl-glucosamine | - | + | - |
| Potassium gluconate | - | + | + |
| Enzymes: | |||
| Alkaline phosphatase | - | + | - |
| Acid phosphatase | - | + | + |
| α-galactosidase | - | - | + |
| β-galactosidase | - | + | + |
| β-glucosidase | - | + | + |
| Acids from carbohydrates: | |||
| Glycerol | - | + | + |
| Galactose | - | + | + |
| Manose | - | + | + |
| Amygdaline | - | + | + |
| Arbutin | - | + | + |
| Salicin | - | + | + |
| Cellobiose | - | + | + |
| Lactose | - | + | + |
| Melibiose | - | + | + |
| Sucrose | - | + | + |
| Trehalose | - | + | + |
| Gentiobiose | - | + | + |
| d-turanose | - | + | + |
| Antimicrobial susceptibility: | |||
| Ampicilin | S | I | S |
| Cloramphenicol | S | I | S |
| Kanamycin | R | R | S |
| Nalidixic acid | S | R | S |
| Rifampicin | S | S | R |
| Trimethoprim/sulfamethoxazole | R | S | S |
| DNA G + C content (Tm) (mol %) | 45–48 | 51–53 | 43–46 |
| DNA G + C content (in silico) (mol %) | 49.8 | 52.4 | 45.5 |
| Cellular Fatty Acids | 1 | 2 | 3 |
|---|---|---|---|
| Straight-chain fatty acids: | |||
| C14:0 | 2.50 | 2.49 | - |
| C15:0 | 1.17 | - | - |
| C16:0 | 11.69 | 25.88 | 8.39 |
| Branched-chain fatty acids: | |||
| iso-c14:0 | 1.86 | - | 3.16 |
| iso-c15:0 | 8.46 | 6.26 | 3.44 |
| anteiso-c15:0 | 56.95 | 49.20 | 47.79 |
| iso-c16:0 | 5.98 | 2.34 | 18.80 |
| iso-c17:0 | 2.63 | 4.73 | 3.38 |
| anteiso-c17:0 | 5.82 | 6.76 | 12.24 |
| Unsaturated fatty acids: | |||
| C16:1 ω11c | 1.59 | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, M.; Reina, J.C.; Béjar, V.; Llamas, I. Paenibacillus lutrae sp. nov., A Chitinolytic Species Isolated from A River Otter in Castril Natural Park, Granada, Spain. Microorganisms 2019, 7, 637. https://doi.org/10.3390/microorganisms7120637
Rodríguez M, Reina JC, Béjar V, Llamas I. Paenibacillus lutrae sp. nov., A Chitinolytic Species Isolated from A River Otter in Castril Natural Park, Granada, Spain. Microorganisms. 2019; 7(12):637. https://doi.org/10.3390/microorganisms7120637
Chicago/Turabian StyleRodríguez, Miguel, José Carlos Reina, Victoria Béjar, and Inmaculada Llamas. 2019. "Paenibacillus lutrae sp. nov., A Chitinolytic Species Isolated from A River Otter in Castril Natural Park, Granada, Spain" Microorganisms 7, no. 12: 637. https://doi.org/10.3390/microorganisms7120637
APA StyleRodríguez, M., Reina, J. C., Béjar, V., & Llamas, I. (2019). Paenibacillus lutrae sp. nov., A Chitinolytic Species Isolated from A River Otter in Castril Natural Park, Granada, Spain. Microorganisms, 7(12), 637. https://doi.org/10.3390/microorganisms7120637






