Comparative Genomics Reveals Metabolic Specificity of Endozoicomonas Isolated from a Marine Sponge and the Genomic Repertoire for Host-Bacteria Symbioses
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Endozoicomonas
2.2. Whole-Genome Sequencing (WGS) and Genome Analyses
2.3. Clusters of Orthologous Groups of Proteins
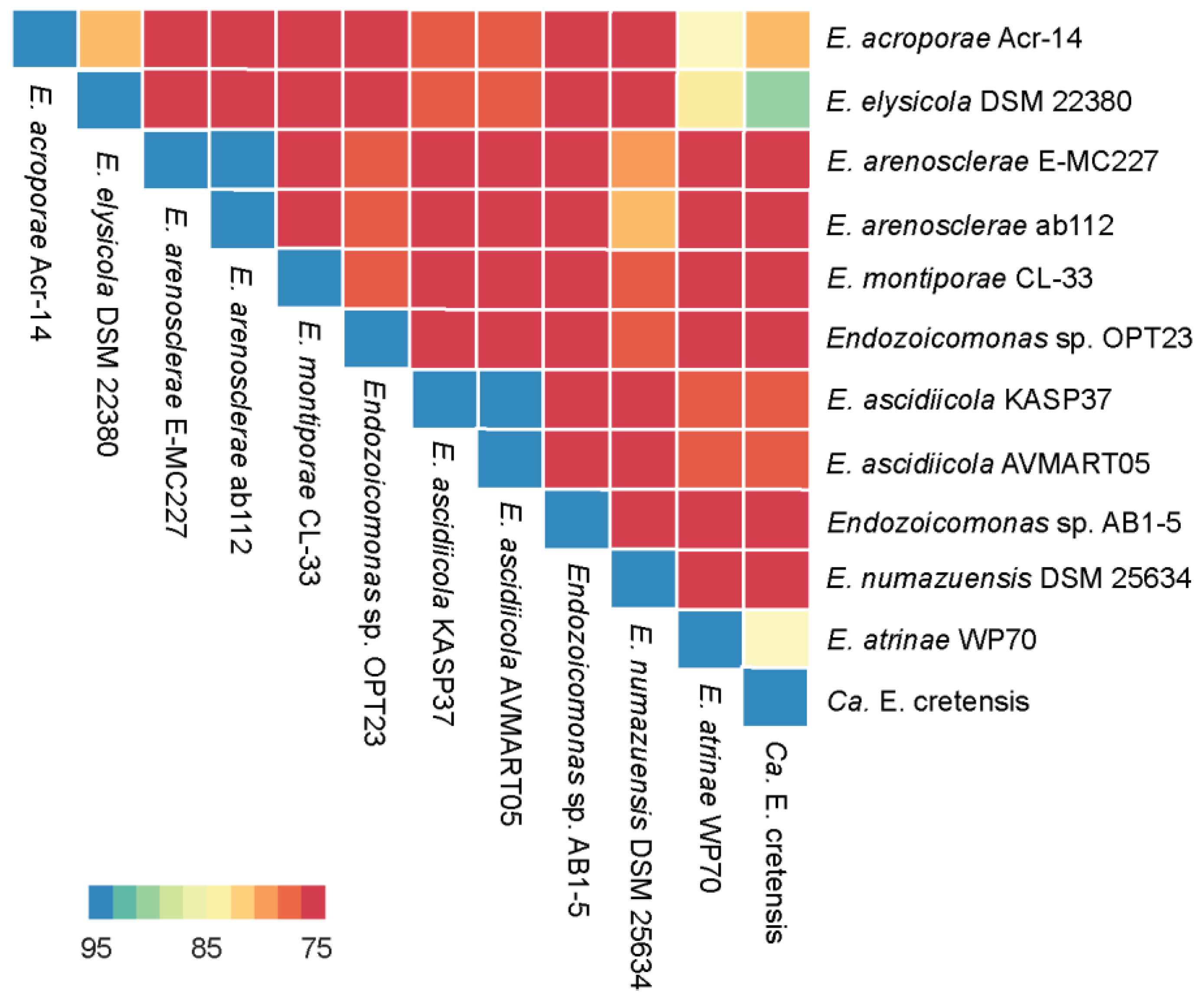
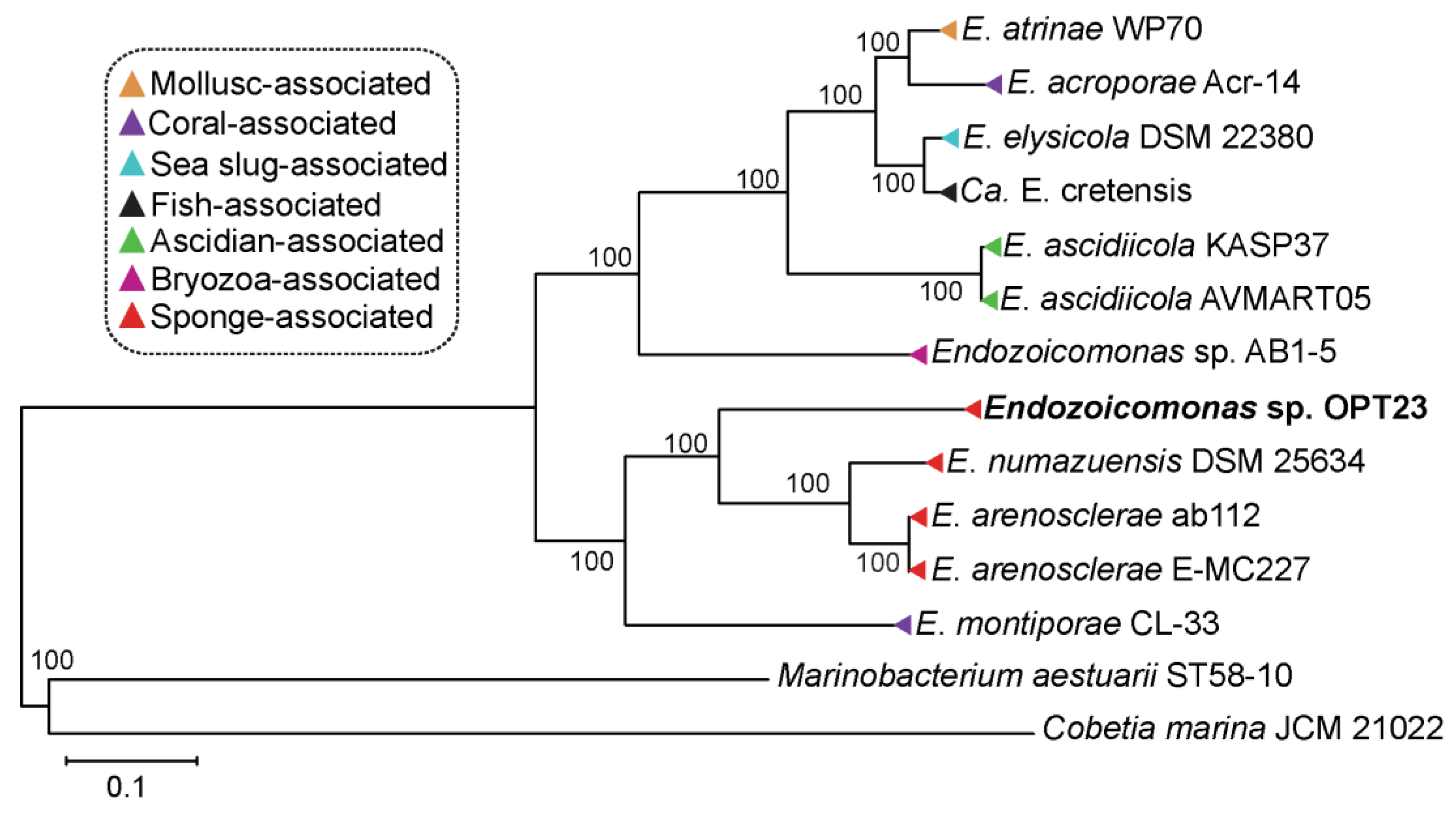
2.4. Phylogenetic Analyses and Average Nucleotide Identity
2.5. Homolog Clustering and Calculation of Genome-Specific Genes
2.6. Prediction of Symbioses Factors and Secretion Systems
2.7. Data Deposition
3. Results and Discussion
3.1. Genome Summary and Phylogeny of the Endozoicomonas sp. OPT23
3.2. Core- and Pan-Genome of Endozoicomonas
3.3. Estimation of Endozoicomonas sp. OPT23-Specific Genes
3.4. Metabolic Specificity of Endozoicomonas sp. OPT23
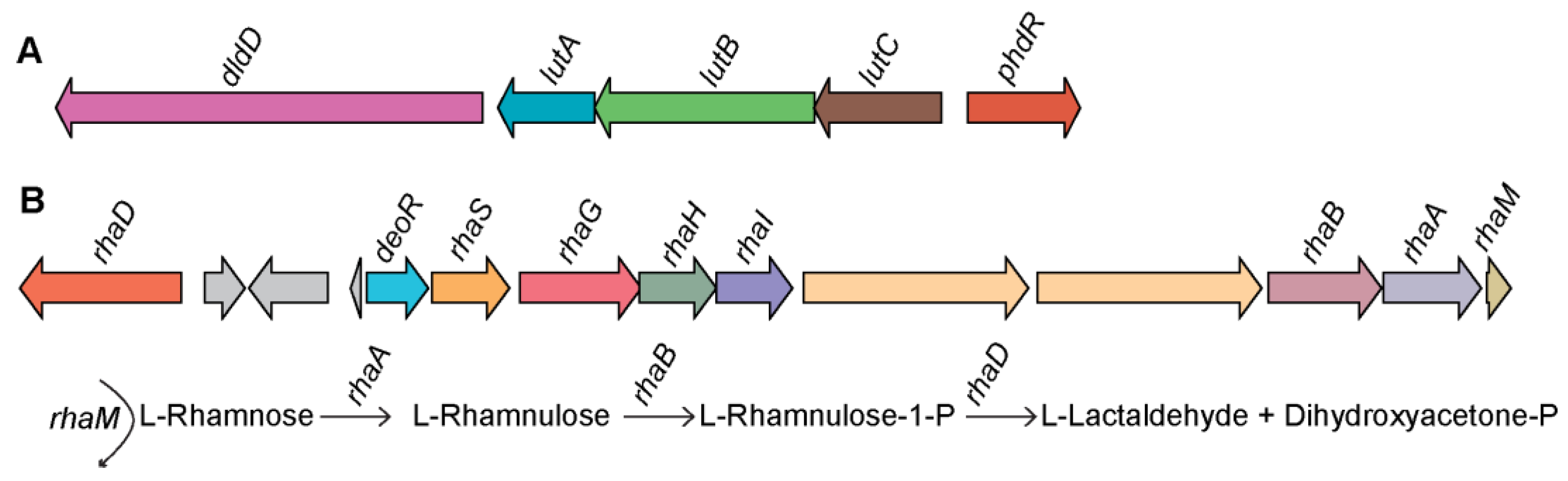
3.4.1. Lactate Utilization Pathway
3.4.2. L-Rhamnose Utilization Pathway
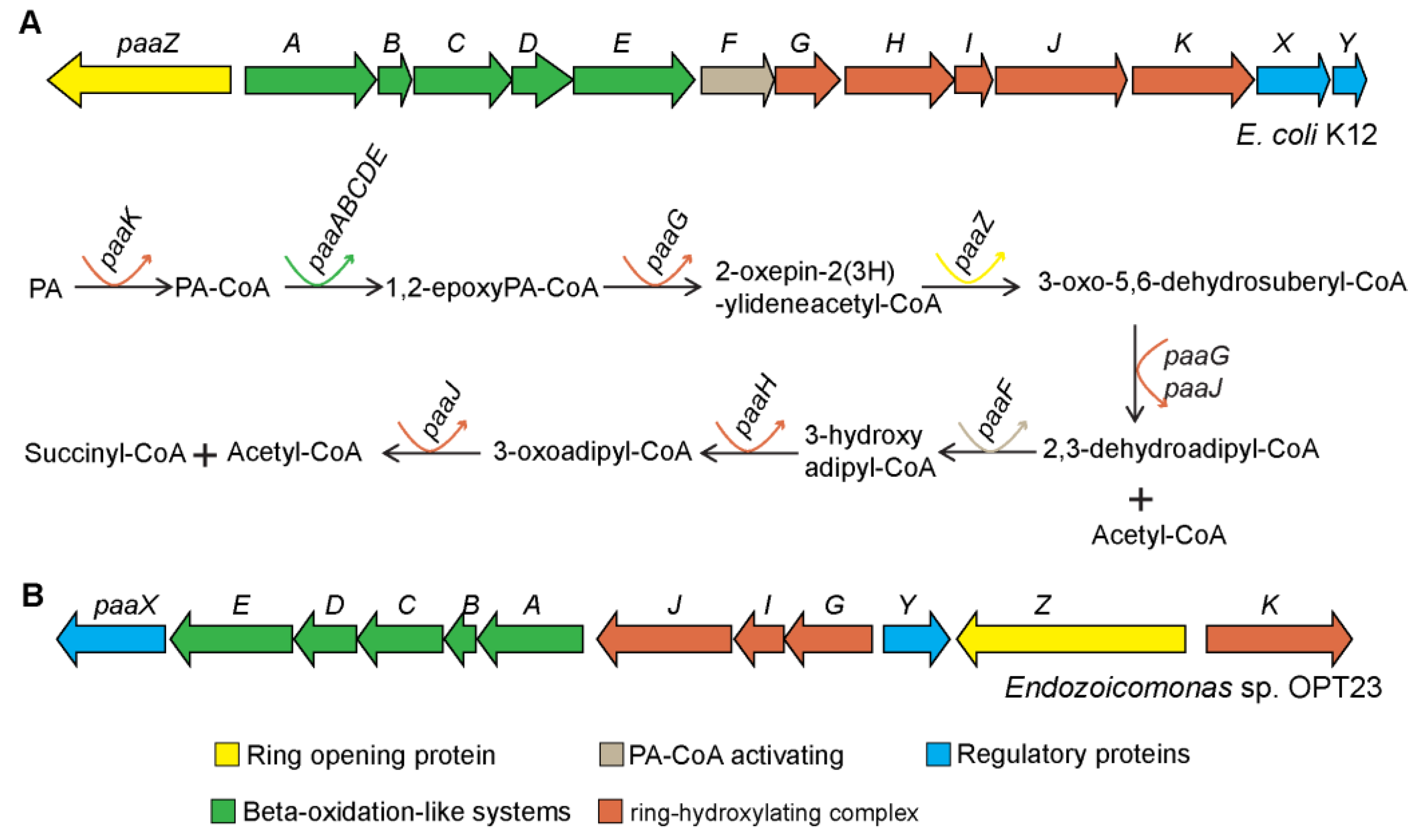
3.4.3. Pheynylacetic Acid Degradation Pathway
3.5. Symbioses Factors and Secretion Systems
3.5.1. Symbioses-Related Genes in Endozoicomonas sp. OPT23
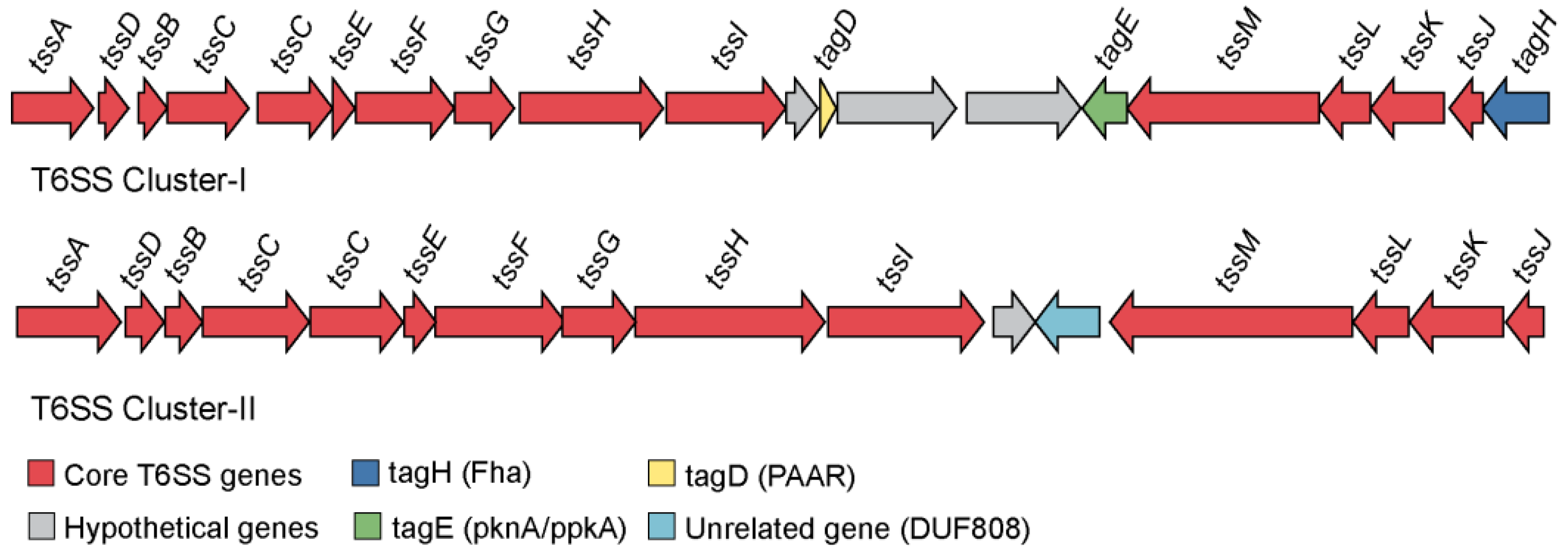
3.5.2. Role of Secretion Systems in Host-Bacterial Relationship
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.-M.; Wang, Y.; Tian, R.-M.; Wong, Y.H.; Batang, Z.B.; Al-Suwailem, A.M.; Bajic, V.B.; Qian, P.-Y. Symbiotic adaptation drives genome streamlining of the cyanobacterial sponge symbiont “Candidatus Synechococcus spongiarum”. MBio 2014, 5, e00079-14. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Reynolds, D.; Liu, M.; Stark, M.; Kjelleberg, S.; Webster, N.S.; Thomas, T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. USA 2012, 109, E1878–E1887. [Google Scholar] [CrossRef] [PubMed]
- Alex, A.; Antunes, A. Whole genome sequencing of the symbiont pseudovibrio sp. from the intertidal marine sponge polymastia penicillus revealed a gene repertoire for host-switching permissive lifestyle. Genome Biol. Evol. 2015, 7, 3022–3032. [Google Scholar] [CrossRef]
- Alex, A.; Antunes, A. Genus-wide comparison of Pseudovibrio bacterial genomes reveal diverse adaptations to different marine invertebrate hosts. PLoS ONE 2018, 13, e0194368. [Google Scholar] [CrossRef]
- Alex, A.; Antunes, A. Whole-genome comparisons among the genus shewanella reveal the enrichment of genes encoding ankyrin-repeats containing proteins in sponge-associated bacteria. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Kurahashi, M.; Yokota, A. Endozoicomonas elysicola gen. nov., sp. nov., a gamma-proteobacterium isolated from the sea slug Elysia ornata. Syst. Appl. Microbiol. 2007, 30, 202–206. [Google Scholar] [CrossRef]
- Neave, M.J.; Rachmawati, R.; Xun, L.; Michell, C.T.; Bourne, D.G.; Apprill, A.; Voolstra, C.R. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 2017, 11, 186–200. [Google Scholar] [CrossRef]
- Schreiber, L.; Kjeldsen, K.U.; Funch, P.; Jensen, J.; Obst, M.; López-Legentil, S.; Schramm, A. Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 2016, 7, 1042. [Google Scholar] [CrossRef]
- Yang, C.-S.; Chen, M.-H.; Arun, A.B.; Chen, C.A.; Wang, J.-T.; Chen, W.-M. Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int. J. Syst. Evol. Microbiol. 2010, 60, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L.; Kjeldsen, K.U.; Obst, M.; Funch, P.; Schramm, A. Description of endozoicomonas ascidiicola sp. nov., isolated from Scandinavian ascidians. Syst. Appl. Microbiol. 2016, 39, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Hyun, D.-W.; Shin, N.-R.; Kim, M.-S.; Oh, S.J.; Kim, P.S.; Whon, T.W.; Bae, J.-W. Endozoicomonas atrinae sp. nov., isolated from the intestine of a comb pen shell Atrina pectinata. Int. J. Syst. Evol. Microbiol. 2014, 64, 2312–2318. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Cascarano, M.C.; Schlapbach, R.; Katharios, P.; Vaughan, L.; Seth-Smith, H.M.B. Ca. Endozoicomonas cretensis: A novel fish pathogen characterized by genome plasticity. Genome Biol. Evol. 2018, 10, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Neave, M.J.; Apprill, A.; Ferrier-Pagès, C.; Voolstra, C.R. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 2016, 100, 8315–8324. [Google Scholar] [CrossRef]
- Vezzulli, L.; Pezzati, E.; Huete-Stauffer, C.; Pruzzo, C.; Cerrano, C. 16SrDNA Pyrosequencing of the Mediterranean gorgonian paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Meyer, J.L.; Paul, V.J.; Teplitski, M. Community shifts in the surface microbiomes of the coral porites astreoides with unusual lesions. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Ding, J.-Y.; Shiu, J.-H.; Chen, W.-M.; Chiang, Y.-R.; Tang, S.-L. Genomic insight into the host-endosymbiont relationship of endozoicomonas montiporae CL-33(T) with its Coral Host. Front. Microbiol. 2016, 7, 251. [Google Scholar] [CrossRef]
- Neave, M.J.; Michell, C.T.; Apprill, A.; Voolstra, C.R. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 2017, 7, 40579. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Nadalin, F.; Vezzi, F.; Policriti, A. GapFiller: A de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 2012, 13, S8. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Bacteria Genomes—NCBI FTP Site. Available online: Ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/ (accessed on 30 November 2018).
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. gplots: Various R Programming Tools for Plotting Data. Available online: https://CRAN.R-project.org/package=gplots (accessed on 15 July 2018).
- R Core Team R: A Language and Environment for Statistical Computing; Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 15 July 2018).
- Emms, D.M.; Kelly, S. OrthoFinder2: Fast and accurate phylogenomic orthology analysis from gene sequences. bioRxiv 2018. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Meusemann, K. FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 2010, 56, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Luo, H.; Lin, Y.; Gao, F.; Zhang, C.-T.; Zhang, R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014, 42, D574–D580. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Versluis, D.; Nijsse, B.; Naim, M.A.; Koehorst, J.J.; Wiese, J.; Imhoff, J.F.; Schaap, P.J.; van Passel, M.W.J.; Smidt, H.; Sipkema, D. Comparative Genomics Highlights Symbiotic Capacities and High Metabolic Flexibility of the Marine Genus Pseudovibrio. Genome Biol. Evol. 2018, 10, 125–142. [Google Scholar] [CrossRef]
- Karpenahalli, M.R.; Lupas, A.N.; Söding, J. TPRpred: A tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinform. 2007, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for functional characterization of genome and metagenome Aequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yao, Y.; Xu, H.H.; Hao, L.; Deng, Z.; Rajakumar, K.; Ou, H.-Y. SecReT6: A web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 2015, 17, 2196–2202. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Wang, J.; Li, C.; Revote, J.; Zhang, Y.; Naderer, T.; Hayashida, M.; Akutsu, T.; Webb, G.I.; Lithgow, T.; et al. SecretEPDB: A comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems. Sci. Rep. 2017, 7, 41031. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.E.; Higgs, P.G. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol. Biol. Evol. 2012, 29, 3413–3425. [Google Scholar] [CrossRef]
- Da Costa, W.L.O.; de Aragao Araujo, C.L.; Dias, L.M.; de Sousa Pereira, L.C.; Alves, J.T.C.; Araújo, F.A.; Folador, E.L.; Henriques, I.; Silva, A.; Folador, A.R.C. Functional annotation of hypothetical proteins from the Exiguobacterium antarcticum strain B7 reveals proteins involved in adaptation to extreme environments, including high arsenic resistance. PLoS ONE 2018, 13, e0198965. [Google Scholar] [CrossRef]
- Lesniak, J.; Barton, W.A.; Nikolov, D.B. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 2003, 12, 2838–2843. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Garvie, E.I. Bacterial lactate dehydrogenases. Microbiol. Rev. 1980, 44, 106–139. [Google Scholar]
- Gibello, A.; Collins, M.D.; Domínguez, L.; Fernández-Garayzábal, J.F.; Richardson, P.T. Cloning and analysis of the l-lactate utilization genes from streptococcus iniae. Appl. Environ. Microbiol. 1999, 65, 4346–4350. [Google Scholar] [PubMed]
- Chai, Y.; Kolter, R.; Losick, R. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 2009, 191, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Vita, N.; Valette, O.; Brasseur, G.; Lignon, S.; Denis, Y.; Ansaldi, M.; Dolla, A.; Pieulle, L. The primary pathway for lactate oxidation in Desulfovibrio vulgaris. Front. Microbiol. 2015, 6, 606. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.T.; Shepherd, M.; Poole, R.K.; van Vliet, A.H.M.; Kelly, D.J.; Pearson, B.M. Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 2011, 13, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, G.E.; Rodionov, D.A.; Yang, C.; Li, X.; Osterman, A.L.; Dervyn, E.; Geydebrekht, O.V.; Reed, S.B.; Romine, M.F.; Collart, F.R.; et al. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc. Natl. Acad. Sci. USA 2009, 106, 2874–2879. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Gao, C.; Ma, C.; Xu, P. Microbial lactate utilization: Enzymes, pathogenesis, and regulation. Trends Microbiol. 2014, 22, 589–599. [Google Scholar] [CrossRef]
- Smith, H.; Yates, E.A.; Cole, J.A.; Parsons, N.J. Lactate stimulation of gonococcal metabolism in media containing glucose: Mechanism, impact on pathogenicity, and wider implications for other pathogens. Infect Immun. 2001, 69, 6565–6572. [Google Scholar] [CrossRef]
- Fuller, J.R.; Vitko, N.P.; Perkowski, E.F.; Scott, E.; Khatri, D.; Spontak, J.S.; Thurlow, L.R.; Richardson, A.R. Identification of a Lactate-quinone oxidoreductase in staphylococcus aureus that is essential for virulence. Front. Cell Infect. Microbiol. 2011, 1. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Cornell, W.C.; Jo, J.; Price-Whelan, A.; Dietrich, L.E.P. The pseudomonas aeruginosa complement of lactate dehydrogenases enables use of d- and l-lactate and metabolic cross-feeding. MBio 2018, 9. [Google Scholar] [CrossRef]
- Giraud, M.F.; Naismith, J.H. The rhamnose pathway. Curr. Opin. Struct. Biol. 2000, 10, 687–696. [Google Scholar] [CrossRef]
- Peng, F.; Peng, P.; Xu, F.; Sun, R.-C. Fractional purification and bioconversion of hemicelluloses. Biotechnol. Adv. 2012, 30, 879–903. [Google Scholar] [CrossRef] [PubMed]
- Eagon, R.G. Bacterial dissimilation of L-fucose and L-rhamnose. J. Bacteriol. 1961, 82, 548–550. [Google Scholar] [PubMed]
- Reinhardt, A.; Johnsen, U.; Schönheit, P. l-Rhamnose catabolism in archaea. Mol. Microbiol. 2019, 111, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, I.A.; Li, X.; Thiel, V.; Stolyar, S.; Stanton, K.; Fredrickson, J.K.; Bryant, D.A.; Osterman, A.L.; Best, A.A.; Rodionov, D.A. Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front. Microbiol. 2013, 4, 407. [Google Scholar] [CrossRef] [PubMed]
- Moralejo, P.; Egan, S.M.; Hidalgo, E.; Aguilar, J. Sequencing and characterization of a gene cluster encoding the enzymes for L-rhamnose metabolism in Escherichia coli. J. Bacteriol. 1993, 175, 5585–5594. [Google Scholar] [CrossRef] [PubMed]
- Hirooka, K.; Kodoi, Y.; Satomura, T.; Fujita, Y. Regulation of the rhaEWRBMA Operon Involved in l-Rhamnose Catabolism through Two Transcriptional Factors, RhaR and CcpA, in Bacillus subtilis. J. Bacteriol. 2015, 198, 830–845. [Google Scholar] [CrossRef]
- Richardson, J.S.; Hynes, M.F.; Oresnik, I.J. A Genetic Locus Necessary for Rhamnose Uptake and Catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 2004, 186, 8433–8442. [Google Scholar] [CrossRef]
- Olivera, E.R.; Miñambres, B.; García, B.; Muñiz, C.; Moreno, M.A.; Ferrández, A.; Díaz, E.; García, J.L.; Luengo, J.M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: The phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 1998, 95, 6419–6424. [Google Scholar] [CrossRef]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef]
- Ferrández, A.; Miñambres, B.; García, B.; Olivera, E.R.; Luengo, J.M.; García, J.L.; Díaz, E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 1998, 273, 25974–25986. [Google Scholar] [CrossRef]
- Buchan, A.; González, J.M.; Chua, M.J. Aerobic Hydrocarbon-Degrading Alphaproteobacteria: Rhodobacteraceae (Roseobacter). Taxon.Genom. Ecophysiol. Hydrocarb. Degrad. Microbes 2019. [Google Scholar] [CrossRef]
- Law, R.J.; Hamlin, J.N.R.; Sivro, A.; McCorrister, S.J.; Cardama, G.A.; Cardona, S.T. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 2008, 190, 7209–7218. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.; Brock, N.L.; Liesegang, H.; Dogs, M.; Preuth, I.; Simon, M.; Dickschat, J.S.; Brinkhoff, T. Genetic analysis of the upper phenylacetate catabolic pathway in the production of tropodithietic acid by Phaeobacter gallaeciensis. Appl. Environ. Microbiol. 2012, 78, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, F.; Pasek, S.; Schenowitz, C.; Dossat, C.; Barbe, V.; Rottman, M.; Macheras, E.; Heym, B.; Herrmann, J.-L.; Daffé, M.; et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS ONE 2009, 4, e5660. [Google Scholar] [CrossRef]
- Gomez-Valero, L.; Rusniok, C.; Cazalet, C.; Buchrieser, C. Comparative and functional genomics of legionella identified eukaryotic like proteins as key players in host–pathogen interactions. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- Cerveny, L.; Straskova, A.; Dankova, V.; Hartlova, A.; Ceckova, M.; Staud, F.; Stulik, J. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 2013, 81, 629–635. [Google Scholar] [CrossRef]
- Kamke, J.; Rinke, C.; Schwientek, P.; Mavromatis, K.; Ivanova, N.; Sczyrba, A.; Woyke, T.; Hentschel, U. The candidate phylum Poribacteria by single-cell genomics: New insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS ONE 2014, 9, e87353. [Google Scholar] [CrossRef]
- Nguyen, M.T.H.D.; Liu, M.; Thomas, T. Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 2014, 23, 1635–1645. [Google Scholar] [CrossRef]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef]
- Basler, M. Type VI secretion system: Secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Drebes Dörr, N.C.; Blokesch, M. Bacterial type VI secretion system facilitates niche domination. Proc. Natl. Acad. Sci. USA 2018, 115, 8855–8857. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Mobley, H.L.T. The Versatile Type VI Secretion System. Microbiol. Spectr. 2016, 4. [Google Scholar]
- Speare, L.; Cecere, A.G.; Guckes, K.R.; Smith, S.; Wollenberg, M.S.; Mandel, M.J.; Miyashiro, T.; Septer, A.N. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. USA 2018, 115, E8528–E8537. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Russell, A.B.; LeRoux, M.; Hathazi, K.; Agnello, D.M.; Ishikawa, T.; Wiggins, P.A.; Wai, S.N.; Mougous, J.D. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 2013, 496, 508–512. [Google Scholar] [CrossRef]
- Park, Y.; Cho, Y.J.; Ahn, T.; Park, C. Molecular interactions in ribose transport: The binding protein module symmetrically associates with the homodimeric membrane transporter. EMBO J. 1999, 18, 4149–4156. [Google Scholar] [CrossRef]
- Shao, H.; James, D.; Lamont, R.J.; Demuth, D.R. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J. Bacteriol. 2007, 189, 5559–5565. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant. Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, H.; Cheng, X.; Shu, X.; White, A.P.; Stavrinides, J.; Köster, W.; Zhu, G.; Zhao, Z.; Wang, Y. A global survey of bacterial type III secretion systems and their effectors. Environ. Microbiol. 2017, 19, 3879–3895. [Google Scholar] [CrossRef]
- Bartra, S.; Cherepanov, P.; Forsberg, A.; Schesser, K. The Yersinia YopE and YopH type III effector proteins enhance bacterial proliferation following contact with eukaryotic cells. BMC Microbiol. 2001, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, K.; Jouihri, N.; Allaoui, A.; Gounon, P.; Sansonetti, P.J.; Parsot, C. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol. Microbiol. 2000, 38, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Henry, T.; Couillault, C.; Rockenfeller, P.; Boucrot, E.; Dumont, A.; Schroeder, N.; Hermant, A.; Knodler, L.A.; Lecine, P.; Steele-Mortimer, O.; et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc. Natl. Acad. Sci. USA 2006, 103, 13497–13502. [Google Scholar] [CrossRef] [PubMed]
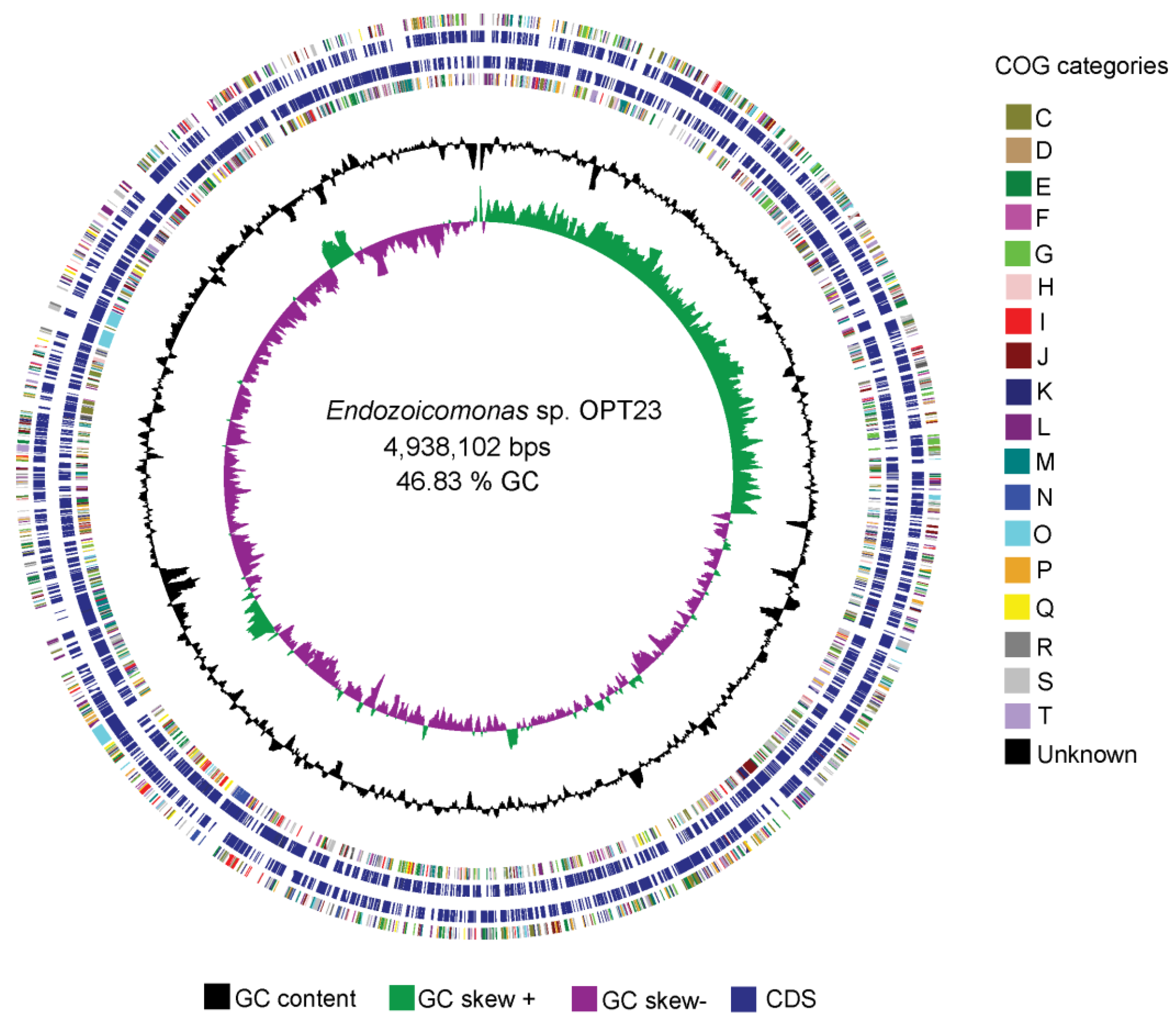

| Organism | Habitat | Bioproject | Assembly Version | # Contigs | Genome Size (bp) | %G+C Content | # Genes | # CDS | %COGs |
|---|---|---|---|---|---|---|---|---|---|
| Endozoicomonas sp. OPT23 * | Sponge | PRJNA430358 | ASM965363v1 | 30 | 4,938,102 | 46.84 | 4304 | 4175 | 79.49 |
| E. arenosclerae ab112 | Sponge | PRJNA279233 | ASM156201V1 | 328 | 6,453,554 | 47.65 | 5752 | 5571 | 72.15 |
| E. arenosclerae E-MC227 | Sponge | PRJNA279233 | ASM156200v1 | 2501 | 6,216,773 | 47.15 | 6025 | 5874 | 61.52 |
| E. numazuensis DSM 25634 | Sponge | PRJNA224116 | ASM72263v1 | 31 | 6,342,227 | 47.02 | 5647 | 5468 | 70.90 |
| E. ascidiicola AVMART05 | Ascidian | PRJNA291958 | AVMART05_1.0 | 36 | 6,130,497 | 46.70 | 5423 | 5282 | 68.61 |
| E. ascidiicola KASP37 | Ascidian | PRJNA291960 | KASP37_1.0 | 34 | 6,512,467 | 46.65 | 5768 | 5629 | 66.69 |
| E. montiporae CL-33 | Coral | PRJNA66389 | ASM158343v1 | 1 | 5,430,256 | 48.46 | 5125 | 4935 | 74.59 |
| E. acroporae Acr14 | Coral | PRJNA422318 | ASM286404v1 | 309 | 6,048,850 | 49.16 | 5274 | 5015 | 64.85 |
| Endozoicomonas sp. AB1-5 | Bryozoa | PRJNA322176 | -NA- | 272 | 4,049,356 | 45.28 | 3694 | 3525 | 78.27 |
| E. elysicola DSM 22380 | Sea slug | PRJNA252578 | ASM71077v1 | 2 | 5,606,375 | 46.75 | 4785 | 4652 | 76.13 |
| Ca. E. cretensis | Fish | PRJEB7440 | -NA- | 638 | 5,876,352 | 46.80 | 5641 | 5505 | 68.30 |
| E. atrinae WP70 | Bivalve | PRJNA224116 | -NA- | 980 | 6,687,418 | 47.94 | 6288 | 6166 | 65.65 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alex, A.; Antunes, A. Comparative Genomics Reveals Metabolic Specificity of Endozoicomonas Isolated from a Marine Sponge and the Genomic Repertoire for Host-Bacteria Symbioses. Microorganisms 2019, 7, 635. https://doi.org/10.3390/microorganisms7120635
Alex A, Antunes A. Comparative Genomics Reveals Metabolic Specificity of Endozoicomonas Isolated from a Marine Sponge and the Genomic Repertoire for Host-Bacteria Symbioses. Microorganisms. 2019; 7(12):635. https://doi.org/10.3390/microorganisms7120635
Chicago/Turabian StyleAlex, Anoop, and Agostinho Antunes. 2019. "Comparative Genomics Reveals Metabolic Specificity of Endozoicomonas Isolated from a Marine Sponge and the Genomic Repertoire for Host-Bacteria Symbioses" Microorganisms 7, no. 12: 635. https://doi.org/10.3390/microorganisms7120635
APA StyleAlex, A., & Antunes, A. (2019). Comparative Genomics Reveals Metabolic Specificity of Endozoicomonas Isolated from a Marine Sponge and the Genomic Repertoire for Host-Bacteria Symbioses. Microorganisms, 7(12), 635. https://doi.org/10.3390/microorganisms7120635






